

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.10 + phase: 0
(449 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
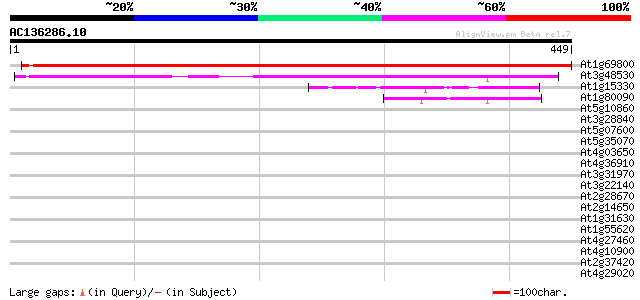
Score E
Sequences producing significant alignments: (bits) Value
At1g69800 Unknown protein 590 e-169
At3g48530 unknown protein 254 7e-68
At1g15330 hypothetical protein 55 8e-08
At1g80090 unknown protein 49 4e-06
At5g10860 unknown protein 37 0.029
At3g28840 unknown protein 36 0.038
At5g07600 unknown protein 36 0.049
At5g35070 putative protein 35 0.084
At4g03650 putative reverse transcriptase 33 0.32
At4g36910 unknown protein 32 0.54
At3g31970 hypothetical protein 32 0.54
At3g22140 hypothetical protein 32 0.54
At2g28670 putative disease resistance response protein 32 0.71
At2g14650 putative retroelement pol polyprotein 31 1.6
At1g31630 putative protein 31 1.6
At1g55620 CLC-f chloride channel protein 30 2.7
At4g27460 unknown protein 30 3.5
At4g10900 putative protein 30 3.5
At2g37420 putative kinesin heavy chain 30 3.5
At4g29020 glycine-rich protein like 29 6.0
>At1g69800 Unknown protein
Length = 447
Score = 590 bits (1520), Expect = e-169
Identities = 298/441 (67%), Positives = 369/441 (83%), Gaps = 3/441 (0%)
Query: 10 EQEVRTSTQLSKCDRYFETIQSRKKLPQTLQETLTDSFAKIPVSSFPGVPGGKVVEILAD 69
++E++++ +S C+ YFE +QSRK LP++LQETL +FA IPVSSFP VPGG+V+EI A+
Sbjct: 9 DKEIKSA--VSSCEAYFEKVQSRKNLPKSLQETLNSAFAGIPVSSFPQVPGGRVIEIQAE 66
Query: 70 TPVGEAVKILSESNILAAPVKDPDAGIGSDWRDRYLGIIDYSAIILWVMESAELAAVALS 129
TPV EAVKILS+S IL+APV + D DWR+RYLGIIDYS+IILWV+ESAELAA+ALS
Sbjct: 67 TPVSEAVKILSDSKILSAPVINTDHESSLDWRERYLGIIDYSSIILWVLESAELAAIALS 126
Query: 130 AGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKDAPQAA 189
A +ATAAGVGAG VGALG ALG TGP A AGL AAAVGAAV GGVAA++ + KDAP AA
Sbjct: 127 ATSATAAGVGAGAVGALGVAALGMTGPVAAAGLAAAAVGAAVAGGVAAERGIGKDAPTAA 186
Query: 190 NNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRLRNVPV 249
+ LG+DFY+VILQEEPFKSTTVR+ILKS+RWAPF+PV+ S+ML+V+LLLSKYRLRNVPV
Sbjct: 187 DKLGKDFYEVILQEEPFKSTTVRTILKSFRWAPFLPVSTESSMLSVMLLLSKYRLRNVPV 246
Query: 250 IEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELI 309
I+ G+ DI N++TQSAV+ GLEGC+GRDWFD I+A P++DLGLPFMS ++VISI+S ELI
Sbjct: 247 IKTGEPDIKNYVTQSAVVHGLEGCKGRDWFDHISALPISDLGLPFMSPNEVISIESEELI 306
Query: 310 LEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDFMKKIV 369
LEAFK MRDN IGGLPVVEG K IVGN+S+RDIRYLLL+PE+FSNFR LTV F KI
Sbjct: 307 LEAFKRMRDNNIGGLPVVEGLNKKIVGNISMRDIRYLLLQPEVFSNFRQLTVKSFATKIA 366
Query: 370 SASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNG-QDQVVGVITLRDVISCFI 428
+A E G ITC+PD+TL SVI++LAS+S+HR+Y G ++++ GVITLRDVISCF+
Sbjct: 367 TAGEEYGLAIPAITCRPDSTLGSVINSLASRSVHRVYVAAGDENELYGVITLRDVISCFV 426
Query: 429 TEPDYHFDDYYGFAVKEMLNQ 449
+EP +F++ GF+VKEMLN+
Sbjct: 427 SEPPNYFENCLGFSVKEMLNR 447
>At3g48530 unknown protein
Length = 424
Score = 254 bits (649), Expect = 7e-68
Identities = 147/440 (33%), Positives = 242/440 (54%), Gaps = 45/440 (10%)
Query: 5 QNMAHEQEVRTSTQLSKCDRYFETIQSRKKLPQTLQETLTDSFAKIPVSSFPGVPGGKVV 64
+++ H +V S+ +K E + +K + E L F IPVS+FP + +
Sbjct: 13 ESLGHRSDV--SSPEAKLGMRVEDLWDEQKPQLSPNEKLNACFESIPVSAFPLSSDSQDI 70
Query: 65 EILADTPVGEAVKILSESNILAAPVKDPDAGIGSDWRDRYLGIIDYSAIILWVMESAELA 124
EI +DT + EAV+ LS+ +L+APV D DA + W DRY+GI+++ I++W++ E
Sbjct: 71 EIRSDTSLAEAVQTLSKFKVLSAPVVDVDAPEDASWIDRYIGIVEFPGIVVWLLHQLEPP 130
Query: 125 AVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKD 184
+ A V A + T G +A G
Sbjct: 131 SPRSPA------------VAASNGFSHDFTTDVLDNGDSAVTSG---------------- 162
Query: 185 APQAANNLGEDFYKVILQEEPFKSTTVRSILKSYRWAPFVPVAKNSAMLTVLLLLSKYRL 244
+F++V+ E +K+T VR I ++RWAPF+ + K ++ LT+LLLLSKY++
Sbjct: 163 ----------NFFEVLTSSELYKNTKVRDISGTFRWAPFLALQKENSFLTMLLLLSKYKM 212
Query: 245 RNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQ 304
+++PV++ G A I N ITQS VI L C G WF+ + ++++GLP MS D +I I
Sbjct: 213 KSIPVVDLGVAKIENIITQSGVIHMLAECAGLLWFEDWGIKTLSEVGLPIMSKDHIIKIY 272
Query: 305 SNELILEAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNFRNLTVMDF 364
+E +L+AFK+MR +IGG+PV+E ++ VGN+S+RD+++LL PEI+ ++R++T +F
Sbjct: 273 EDEPVLQAFKLMRRKRIGGIPVIERNSEKPVGNISLRDVQFLLTAPEIYHDYRSITTKNF 332
Query: 365 MKKIVSASYESGKVTRP-----ITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVIT 419
+ + + G + P I C + TL+ +I L ++ IHRIY V+ + G+IT
Sbjct: 333 LVSVREHLEKCGDTSAPIMSGVIACTKNHTLKELILMLDAEKIHRIYVVDDFGNLEGLIT 392
Query: 420 LRDVISCFITEPDYHFDDYY 439
LRD+I+ + EP +F D++
Sbjct: 393 LRDIIARLVHEPSGYFGDFF 412
>At1g15330 hypothetical protein
Length = 352
Score = 55.1 bits (131), Expect = 8e-08
Identities = 45/199 (22%), Positives = 91/199 (45%), Gaps = 28/199 (14%)
Query: 240 SKYRLRNVPVIEPGKADIVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADK 299
S + V +IE A +TQ +++ L+ D + +R ++DLG D
Sbjct: 160 SNNTIAGVELIESASA--YKMLTQMDLLRFLKDHHFDD-LKTVLSRSISDLGAV---NDS 213
Query: 300 VISIQSNELILEAFKIMRDNQIGGLPVVEGPA--------------KTIVGNLSIRDIRY 345
V +I + A +M+ + +P+V P + ++G S D++
Sbjct: 214 VYAITERTTVSNAINVMKGALLNAVPIVHAPDIAQEDHLQLVNGRHRKVIGTFSATDLKG 273
Query: 346 LLLKPEIFSNFRNLTVMDFMKKIVSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRI 405
L PE+ + LT ++F +K SGK ++C ++T++ I + ++ +HR+
Sbjct: 274 CRL-PEL-QTWLPLTALEFTEKT------SGKEREVVSCGVESTMEEAIEKVVTRGVHRV 325
Query: 406 YTVNGQDQVVGVITLRDVI 424
+ ++ Q + GV++L D+I
Sbjct: 326 WVMDQQGLLQGVVSLTDII 344
>At1g80090 unknown protein
Length = 357
Score = 49.3 bits (116), Expect = 4e-06
Identities = 32/143 (22%), Positives = 68/143 (47%), Gaps = 19/143 (13%)
Query: 300 VISIQSNELILEAFKIMRDNQIGGLPVVE-------------GPAKTIVGNLSIRDIRYL 346
V+++ S + +A + M + +P+VE G + +VG S D++
Sbjct: 203 VLALTSQARVKDAIQCMSIAMLNAVPIVEASGEGEDHKQLVDGKNRRVVGTFSASDLKGC 262
Query: 347 LLKPEIFSNFRNLTVMDFMKKIVSASYESGKVTRP----ITCKPDATLQSVIHTLASQSI 402
L ++ L ++F++KI + + P +TC +TL VIH + ++ +
Sbjct: 263 HLAT--LRSWLPLNALEFVEKIPRTLLFTAATSTPGRELVTCHVTSTLAQVIHMVTTKRV 320
Query: 403 HRIYTVNGQDQVVGVITLRDVIS 425
HR++ V+ + G+++L D+I+
Sbjct: 321 HRVWVVDQNGGLQGLVSLTDIIA 343
>At5g10860 unknown protein
Length = 206
Score = 36.6 bits (83), Expect = 0.029
Identities = 30/147 (20%), Positives = 74/147 (49%), Gaps = 14/147 (9%)
Query: 201 LQEEPFKSTTVRSILKSYRWAP---FVPVAKNSAMLTVLLLLSKYRLRNVPVIEPGKAD- 256
++E F+STT+ ++KS + ++ + + + ++++ + + V++PG+
Sbjct: 47 MEESGFESTTISDVMKSKGKSADGSWLWCTTDDTVYDAVKSMTQHNVGALVVVKPGEQQA 106
Query: 257 IVNFITQSAVIQGLEGCRGRDWFDCIAARPMADLGLPFMSADKVISIQSNELILEAFKIM 316
+ IT+ ++ + +GR + +G +K+I++ +L A ++M
Sbjct: 107 LAGIITERDYLRKII-VQGR-------SSKSTKVGDIMTEENKLITVTPETKVLRAMQLM 158
Query: 317 RDNQIGGLPVVEGPAKTIVGNLSIRDI 343
DN+I +PV++ K ++G +SI D+
Sbjct: 159 TDNRIRHIPVIKD--KGMIGMVSIGDV 183
>At3g28840 unknown protein
Length = 391
Score = 36.2 bits (82), Expect = 0.038
Identities = 24/53 (45%), Positives = 26/53 (48%), Gaps = 3/53 (5%)
Query: 127 ALSAGTA---TAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVA 176
A AGTA TA+G G G A A A G A AG AAA G GGV+
Sbjct: 279 ASDAGTAAGTTASGAGTAAGGTTAAGAGAAAGAGAAAGAGAAAGGTTAAGGVS 331
Score = 35.8 bits (81), Expect = 0.049
Identities = 28/82 (34%), Positives = 33/82 (40%), Gaps = 6/82 (7%)
Query: 121 AELAAVALSAGTATAAGVG---AGTVGALGAIALGATGP---AAIAGLTAAAVGAAVVGG 174
A A +AG ATA+G G GT + A G T A G TAA GAA G
Sbjct: 253 AGTAGYGATAGGATASGAGTAAGGTTASDAGTAAGTTASGAGTAAGGTTAAGAGAAAGAG 312
Query: 175 VAADKTMAKDAPQAANNLGEDF 196
AA A AA + +
Sbjct: 313 AAAGAGAAAGGTTAAGGVSGSY 334
Score = 35.0 bits (79), Expect = 0.084
Identities = 24/61 (39%), Positives = 27/61 (43%), Gaps = 3/61 (4%)
Query: 120 SAELAAVALSAGTATAAGVGAGTVGALGAIALGAT---GPAAIAGLTAAAVGAAVVGGVA 176
+A A A TA+ AG AGT + A G T G A AG AAA A GG
Sbjct: 266 TASGAGTAAGGTTASDAGTAAGTTASGAGTAAGGTTAAGAGAAAGAGAAAGAGAAAGGTT 325
Query: 177 A 177
A
Sbjct: 326 A 326
Score = 34.3 bits (77), Expect = 0.14
Identities = 29/64 (45%), Positives = 31/64 (48%), Gaps = 6/64 (9%)
Query: 129 SAGTATAAGVGAGTVGALGAIALGAT--GPAAIAGLTAAAVGAAVVG-GVAADKTMAKDA 185
+AG TA G GT A G+ A G T A AG A A GA G G AA T A DA
Sbjct: 226 AAGGTTATG---GTTAAGGSTAAGGTTASGAGTAGYGATAGGATASGAGTAAGGTTASDA 282
Query: 186 PQAA 189
AA
Sbjct: 283 GTAA 286
Score = 32.0 bits (71), Expect = 0.71
Identities = 26/74 (35%), Positives = 36/74 (48%), Gaps = 4/74 (5%)
Query: 108 IDYSAI-ILWVMESAELAAVALSAGTATAAGVGAGTVGALGAIALG---ATGPAAIAGLT 163
ID S++ I + SA A + ++G +A G G+ GA G+ A G A G G T
Sbjct: 177 IDISSLGIDGIDASAAGAGDSTASGGVSATGTGSYGAGAGGSSASGSDTAAGGTTATGGT 236
Query: 164 AAAVGAAVVGGVAA 177
AA G+ GG A
Sbjct: 237 TAAGGSTAAGGTTA 250
Score = 32.0 bits (71), Expect = 0.71
Identities = 31/79 (39%), Positives = 34/79 (42%), Gaps = 19/79 (24%)
Query: 129 SAGTATAAG----VGAGTVGALGAIALGATGPAA--------------IAGLTAAAVGAA 170
+AG +TAAG GAGT G GA A GAT A AG TA+ G A
Sbjct: 238 AAGGSTAAGGTTASGAGTAG-YGATAGGATASGAGTAAGGTTASDAGTAAGTTASGAGTA 296
Query: 171 VVGGVAADKTMAKDAPQAA 189
G AA A A AA
Sbjct: 297 AGGTTAAGAGAAAGAGAAA 315
Score = 29.3 bits (64), Expect = 4.6
Identities = 21/60 (35%), Positives = 29/60 (48%), Gaps = 6/60 (10%)
Query: 129 SAGTATAAGVGA----GTVGALGAIALGATGPAAIAGLTAAAVGAA--VVGGVAADKTMA 182
+AG TAAG GA G GA A G T ++G ++ G+A GG ++ T A
Sbjct: 296 AAGGTTAAGAGAAAGAGAAAGAGAAAGGTTAAGGVSGSYGSSGGSASNTNGGTSSCGTSA 355
Score = 29.3 bits (64), Expect = 4.6
Identities = 20/70 (28%), Positives = 33/70 (46%), Gaps = 2/70 (2%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAA--VGAAVVGGVAADKTM 181
A +AG TAAG +G+ G+ G A G + G +A + GA+ G +
Sbjct: 315 AGAGAAAGGTTAAGGVSGSYGSSGGSASNTNGGTSSCGTSACSNGGGASFKGAMNFQGKA 374
Query: 182 AKDAPQAANN 191
+ + QAA++
Sbjct: 375 SSQSQQAASS 384
>At5g07600 unknown protein
Length = 230
Score = 35.8 bits (81), Expect = 0.049
Identities = 23/46 (50%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query: 121 AELAAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAA 166
A A A AG A AAG A V LGA + GA GP A AG A A
Sbjct: 173 AGAGAAAAGAGAAAAAGAAAAAVPGLGAASAGA-GPGAGAGTPAPA 217
Score = 32.7 bits (73), Expect = 0.42
Identities = 20/44 (45%), Positives = 22/44 (49%), Gaps = 1/44 (2%)
Query: 132 TATAAGVGAGTVGA-LGAIALGATGPAAIAGLTAAAVGAAVVGG 174
T AAG GAG A GA A AA+ GL AA+ GA G
Sbjct: 167 TPEAAGAGAGAAAAGAGAAAAAGAAAAAVPGLGAASAGAGPGAG 210
Score = 29.6 bits (65), Expect = 3.5
Identities = 24/48 (50%), Positives = 26/48 (54%), Gaps = 8/48 (16%)
Query: 129 SAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAV---GAAVVG 173
+AG +T GAG GA A GA G AA AG AAAV GAA G
Sbjct: 162 TAGGSTPEAAGAGA----GAAAAGA-GAAAAAGAAAAAVPGLGAASAG 204
>At5g35070 putative protein
Length = 356
Score = 35.0 bits (79), Expect = 0.084
Identities = 18/39 (46%), Positives = 21/39 (53%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGL 162
AA+ +AGTAT A G G VG G +G G AA L
Sbjct: 314 AAIGAAAGTATGAAAGGGVVGRAGGGIVGRAGGAARGAL 352
>At4g03650 putative reverse transcriptase
Length = 839
Score = 33.1 bits (74), Expect = 0.32
Identities = 23/46 (50%), Positives = 25/46 (54%), Gaps = 5/46 (10%)
Query: 128 LSAGTATAAGVGAGTVG-ALGAIALGATGPAAIAGLTAAAVGAAVV 172
L A A AAGVGAG G G A+GA GP G+ AA G A V
Sbjct: 54 LGADIAGAAGVGAGGAGVGTGVHAVGAEGP----GVMGAAAGGAQV 95
>At4g36910 unknown protein
Length = 236
Score = 32.3 bits (72), Expect = 0.54
Identities = 27/126 (21%), Positives = 55/126 (43%), Gaps = 13/126 (10%)
Query: 311 EAFKIMRDNQIGGLPVVEGPAKTIVGNLSIRDIRYL-----------LLKPEIFSNFRNL 359
EA +++ +N+I G PV++ K +VG +S D+ L + PE+ S ++
Sbjct: 98 EALELLVENRITGFPVIDEDWK-LVGLVSDYDLLALDSISGSGRTENSMFPEVDSTWKTF 156
Query: 360 TVMDFMKKIVSASYESGKVT-RPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVI 418
+ + + +T P+ + L+ L R+ V+ ++VG+I
Sbjct: 157 NAVQKLLSKTNGKLVGDLMTPAPLVVEEKTNLEDAAKILLETKYRRLPVVDSDGKLVGII 216
Query: 419 TLRDVI 424
T +V+
Sbjct: 217 TRGNVV 222
>At3g31970 hypothetical protein
Length = 1329
Score = 32.3 bits (72), Expect = 0.54
Identities = 26/48 (54%), Positives = 28/48 (58%), Gaps = 6/48 (12%)
Query: 128 LSAGTATAAGVGAG--TVGALGAIALGATGPAAIAGLTAAAVGAAVVG 173
L A A AAGVGAG VGA G A+GA GP + AAA GA V G
Sbjct: 56 LGADIAGAAGVGAGGADVGA-GVHAVGAEGPRVMG---AAAGGAQVPG 99
>At3g22140 hypothetical protein
Length = 1237
Score = 32.3 bits (72), Expect = 0.54
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALG--ATGPAAIAGLT--AAAVGAAVVGGVA 176
A ++ G GV G V +G A+G ATG A+ G+T A+G +GGVA
Sbjct: 391 AVGGVAIGGIATGGVAVGGV-VVGGFAIGGVATGEVAVGGVTTGGVAIGEVAIGGVA 446
Score = 32.3 bits (72), Expect = 0.54
Identities = 25/56 (44%), Positives = 32/56 (56%), Gaps = 6/56 (10%)
Query: 123 LAAVALSAGTATAAGVGAGTVGALGAIALG--ATGPAAIAGLTAAAVGAAVVGGVA 176
+AA ++ G GV G A+G +A+G ATG A G+ AVG AVVGGVA
Sbjct: 530 VAAGGVATGGFEIGGVKIGGF-AVGGVAIGGVATGGFATGGV---AVGGAVVGGVA 581
Score = 32.0 bits (71), Expect = 0.71
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALG--ATGPAAIAGLT--AAAVGAAVVGGVA 176
A ++ G GV G + +G A+G ATG A+ G+T A+G +GGVA
Sbjct: 641 AVGGVAIGGIATGGVAVGGI-VVGGFAIGGVATGGVAVGGVTTGGVAIGGVAIGGVA 696
Score = 30.0 bits (66), Expect = 2.7
Identities = 19/59 (32%), Positives = 25/59 (42%), Gaps = 10/59 (16%)
Query: 123 LAAVALSAGTATAAGVGAGTVG----ALGAIALGATGPAAIAGLTAAAVGAAVVGGVAA 177
+A + G GV G V A+G +A+G AG G V+GGVAA
Sbjct: 730 VATGGFATGEVAIGGVAVGGVATGGFAMGGVAIGGVATGGFAG------GGVVIGGVAA 782
Score = 29.6 bits (65), Expect = 3.5
Identities = 23/56 (41%), Positives = 31/56 (55%), Gaps = 6/56 (10%)
Query: 123 LAAVALSAGTATAAGVGAGTVGALGAIALG--ATGPAAIAGLTAAAVGAAVVGGVA 176
+AA ++ G GV G A+G +A+G +TG A G+ AVG VVGGVA
Sbjct: 900 VAAGGVATGGFEIGGVEIGGF-AVGGVAIGGVSTGGFATGGV---AVGGVVVGGVA 951
Score = 28.9 bits (63), Expect = 6.0
Identities = 20/50 (40%), Positives = 25/50 (50%), Gaps = 4/50 (8%)
Query: 131 GTATAAGVGAGTVGALGAIALG--ATGPAAIAGLTAAAVGAAVVGGVAAD 178
G AT V G A+G +A+G ATG A+ G G VVGG+ D
Sbjct: 1137 GVATGGIVMGGV--AVGGVAIGGVATGDLAMGGGWGVDGGGLVVGGLGVD 1184
Score = 28.5 bits (62), Expect = 7.9
Identities = 17/56 (30%), Positives = 27/56 (47%), Gaps = 3/56 (5%)
Query: 122 ELAAVALSAGTATAAGVGAGTVGALGAIALG--ATGPAAIAGLTAAAVGAAVVGGV 175
++A ++ G + GV G +G +A+G ATG A+ G+ V GGV
Sbjct: 994 DVAVGGVATGDSEIGGVEIGDF-EVGGVAIGGVATGGVAVGGVATGGCAIGVTGGV 1048
>At2g28670 putative disease resistance response protein
Length = 447
Score = 32.0 bits (71), Expect = 0.71
Identities = 25/70 (35%), Positives = 29/70 (40%), Gaps = 5/70 (7%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAK 183
+A AGT +A G G G ALG A GPA G A G A+ GG A +
Sbjct: 121 SATGAGAGTGSALGGGPGAGSALGGGA--GAGPALGGG---AGAGPALGGGAGAGSALGG 175
Query: 184 DAPQAANNLG 193
A LG
Sbjct: 176 GGAGAGPALG 185
Score = 31.6 bits (70), Expect = 0.93
Identities = 18/54 (33%), Positives = 24/54 (44%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAA 177
A AL G + +G G GA A+ G G G A G+A+ GG +A
Sbjct: 159 AGPALGGGAGAGSALGGGGAGAGPALGGGGAGAGPALGGGVAGSGSALGGGASA 212
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 30.8 bits (68), Expect = 1.6
Identities = 22/48 (45%), Positives = 24/48 (49%), Gaps = 5/48 (10%)
Query: 128 LSAGTATAAGVGAGTVG-ALGAIALGATGP----AAIAGLTAAAVGAA 170
L A AAGVGAG G G A+GA GP AA G+ VG A
Sbjct: 54 LGTDIAGAAGVGAGGAGVGTGVHAVGAEGPGVMGAAAGGVQVPEVGLA 101
Score = 30.0 bits (66), Expect = 2.7
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 4/50 (8%)
Query: 58 VPGGKVVEILADTPVGEAVKILSES---NILAAPVKDPDAGIGSDWRDRY 104
V GG+ V +L T G ++I ES +++ +PV+ D +G DW D Y
Sbjct: 349 VAGGQFVAVLGRTK-GVDIQIAGESMPADLIISPVELYDVILGMDWLDHY 397
>At1g31630 putative protein
Length = 339
Score = 30.8 bits (68), Expect = 1.6
Identities = 21/58 (36%), Positives = 27/58 (46%), Gaps = 2/58 (3%)
Query: 134 TAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKDAPQAANN 191
+ AG GAG A +A P A+AG A+ + A VG AA +A P A N
Sbjct: 171 SVAGAGAGAGAAPLVVAGAGAAPLAVAGAGASPLAVAGVG--AAPLAVAGAGPPMAQN 226
>At1g55620 CLC-f chloride channel protein
Length = 781
Score = 30.0 bits (66), Expect = 2.7
Identities = 21/62 (33%), Positives = 29/62 (45%), Gaps = 2/62 (3%)
Query: 123 LAAVALSAGTATAAGVGAGTVGALGA--IALGATGPAAIAGLTAAAVGAAVVGGVAADKT 180
LA +A + ATA G+G VG L A + +GA A G A + A+ G A +
Sbjct: 439 LAQLAAAKVVATALCKGSGLVGGLYAPSLMIGAAVGAVFGGSAAEIINRAIPGNAAVAQP 498
Query: 181 MA 182
A
Sbjct: 499 QA 500
>At4g27460 unknown protein
Length = 391
Score = 29.6 bits (65), Expect = 3.5
Identities = 13/59 (22%), Positives = 29/59 (49%)
Query: 369 VSASYESGKVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVGVITLRDVISCF 427
+S+S + + I C P ++L +V+ + ++ + V VG++T D++ F
Sbjct: 325 MSSSARMARKSEAIVCNPKSSLMAVMIQAVAHRVNYAWVVEKDGCFVGMVTFVDILKVF 383
>At4g10900 putative protein
Length = 169
Score = 29.6 bits (65), Expect = 3.5
Identities = 15/39 (38%), Positives = 21/39 (53%)
Query: 151 LGATGPAAIAGLTAAAVGAAVVGGVAADKTMAKDAPQAA 189
LG I L AAAVGA ++GG+ +K P+A+
Sbjct: 109 LGQRKSLGICPLRAAAVGAVIIGGIGYVVLYSKKKPEAS 147
>At2g37420 putative kinesin heavy chain
Length = 1022
Score = 29.6 bits (65), Expect = 3.5
Identities = 25/100 (25%), Positives = 46/100 (46%), Gaps = 16/100 (16%)
Query: 323 GLPVVEGPAKTIVGNLSIRDIRYLLLKPEIFSNF-----RNLTVMDFMK-KIVSASYESG 376
G+ V+ PA + L R +P+ FSN + + V ++ K +S +
Sbjct: 13 GVGVIPSPAPFLTPRLERR-------RPDSFSNRLDRDNKEVNVQVILRCKPLSEEEQKS 65
Query: 377 KVTRPITCKPDATLQSVIHTLASQSIHRIYTVNGQDQVVG 416
V R I+C +V+HT+A++ + R++ D+V G
Sbjct: 66 SVPRVISCNEMRREVNVLHTIANKQVDRLFNF---DKVFG 102
>At4g29020 glycine-rich protein like
Length = 158
Score = 28.9 bits (63), Expect = 6.0
Identities = 19/51 (37%), Positives = 24/51 (46%), Gaps = 4/51 (7%)
Query: 124 AAVALSAGTATAAGVGAGTVGALGAIALGATGPAAIAGLTAAAVGAAVVGG 174
A V +AG A GVGAG LG +A G G A + + G +GG
Sbjct: 44 AGVGGAAGIGGAGGVGAG----LGGVAGGVGGVAGVLPVGGVGGGIGGLGG 90
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,926,730
Number of Sequences: 26719
Number of extensions: 351368
Number of successful extensions: 1161
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 1077
Number of HSP's gapped (non-prelim): 72
length of query: 449
length of database: 11,318,596
effective HSP length: 103
effective length of query: 346
effective length of database: 8,566,539
effective search space: 2964022494
effective search space used: 2964022494
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC136286.10


