

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136140.11 - phase: 0 /pseudo
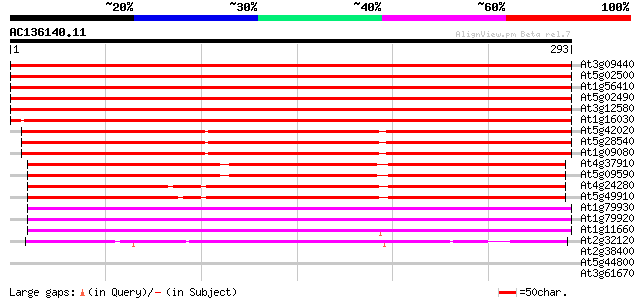
(293 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g09440 heat-shock protein (At-hsc70-3) 553 e-158
At5g02500 dnaK-type molecular chaperone hsc70.1 550 e-157
At1g56410 hypothetical protein 547 e-156
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 547 e-156
At3g12580 putative protein 545 e-155
At1g16030 unknown protein 501 e-142
At5g42020 luminal binding protein (dbj|BAA13948.1) 381 e-106
At5g28540 luminal binding protein 381 e-106
At1g09080 putative luminal binding protein 375 e-104
At4g37910 heat shock protein 70 like protein 290 5e-79
At5g09590 heat shock protein 70 (Hsc70-5) 283 6e-77
At4g24280 hsp 70-like protein 262 1e-70
At5g49910 heat shock protein 70 (Hsc70-7) 259 2e-69
At1g79930 putative heat-shock protein 210 6e-55
At1g79920 putative heat-shock protein (At1g79920) 208 2e-54
At1g11660 heat-shock protein like 192 2e-49
At2g32120 70kD heat shock protein 125 2e-29
At2g38400 putative beta-alanine-pyruvate aminotransferase 31 0.69
At5g44800 helicase-like protein 28 5.8
At3g61670 putative protein 28 7.6
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 553 bits (1424), Expect = e-158
Identities = 276/293 (94%), Positives = 288/293 (98%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP+NTVFDAKRLIGRR +D+SVQSD+KLWPF + +GP EKPMI VNYKGEDK+F+A
Sbjct: 61 QVAMNPINTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKSGPAEKPMIVVNYKGEDKEFSA 120
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSM+LIKMREIAEAYLG+T+KNAVVTVPAYFNDSQRQATKDAGVIAGLNV+RIINEP
Sbjct: 121 EEISSMILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEP 180
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 550 bits (1417), Expect = e-157
Identities = 276/293 (94%), Positives = 287/293 (97%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
M+GKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MSGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNPVNTVFDAKRLIGRR SD+SVQSDMKLWPFK+ AGP +KPMI V YKGE+K+FAA
Sbjct: 61 QVAMNPVNTVFDAKRLIGRRFSDSSVQSDMKLWPFKIQAGPADKPMIYVEYKGEEKEFAA 120
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVLIKMREIAEAYLG T+KNAVVTVPAYFNDSQRQATKDAGVIAGLNV+RIINEP
Sbjct: 121 EEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEP 180
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRK+KKDI+GNPRALRRLRT+CERAKRTLSSTAQTTIEIDSL
Sbjct: 241 NRMVNHFVQEFKRKSKKDITGNPRALRRLRTSCERAKRTLSSTAQTTIEIDSL 293
>At1g56410 hypothetical protein
Length = 617
Score = 547 bits (1410), Expect = e-156
Identities = 277/293 (94%), Positives = 284/293 (96%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNPVNTVFDAKRLIGRR SDASVQSDMK WPFKV G +KPMI VNYKGE+KQFAA
Sbjct: 61 QVAMNPVNTVFDAKRLIGRRFSDASVQSDMKFWPFKVTPGQADKPMIFVNYKGEEKQFAA 120
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVLIKMREIAEAYLGS++KNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP
Sbjct: 121 EEISSMVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATSVG KNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 181 TAAAIAYGLDKKATSVGIKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRKNKKDISG+ RALRRLRTACERAKRTLSSTAQTT+E+DSL
Sbjct: 241 NRMVNHFVQEFKRKNKKDISGDARALRRLRTACERAKRTLSSTAQTTVEVDSL 293
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 547 bits (1409), Expect = e-156
Identities = 277/293 (94%), Positives = 285/293 (96%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNPVNTVFDAKRLIGRR SDASVQSD +LWPF +I+G EKPMI V YKGE+KQFAA
Sbjct: 61 QVAMNPVNTVFDAKRLIGRRFSDASVQSDRQLWPFTIISGTAEKPMIVVEYKGEEKQFAA 120
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVLIKMREIAEA+LG+TVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP
Sbjct: 121 EEISSMVLIKMREIAEAFLGTTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRKNK+DI+G PRALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 241 NRMVNHFVQEFKRKNKQDITGQPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
>At3g12580 putative protein
Length = 650
Score = 545 bits (1404), Expect = e-155
Identities = 274/293 (93%), Positives = 285/293 (96%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP NTVFDAKRLIGRR SD SVQ+D WPFKV++GPGEKPMI VN+KGE+KQF+A
Sbjct: 61 QVAMNPTNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQFSA 120
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVLIKMREIAEA+LGS VKNAVVTVPAYFNDSQRQATKDAGVI+GLNV+RIINEP
Sbjct: 121 EEISSMVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEP 180
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKKA+SVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD
Sbjct: 181 TAAAIAYGLDKKASSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NRMVNHFVQEFKRKNKKDI+GNPRALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 241 NRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
>At1g16030 unknown protein
Length = 646
Score = 501 bits (1289), Expect = e-142
Identities = 252/293 (86%), Positives = 272/293 (92%), Gaps = 1/293 (0%)
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MA K E AIGIDLGTTYSCVGVW +DRVEII NDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 1 MATKSE-KAIGIDLGTTYSCVGVWMNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKN 59
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVA+NP NTVFDAKRLIGR+ SD SVQSD+ WPFKV++GPGEKPMI V+YK E+KQF+
Sbjct: 60 QVALNPQNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQFSP 119
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEP 180
EEISSMVL+KM+E+AEA+LG TVKNAVVTVPAYFNDSQRQATKDAG I+GLNVLRIINEP
Sbjct: 120 EEISSMVLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEP 179
Query: 181 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 240
TAAAIAYGLDKK T GEKNVLIFDLGGGTFDVSLLTIEEG+FEVKATAGDTHLGGEDFD
Sbjct: 180 TAAAIAYGLDKKGTKAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGEDFD 239
Query: 241 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
NR+VNHFV EF+RK+KKDI+GN RALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 240 NRLVNHFVAEFRRKHKKDIAGNARALRRLRTACERAKRTLSSTAQTTIEIDSL 292
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 381 bits (978), Expect = e-106
Identities = 196/288 (68%), Positives = 234/288 (81%), Gaps = 5/288 (1%)
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V FTDSERLIG+AAKNQ A+NP
Sbjct: 35 GSVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAVNP 94
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
TVFD KRLIGR+ D VQ D KL P++++ G KP I V K GE K F+ EEIS+
Sbjct: 95 ERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVNKDG-KPYIQVKIKDGETKVFSPEEISA 153
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AEAYLG +K+AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 154 MILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 213
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV +T GDTHLGGEDFD+R++
Sbjct: 214 AYGLDKKG---GEKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRIME 270
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+F++ K+K++KDIS + +AL +LR CERAKR LSS Q +EI+SL
Sbjct: 271 YFIKLIKKKHQKDISKDNKALGKLRRECERAKRALSSQHQVRVEIESL 318
>At5g28540 luminal binding protein
Length = 669
Score = 381 bits (978), Expect = e-106
Identities = 196/288 (68%), Positives = 234/288 (81%), Gaps = 5/288 (1%)
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+V FTDSERLIG+AAKNQ A+NP
Sbjct: 35 GSVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAVNP 94
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
TVFD KRLIGR+ D VQ D KL P++++ G KP I V K GE K F+ EEIS+
Sbjct: 95 ERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVNKDG-KPYIQVKIKDGETKVFSPEEISA 153
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
M+L KM+E AEAYLG +K+AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPTAAAI
Sbjct: 154 MILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAI 213
Query: 186 AYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVN 245
AYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV +T GDTHLGGEDFD+R++
Sbjct: 214 AYGLDKKG---GEKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRVME 270
Query: 246 HFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+F++ K+K++KDIS + +AL +LR CERAKR LSS Q +EI+SL
Sbjct: 271 YFIKLIKKKHQKDISKDNKALGKLRRECERAKRALSSQHQVRVEIESL 318
>At1g09080 putative luminal binding protein
Length = 678
Score = 375 bits (964), Expect = e-104
Identities = 188/287 (65%), Positives = 232/287 (80%), Gaps = 4/287 (1%)
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+ + VEIIANDQGNR TPS+VAFTD+ERLIG+AAKNQ A NP
Sbjct: 50 GTVIGIDLGTTYSCVGVYHNKHVEIIANDQGNRITPSWVAFTDTERLIGEAAKNQAAKNP 109
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSM 126
T+FD KRLIGR+ D VQ D+K P+KV+ G KP I V KGE+K F+ EEIS+M
Sbjct: 110 ERTIFDPKRLIGRKFDDPDVQRDIKFLPYKVVNKDG-KPYIQVKVKGEEKLFSPEEISAM 168
Query: 127 VLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIA 186
+L KM+E AEA+LG +K+AV+TVPAYFND+QRQATKDAG IAGLNV+RIINEPT AAIA
Sbjct: 169 ILTKMKETAEAFLGKKIKDAVITVPAYFNDAQRQATKDAGAIAGLNVVRIINEPTGAAIA 228
Query: 187 YGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNH 246
YGLDKK GE N+L++DLGGGTFDVS+LTI+ G+FEV +T+GDTHLGGEDFD+R++++
Sbjct: 229 YGLDKKG---GESNILVYDLGGGTFDVSILTIDNGVFEVLSTSGDTHLGGEDFDHRVMDY 285
Query: 247 FVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
F++ K+K KDIS + +AL +LR CE AKR+LS+ Q +EI+SL
Sbjct: 286 FIKLVKKKYNKDISKDHKALGKLRRECELAKRSLSNQHQVRVEIESL 332
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 290 bits (743), Expect = 5e-79
Identities = 153/282 (54%), Positives = 194/282 (68%), Gaps = 10/282 (3%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPVN 68
IGIDLGTT SCV V + +I N +G+RTTPS VA E L+G AK Q NP N
Sbjct: 55 IGIDLGTTNSCVSVMEGKTARVIENAEGSRTTPSVVAMNQKGELLVGTPAKRQAVTNPTN 114
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
T+F +KRLIGRR D Q +MK+ P+K++ P + N ++F+ +I + VL
Sbjct: 115 TIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAN----GQKFSPSQIGANVL 170
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
KM+E AEAYLG ++ AVVTVPAYFND+QRQATKDAG IAGL+V RIINEPTAAA++YG
Sbjct: 171 TKMKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIINEPTAAALSYG 230
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
++ K E + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN ++ + V
Sbjct: 231 MNNK-----EGVIAVFDLGGGTFDVSILEISSGVFEVKATNGDTFLGGEDFDNTLLEYLV 285
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEI 290
EFKR + D++ + AL+RLR A E+AK LSST QT I +
Sbjct: 286 NEFKRSDNIDLTKDNLALQRLREAAEKAKIELSSTTQTEINL 327
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 283 bits (725), Expect = 6e-77
Identities = 153/282 (54%), Positives = 192/282 (67%), Gaps = 10/282 (3%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAF-TDSERLIGDAAKNQVAMNPVN 68
IGIDLGTT SCV V + ++I N +G RTTPS VAF T E L+G AK Q NP N
Sbjct: 60 IGIDLGTTNSCVAVMEGKNPKVIENAEGARTTPSVVAFNTKGELLVGTPAKRQAVTNPTN 119
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
TV KRLIGR+ D Q +MK+ P+K++ P + N +Q++ +I + +L
Sbjct: 120 TVSGTKRLIGRKFDDPQTQKEMKMVPYKIVRAPNGDAWVEAN----GQQYSPSQIGAFIL 175
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
KM+E AEAYLG +V AVVTVPAYFND+QRQATKDAG IAGL+V RIINEPTAAA++YG
Sbjct: 176 TKMKETAEAYLGKSVTKAVVTVPAYFNDAQRQATKDAGRIAGLDVERIINEPTAAALSYG 235
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
+ K E + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 236 MTNK-----EGLIAVFDLGGGTFDVSVLEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 290
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEI 290
EFK D++ + AL+RLR A E+AK LSST+QT I +
Sbjct: 291 NEFKTTEGIDLAKDRLALQRLREAAEKAKIELSSTSQTEINL 332
>At4g24280 hsp 70-like protein
Length = 718
Score = 262 bits (670), Expect = 1e-70
Identities = 143/282 (50%), Positives = 191/282 (67%), Gaps = 9/282 (3%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPVN 68
+GIDLGTT S V + + I+ N +G RTTPS VA+T S +RL+G AK Q +NP N
Sbjct: 81 VGIDLGTTNSAVAAMEGGKPTIVTNAEGQRTTPSVVAYTKSGDRLVGQIAKRQAVVNPEN 140
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
T F KR IGR++++ V + K ++V+ E + + +KQFAAEEIS+ VL
Sbjct: 141 TFFSVKRFIGRKMNE--VDEESKQVSYRVVRD--ENNNVKLECPAINKQFAAEEISAQVL 196
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
K+ + A +L V AV+TVPAYFNDSQR ATKDAG IAGL VLRIINEPTAA++AYG
Sbjct: 197 RKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLAYG 256
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
D+KA + +L+FDLGGGTFDVS+L + +G+FEV +T+GDTHLGG+DFD R+V+
Sbjct: 257 FDRKA----NETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDWLA 312
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEI 290
EFK+ D+ + +AL+RL A E+AK LSS QT + +
Sbjct: 313 AEFKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSL 354
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 259 bits (661), Expect = 2e-69
Identities = 142/282 (50%), Positives = 191/282 (67%), Gaps = 9/282 (3%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDS-ERLIGDAAKNQVAMNPVN 68
+GIDLGTT S V + + I+ N +G RTTPS VA+T S +RL+G AK Q +NP N
Sbjct: 81 VGIDLGTTNSAVAAMEGGKPTIVTNAEGQRTTPSVVAYTKSKDRLVGQIAKRQAVVNPEN 140
Query: 69 TVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVL 128
T F KR IGRR+++ + +S K ++VI E + ++ KQFAAEEIS+ VL
Sbjct: 141 TFFSVKRFIGRRMNEVAEES--KQVSYRVIKD--ENGNVKLDCPAIGKQFAAEEISAQVL 196
Query: 129 IKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
K+ + A +L V AV+TVPAYFNDSQR ATKDAG IAGL VLRIINEPTAA++AYG
Sbjct: 197 RKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLAYG 256
Query: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
++K+ + +L+FDLGGGTFDVS+L + +G+FEV +T+GDTHLGG+DFD R+V+
Sbjct: 257 FERKS----NETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDWLA 312
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEI 290
FK+ D+ + +AL+RL A E+AK LSS QT + +
Sbjct: 313 STFKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSL 354
>At1g79930 putative heat-shock protein
Length = 831
Score = 210 bits (535), Expect = 6e-55
Identities = 110/285 (38%), Positives = 161/285 (55%), Gaps = 1/285 (0%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNT 69
+G D G V V + ++++ ND+ NR TP+ V F D +R IG A MNP N+
Sbjct: 4 VGFDFGNENCLVAVARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPKNS 63
Query: 70 VFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLI 129
+ KRLIGR+ SD +Q D+K PF V GP P+I NY GE + F ++ M+L
Sbjct: 64 ISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEKRAFTPTQVMGMMLS 123
Query: 130 KMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGL 189
++ IAE L + V + + +P YF D QR+A DA IAGL+ LR+I+E TA A+AYG+
Sbjct: 124 NLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLRLIHETTATALAYGI 183
Query: 190 DKKATSVGEK-NVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
K ++ NV D+G + V + ++G ++ + A D LGG DFD + NHF
Sbjct: 184 YKTDLPESDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNHFA 243
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+FK + K D+S N +A RLR CE+ K+ LS+ + I+ L
Sbjct: 244 AKFKDEYKIDVSQNAKASLRLRATCEKLKKVLSANPLAPLNIECL 288
>At1g79920 putative heat-shock protein (At1g79920)
Length = 736
Score = 208 bits (530), Expect = 2e-54
Identities = 109/285 (38%), Positives = 160/285 (55%), Gaps = 1/285 (0%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNT 69
+G D G V V + ++++ ND+ NR TP+ V F D +R IG A MNP N+
Sbjct: 4 VGFDFGNENCLVAVARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPKNS 63
Query: 70 VFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLI 129
+ KRLIGR+ SD +Q D+K PF V GP P+I NY GE + F ++ M+L
Sbjct: 64 ISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEIRAFTPTQVMGMMLS 123
Query: 130 KMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGL 189
++ IAE L + V + + +P YF D QR+A DA IAGL+ L +I+E TA A+AYG+
Sbjct: 124 NLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLHLIHETTATALAYGI 183
Query: 190 DKKATSVGEK-NVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
K ++ NV D+G + V + ++G ++ + A D LGG DFD + NHF
Sbjct: 184 YKTDLPENDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNHFA 243
Query: 249 QEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
+FK + K D+S N +A RLR CE+ K+ LS+ + I+ L
Sbjct: 244 AKFKDEYKIDVSQNAKASLRLRATCEKLKKVLSANPMAPLNIECL 288
>At1g11660 heat-shock protein like
Length = 773
Score = 192 bits (487), Expect = 2e-49
Identities = 100/286 (34%), Positives = 163/286 (56%), Gaps = 2/286 (0%)
Query: 10 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNT 69
+G D+G + V + ++++ ND+ NR P+ V+F + +R +G AA M+P +T
Sbjct: 4 VGFDVGNENCVIAVAKQRGIDVLLNDESNRENPAMVSFGEKQRFMGAAAAASATMHPKST 63
Query: 70 VFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLI 129
+ KRLIGR+ + VQ+D++L+PF+ I + Y GE + F+ +I M+L
Sbjct: 64 ISQLKRLIGRKFREPDVQNDLRLFPFETSEDSDGGIQIRLRYMGEIQSFSPVQILGMLLS 123
Query: 130 KMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGL 189
+++IAE L + V + V+ +P+YF +SQR A DA IAGL LR++++ TA A+ YG+
Sbjct: 124 HLKQIAEKSLKTPVSDCVIGIPSYFTNSQRLAYLDAAAIAGLRPLRLMHDSTATALGYGI 183
Query: 190 DKK--ATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHF 247
K + ++ D+G V + + E G V++ A D +LGG DFD + NHF
Sbjct: 184 YKTDLVANSSPTYIVFIDIGHCDTQVCVASFESGSMRVRSHAFDRNLGGRDFDEVLFNHF 243
Query: 248 VQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
EFK K D+ N +A RLR +CE+ K+ LS+ A+ + I+ L
Sbjct: 244 ALEFKEKYNIDVYTNTKACVRLRASCEKVKKVLSANAEAQLNIECL 289
>At2g32120 70kD heat shock protein
Length = 563
Score = 125 bits (315), Expect = 2e-29
Identities = 93/294 (31%), Positives = 143/294 (48%), Gaps = 26/294 (8%)
Query: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVA----M 64
A+GID+GT+ + VW +V I+ N + + S+V F D G NQ+A M
Sbjct: 30 ALGIDIGTSQCSIAVWNGSQVHILRNTRNQKLIKSFVTFKDEVPAGG--VSNQLAHEQEM 87
Query: 65 NPVNTVFDAKRLIGRRISDASVQSDMKLWPFKV-IAGPGEKPMIGVNYKGEDKQFAAEEI 123
+F+ KRL+GR +D V + L PF V G +P I + EE+
Sbjct: 88 LTGAAIFNMKRLVGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNAWRSTTPEEV 146
Query: 124 SSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAA 183
++ L+++R +AEA L V+N V+TVP F+ Q + A +AGL+VLR++ EPTA
Sbjct: 147 LAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQLTRFERACAMAGLHVLRLMPEPTAI 206
Query: 184 AIAYGLDKKAT------SVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGE 237
A+ Y ++ T S E+ +IF++G G DV++ G+ ++KA AG + +GGE
Sbjct: 207 ALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDVAVTATAGGVSQIKALAG-SPIGGE 265
Query: 238 DFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEID 291
D + H N A LR A + A L+ IE+D
Sbjct: 266 DILQNTIRHIAPP-----------NEEASGLLRVAAQDAIHRLTDQENVQIEVD 308
>At2g38400 putative beta-alanine-pyruvate aminotransferase
Length = 477
Score = 31.2 bits (69), Expect = 0.69
Identities = 31/126 (24%), Positives = 53/126 (41%), Gaps = 12/126 (9%)
Query: 118 FAAEEISSMVLIKMREIAEAY-LGSTVKN----AVVTVPAYFNDSQRQATKDAGVIAGLN 172
F A + ++ + I + LG+ V V+T +YFN AGL
Sbjct: 307 FEAHNVVPDIVTMAKGIGNGFPLGAVVTTPEIAGVLTRRSYFNTF---GGNSVSTTAGLA 363
Query: 173 VLRIIN----EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKAT 228
VL +I + AA + L +K T + EK+ +I D+ G + + + + + AT
Sbjct: 364 VLNVIEKEKLQENAAMVGSYLKEKLTQLKEKHEIIGDVRGRGLMLGVELVSDRKLKTPAT 423
Query: 229 AGDTHL 234
A H+
Sbjct: 424 AETLHI 429
>At5g44800 helicase-like protein
Length = 2228
Score = 28.1 bits (61), Expect = 5.8
Identities = 20/59 (33%), Positives = 32/59 (53%), Gaps = 4/59 (6%)
Query: 233 HLGGEDFDNRMVNHF-VQEFKRKNKKDISGNPRALRRLRTACERAKRTLS---STAQTT 287
H E +N+ V + KRK +K + +L R +T +RAK++LS S++QTT
Sbjct: 253 HASSEIVENKTVAEMETGKGKRKKRKRELNDGESLERCKTDKKRAKKSLSKVGSSSQTT 311
>At3g61670 putative protein
Length = 790
Score = 27.7 bits (60), Expect = 7.6
Identities = 15/46 (32%), Positives = 23/46 (49%)
Query: 27 DRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVNTVFD 72
D + I ND+GN++ S +ERL+ A K + P N +D
Sbjct: 635 DLTKSIQNDEGNKSNVSINGHPLTERLLRKAEKQAGVIQPGNYWYD 680
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,118,344
Number of Sequences: 26719
Number of extensions: 249148
Number of successful extensions: 599
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 556
Number of HSP's gapped (non-prelim): 23
length of query: 293
length of database: 11,318,596
effective HSP length: 99
effective length of query: 194
effective length of database: 8,673,415
effective search space: 1682642510
effective search space used: 1682642510
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136140.11


