

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136140.1 - phase: 0
(115 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
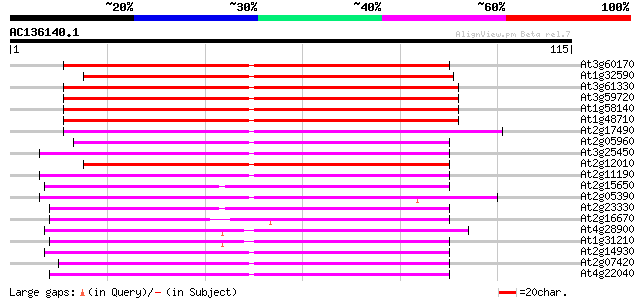
Score E
Sequences producing significant alignments: (bits) Value
At3g60170 putative protein 98 6e-22
At1g32590 hypothetical protein, 5' partial 81 1e-16
At3g61330 copia-type polyprotein 78 6e-16
At3g59720 copia-type reverse transcriptase-like protein 78 6e-16
At1g58140 hypothetical protein 78 6e-16
At1g48710 hypothetical protein 75 5e-15
At2g17490 putative retroelement pol polyprotein 71 8e-14
At2g05960 putative retroelement pol polyprotein 68 8e-13
At3g25450 hypothetical protein 67 1e-12
At2g12010 pseudogene 64 9e-12
At2g11190 putative retroelement pol polyprotein 63 3e-11
At2g15650 putative retroelement pol polyprotein 62 4e-11
At2g05390 putative retroelement pol polyprotein 57 1e-09
At2g23330 putative retroelement pol polyprotein 54 1e-08
At2g16670 putative retroelement pol polyprotein 50 1e-07
At4g28900 putative protein 50 2e-07
At1g31210 putative reverse transcriptase 50 2e-07
At2g14930 pseudogene 49 5e-07
At2g07420 putative retroelement pol polyprotein 49 5e-07
At4g22040 LTR retrotransposon like protein 47 1e-06
>At3g60170 putative protein
Length = 1339
Score = 98.2 bits (243), Expect = 6e-22
Identities = 48/79 (60%), Positives = 59/79 (73%), Gaps = 1/79 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V+ TP EAWS +KP V FR+FG I Y HIP +R K LDD S KCV LGVSE+SKA+R
Sbjct: 643 VEGMTPEEAWSGRKPVVEYFRVFGCIGYVHIPDQKRSK-LDDKSKKCVFLGVSEESKAWR 701
Query: 72 FFDRIIKKIVVSEDVAFDE 90
+D ++KKIV+S+DV FDE
Sbjct: 702 LYDPVMKKIVISKDVVFDE 720
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 80.9 bits (198), Expect = 1e-16
Identities = 41/76 (53%), Positives = 53/76 (68%), Gaps = 1/76 (1%)
Query: 16 TPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFDR 75
TP E WS KP V RIFG +AYA +P+ +R K LD+ S KCV+ GVS++SKAYR +D
Sbjct: 609 TPEEKWSSWKPSVEHLRIFGSLAYALVPYQKRIK-LDEKSIKCVMFGVSKESKAYRLYDP 667
Query: 76 IIKKIVVSEDVAFDEQ 91
KI++S DV FDE+
Sbjct: 668 ATGKILISRDVQFDEE 683
Score = 26.2 bits (56), Expect = 2.8
Identities = 11/23 (47%), Positives = 16/23 (68%)
Query: 51 LDDNSNKCVLLGVSEKSKAYRFF 73
+DD S KC +SEKS+ ++FF
Sbjct: 483 IDDFSRKCWTYLLSEKSETFQFF 505
>At3g61330 copia-type polyprotein
Length = 1352
Score = 78.2 bits (191), Expect = 6e-16
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +KP V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 78.2 bits (191), Expect = 6e-16
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +KP V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRNK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>At1g58140 hypothetical protein
Length = 1320
Score = 78.2 bits (191), Expect = 6e-16
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +KP V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>At1g48710 hypothetical protein
Length = 1352
Score = 75.1 bits (183), Expect = 5e-15
Identities = 35/81 (43%), Positives = 50/81 (61%), Gaps = 1/81 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
V TP EAWS +K V R+FG IA+AH+P +R K LDD S K + +G SK Y+
Sbjct: 675 VSGKTPQEAWSGRKSGVSHLRVFGSIAHAHVPDEKRSK-LDDKSEKYIFIGYDNNSKGYK 733
Query: 72 FFDRIIKKIVVSEDVAFDEQG 92
++ KK ++S ++ FDE+G
Sbjct: 734 LYNPDTKKTIISRNIVFDEEG 754
>At2g17490 putative retroelement pol polyprotein
Length = 822
Score = 71.2 bits (173), Expect = 8e-14
Identities = 37/90 (41%), Positives = 51/90 (56%), Gaps = 1/90 (1%)
Query: 12 VKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYR 71
+K+ TP EAWS KP V ++FG I Y HIP +R+K DD S + + +G S ++K YR
Sbjct: 367 LKNKTPLEAWSDSKPSVSHMKVFGSICYVHIPDEKRRK-WDDKSKRAIFVGYSSQTKGYR 425
Query: 72 FFDRIIKKIVVSEDVAFDEQGMLSLILKEI 101
+ KI +S DV FDE KE+
Sbjct: 426 VYLLKENKIDISRDVIFDEDSKWDWEKKEV 455
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 67.8 bits (164), Expect = 8e-13
Identities = 36/77 (46%), Positives = 45/77 (57%), Gaps = 1/77 (1%)
Query: 14 SSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFF 73
+ TPYEA KKP V R+FG ++YA + F +K LD+ S V LG SKAY
Sbjct: 599 NQTPYEALKKKKPNVEHLRVFGCVSYAKVEFPHLRK-LDERSRILVYLGTETGSKAYTLL 657
Query: 74 DRIIKKIVVSEDVAFDE 90
D +KI+VS DV FDE
Sbjct: 658 DPTTRKIIVSRDVVFDE 674
>At3g25450 hypothetical protein
Length = 1343
Score = 67.4 bits (163), Expect = 1e-12
Identities = 37/84 (44%), Positives = 47/84 (55%), Gaps = 1/84 (1%)
Query: 7 VSSLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEK 66
V + + + TPYE + KKP V R+FG ++YA + KK LDD S V LG
Sbjct: 628 VGTRSLSNQTPYEVFKHKKPNVEHLRVFGCVSYAKVEVPNLKK-LDDRSRMLVYLGTEPG 686
Query: 67 SKAYRFFDRIIKKIVVSEDVAFDE 90
SKAYR D ++I VS DV FDE
Sbjct: 687 SKAYRLLDPTKRRIFVSRDVVFDE 710
>At2g12010 pseudogene
Length = 616
Score = 64.3 bits (155), Expect = 9e-12
Identities = 32/75 (42%), Positives = 46/75 (60%), Gaps = 1/75 (1%)
Query: 16 TPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAYRFFDR 75
TP E WS KP V ++FG I Y HIP +R+K LD+ + + + +G S KSK YR
Sbjct: 416 TPIEKWSGHKPTVEHLKVFGSICYIHIPKEKRRK-LDEKAMRVIFIGYSNKSKGYRVLLL 474
Query: 76 IIKKIVVSEDVAFDE 90
+++ +S DVAF+E
Sbjct: 475 DKERVEISRDVAFEE 489
>At2g11190 putative retroelement pol polyprotein
Length = 469
Score = 62.8 bits (151), Expect = 3e-11
Identities = 39/84 (46%), Positives = 47/84 (55%), Gaps = 1/84 (1%)
Query: 7 VSSLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEK 66
V++ + + TPYEA+ KKP V R+FG I YA + KK LDD S V L
Sbjct: 12 VATRTLVNQTPYEAFIGKKPNVERLRVFGCIGYAKVETVNLKK-LDDCSRMLVHLRTEPG 70
Query: 67 SKAYRFFDRIIKKIVVSEDVAFDE 90
SKAYR D KK+VVS V FDE
Sbjct: 71 SKAYRLLDPTKKKVVVSRYVVFDE 94
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 62.4 bits (150), Expect = 4e-11
Identities = 35/83 (42%), Positives = 45/83 (54%), Gaps = 1/83 (1%)
Query: 8 SSLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKS 67
S I TP E W KP V RIFG I Y HIP Q+++ LD + +L+G S ++
Sbjct: 650 SKAIEDDVTPMEKWCGHKPNVSHLRIFGSICYVHIP-DQKRRKLDAKAKCGILIGYSNQT 708
Query: 68 KAYRFFDRIIKKIVVSEDVAFDE 90
K YR F +K+ VS DV F E
Sbjct: 709 KGYRVFLLEDEKVEVSRDVVFQE 731
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 57.4 bits (137), Expect = 1e-09
Identities = 38/97 (39%), Positives = 49/97 (50%), Gaps = 4/97 (4%)
Query: 7 VSSLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEK 66
V + +++ TPYE + +KP V R+FG I YA I +K LDD S V LG
Sbjct: 596 VGTRSLQNQTPYEVFKQRKPNVEHLRVFGCIGYAKIEGPHLRK-LDDRSKMLVYLGTEPG 654
Query: 67 SKAYRFFDRIIKKIVV---SEDVAFDEQGMLSLILKE 100
SKAYR D +KI+ S+ D G SL L E
Sbjct: 655 SKAYRLLDPTNRKIIKWNNSDSETRDISGTFSLTLGE 691
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 54.3 bits (129), Expect = 1e-08
Identities = 31/82 (37%), Positives = 40/82 (47%), Gaps = 1/82 (1%)
Query: 9 SLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSK 68
S ++K TPYE ++P R FG + YAHI + K S KCV +G K
Sbjct: 700 SAVLKGKTPYELLFGERPSYDMLRSFGCLCYAHIR-PRNKDKFTSRSRKCVFIGYPHGKK 758
Query: 69 AYRFFDRIIKKIVVSEDVAFDE 90
A+R +D KI S DV F E
Sbjct: 759 AWRVYDLETGKIFASRDVRFHE 780
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 50.4 bits (119), Expect = 1e-07
Identities = 30/85 (35%), Positives = 41/85 (47%), Gaps = 7/85 (8%)
Query: 9 SLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLD---DNSNKCVLLGVSE 65
S ++ STPYE K+P R+FG + YAH R + D + S +CV +G
Sbjct: 590 SSVLNDSTPYERLHKKQPRFDHLRVFGSLCYAH----NRNRGGDKFAERSRRCVFVGYPH 645
Query: 66 KSKAYRFFDRIIKKIVVSEDVAFDE 90
K +R FD + VS DV F E
Sbjct: 646 GQKGWRLFDLEQNEFFVSRDVVFSE 670
>At4g28900 putative protein
Length = 1415
Score = 50.1 bits (118), Expect = 2e-07
Identities = 30/88 (34%), Positives = 42/88 (47%), Gaps = 3/88 (3%)
Query: 8 SSLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHI-PFSQRKKTLDDNSNKCVLLGVSEK 66
SS++ +PYE K P R+FG Y ++ P++ K D S CV G +EK
Sbjct: 627 SSVLKDQKSPYEVLMGKAPVYTSLRVFGCACYPNLRPYASNK--FDPKSLLCVFTGYNEK 684
Query: 67 SKAYRFFDRIIKKIVVSEDVAFDEQGML 94
K Y+ F KI ++ V FDE L
Sbjct: 685 YKGYKCFHPPTGKIYINRHVLFDESKFL 712
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 50.1 bits (118), Expect = 2e-07
Identities = 30/83 (36%), Positives = 45/83 (54%), Gaps = 3/83 (3%)
Query: 9 SLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHI-PFSQRKKTLDDNSNKCVLLGVSEKS 67
S ++K+ +PYEA +KP R+FG Y + P +Q K D S +CV LG + +
Sbjct: 657 SSVLKNLSPYEALFGEKPDYSSLRVFGSACYPCLRPLAQNK--FDPRSLQCVFLGYNSQY 714
Query: 68 KAYRFFDRIIKKIVVSEDVAFDE 90
K YR F K+ +S +V F+E
Sbjct: 715 KGYRCFYPPTGKVYISRNVIFNE 737
>At2g14930 pseudogene
Length = 1149
Score = 48.5 bits (114), Expect = 5e-07
Identities = 29/83 (34%), Positives = 40/83 (47%), Gaps = 1/83 (1%)
Query: 8 SSLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKS 67
+S + S+TPY+ K P R FG + + R K D S KC+ LG +EK
Sbjct: 649 TSALDSSTTPYQVLFGKAPDYSALRTFGCACFPTLRAYARNK-FDPRSLKCIFLGYTEKY 707
Query: 68 KAYRFFDRIIKKIVVSEDVAFDE 90
K YR F ++ +S V FDE
Sbjct: 708 KGYRCFFPPTNRVYLSRHVLFDE 730
>At2g07420 putative retroelement pol polyprotein
Length = 1664
Score = 48.5 bits (114), Expect = 5e-07
Identities = 26/80 (32%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Query: 11 IVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSKAY 70
I+ S+P+E + KP++ R+FG + + I QR K L S K + +G S K Y
Sbjct: 640 ILLDSSPFEVLNKVKPFINHLRVFGCVCFVLISGEQRNK-LQPKSTKGMFIGYSINQKGY 698
Query: 71 RFFDRIIKKIVVSEDVAFDE 90
+ + +K+++S DV F E
Sbjct: 699 KCYVLETRKVLISRDVKFLE 718
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 47.4 bits (111), Expect = 1e-06
Identities = 27/82 (32%), Positives = 39/82 (46%), Gaps = 1/82 (1%)
Query: 9 SLIVKSSTPYEAWSVKKPYVMPFRIFGFIAYAHIPFSQRKKTLDDNSNKCVLLGVSEKSK 68
S++++ +PYE P R+FG + YAH + K S +CV +G K
Sbjct: 490 SMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYAHNQNHKGDK-FAARSRRCVFVGYPHGQK 548
Query: 69 AYRFFDRIIKKIVVSEDVAFDE 90
+R FD +K VS DV F E
Sbjct: 549 GWRLFDLEEQKFFVSRDVIFQE 570
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,520,547
Number of Sequences: 26719
Number of extensions: 93173
Number of successful extensions: 381
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 267
Number of HSP's gapped (non-prelim): 78
length of query: 115
length of database: 11,318,596
effective HSP length: 91
effective length of query: 24
effective length of database: 8,887,167
effective search space: 213292008
effective search space used: 213292008
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC136140.1


