

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135848.10 - phase: 0
(346 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
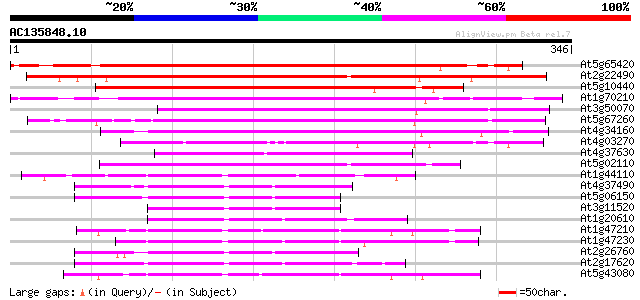
Score E
Sequences producing significant alignments: (bits) Value
At5g65420 D-type cyclin (emb|CAB41347.1) 278 4e-75
At2g22490 putative cyclin D 269 1e-72
At5g10440 cyclin protein - like 242 2e-64
At1g70210 unknown protein 178 4e-45
At3g50070 cyclin D3-like protein 176 1e-44
At5g67260 cyclin D3-like protein 161 6e-40
At4g34160 cyclin delta-3 156 2e-38
At4g03270 putative D-type cyclin 109 2e-24
At4g37630 putative protein 100 1e-21
At5g02110 putative protein 77 1e-14
At1g44110 mitotic cyclin a2-type, putative 69 3e-12
At4g37490 cyclin cyc1 67 1e-11
At5g06150 mitosis-specific cyclin 1b 62 6e-10
At3g11520 cyclin box 60 2e-09
At1g20610 hypothetical protein 60 2e-09
At1g47210 cyclin like protein 59 4e-09
At1g47230 cyclin like protein 58 7e-09
At2g26760 putative cyclin 57 1e-08
At2g17620 putative cyclin 2 56 3e-08
At5g43080 cyclin A-type 55 7e-08
>At5g65420 D-type cyclin (emb|CAB41347.1)
Length = 308
Score = 278 bits (710), Expect = 4e-75
Identities = 160/323 (49%), Positives = 218/323 (66%), Gaps = 31/323 (9%)
Query: 1 MAESFDSAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEE 60
MAE ++ E +LLC+E+N VDD G+ +++ +G ++SEE
Sbjct: 1 MAE--ENLELSLLCTESN--------VDDEGMIVDETPIEISIPQMGFSQ-----SESEE 45
Query: 61 IVKVMVEKEKDHLPREDYLIRLRGGDLDLSV-RREALDWIWKAHAYYGFGPLSLCLSVNY 119
I+ MVEKEK HLP +DY+ RLR GDLDL+V RR+AL+WIWKA + FGPL CL++NY
Sbjct: 46 IIMEMVEKEKQHLPSDDYIKRLRSGDLDLNVGRRDALNWIWKACEVHQFGPLCFCLAMNY 105
Query: 120 LDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRM 179
LDRFLSV P G W +QLLAVAC SLAAK+EE +VP +DLQVG+P+FVF+AK++QRM
Sbjct: 106 LDRFLSVHDLPSGKGWILQLLAVACLSLAAKIEETEVPMLIDLQVGDPQFVFEAKSVQRM 165
Query: 180 ELMILSSLGWKMRALTPCSFIDYFLAKIS-CEKYPDKSLIARSVQLILNIIKGIDFLEFR 238
EL++L+ L W++RA+TPCS+I YFL K+S C++ P +LI+RS+Q+I + KGIDFLEFR
Sbjct: 166 ELLVLNKLKWRLRAITPCSYIRYFLRKMSKCDQEPSNTLISRSLQVIASTTKGIDFLEFR 225
Query: 239 SSEIAAAVAISLK-ELPTQEVDKAITD--FFIVDKERVLKCVELIRDLSLIKVGGNNFAS 295
SE+AAAVA+S+ EL D + F ++ KERV K E+I G++ S
Sbjct: 226 PSEVAAAVALSVSGELQRVHFDNSSFSPLFSLLQKERVKKIGEMIES------DGSDLCS 279
Query: 296 FVPQSPIGVLD--AGCMSFKSDE 316
Q+P GVL+ A C SFK+ +
Sbjct: 280 ---QTPNGVLEVSACCFSFKTHD 299
>At2g22490 putative cyclin D
Length = 361
Score = 269 bits (688), Expect = 1e-72
Identities = 156/345 (45%), Positives = 216/345 (62%), Gaps = 26/345 (7%)
Query: 11 NLLCSENNSTCFDDVVVDD----SGISPSWDHTN---VNLDNVGSDSFLCFVAQS----- 58
NL C E + + D DD G + D+ + DN G + + + S
Sbjct: 4 NLACGETSESWIIDNDDDDINYGGGFTNEIDYNHQLFAKDDNFGGNGSIPMMGSSSSSLS 63
Query: 59 EEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVN 118
E+ +K M+ +E + P DY+ RL GDLDLSVR +ALDWI K A+Y FG L +CLS+N
Sbjct: 64 EDRIKEMLVREIEFCPGTDYVKRLLSGDLDLSVRNQALDWILKVCAHYHFGHLCICLSMN 123
Query: 119 YLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQR 178
YLDRFL+ ++ P+ W QLLAV+C SLA+KMEE VP VDLQV +PKFVF+AKTI+R
Sbjct: 124 YLDRFLTSYELPKDKDWAAQLLAVSCLSLASKMEETDVPHIVDLQVEDPKFVFEAKTIKR 183
Query: 179 MELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFR 238
MEL+++++L W+++ALTP SFIDYF+ KIS + ++LI RS + ILN K I+FL+FR
Sbjct: 184 MELLVVTTLNWRLQALTPFSFIDYFVDKIS--GHVSENLIYRSSRFILNTTKAIEFLDFR 241
Query: 239 SSEIAAAVAISLK---ELPTQEVDKAITDFFIVDKERVLKCVELIRDL---------SLI 286
SEIAAA A+S+ E + +KA++ V +ERV +C+ L+R L SL
Sbjct: 242 PSEIAAAAAVSVSISGETECIDEEKALSSLIYVKQERVKRCLNLMRSLTGEENVRGTSLS 301
Query: 287 KVGGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSPN 331
+ VP SP+GVL+A C+S++S+E T SC NSS SSP+
Sbjct: 302 QEQARVAVRAVPASPVGVLEATCLSYRSEERTVESCTNSSQSSPD 346
>At5g10440 cyclin protein - like
Length = 317
Score = 242 bits (617), Expect = 2e-64
Identities = 131/249 (52%), Positives = 172/249 (68%), Gaps = 25/249 (10%)
Query: 54 FVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSL 113
F +SEEIV+ M+EKE+ H PR+DYL RLR GDLD +VR +AL WIWKA FGPL +
Sbjct: 31 FPLESEEIVREMIEKERQHSPRDDYLKRLRNGDLDFNVRIQALGWIWKACEELQFGPLCI 90
Query: 114 CLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQA 173
CL++NYLDRFLSV P G WTVQLLAVAC SLAAK+EE VP+ + LQVG P FVF+A
Sbjct: 91 CLAMNYLDRFLSVHDLPSGKAWTVQLLAVACLSLAAKIEETNVPELMQLQVGAPMFVFEA 150
Query: 174 KTIQRMELMILSSLGWKMRALTPCSFIDYFLAKIS-CEKYPDKSLIARSVQ--------- 223
K++QRMEL++L+ L W++RA+TPCS++ YFL+KI+ ++ P L+ RS+Q
Sbjct: 151 KSVQRMELLVLNVLRWRLRAVTPCSYVRYFLSKINGYDQEPHSRLVTRSLQVIASTTKGD 210
Query: 224 ----------LILNIIKGIDFLEFRSSEIAAAVAISLKELPTQEVDK--AITDFFIVDKE 271
LI+++ GIDFLEFR+SEIAAAVA+S+ + DK + F ++KE
Sbjct: 211 RLGLFFFKGVLIVDVWAGIDFLEFRASEIAAAVALSVS---GEHFDKFSFSSSFSSLEKE 267
Query: 272 RVLKCVELI 280
RV K E+I
Sbjct: 268 RVKKIGEMI 276
>At1g70210 unknown protein
Length = 339
Score = 178 bits (451), Expect = 4e-45
Identities = 123/349 (35%), Positives = 183/349 (52%), Gaps = 39/349 (11%)
Query: 1 MAESFDSAESNLLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEE 60
M+ SF S + +L C E++ + VD S + +D+ DS CF+
Sbjct: 12 MSVSF-SNDMDLFCGEDSGVFSGESTVDFS---------SSEVDSWPGDSIACFI----- 56
Query: 61 IVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYL 120
E E+ +P DYL R + LD S R +++ WI K AYY F PL+ L+VNY+
Sbjct: 57 ------EDERHFVPGHDYLSRFQTRSLDASAREDSVAWILKVQAYYNFQPLTAYLAVNYM 110
Query: 121 DRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRME 180
DRFL + P W +QLLAVAC SLAAKMEE+ VP D QV K++F+AKTI+RME
Sbjct: 111 DRFLYARRLPETSGWPMQLLAVACLSLAAKMEEILVPSLFDFQVAGVKYLFEAKTIKRME 170
Query: 181 LMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFRSS 240
L++LS L W++R++TP FI +F KI I+ + ++IL+ IK FLE+ S
Sbjct: 171 LLVLSVLDWRLRSVTPFDFISFFAYKIDPSGTFLGFFISHATEIILSNIKEASFLEYWPS 230
Query: 241 EIAAAVAISL-KELPT-------QEVDKAITDFFIVDKERVLKCVELIRDLSLIKVGGNN 292
IAAA + + ELP+ E + D + KE++++C L++ ++ I+ N
Sbjct: 231 SIAAAAILCVANELPSLSSVVNPHESPETWCDG--LSKEKIVRCYRLMKAMA-IENNRLN 287
Query: 293 FASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSPNAKRMKFDGPS 341
+ + + V + ++ SDE +S SS KR K G S
Sbjct: 288 TPKVIAKLRVSVRASSTLTRPSDE-------SSFSSSSPCKRRKLSGYS 329
>At3g50070 cyclin D3-like protein
Length = 361
Score = 176 bits (447), Expect = 1e-44
Identities = 102/246 (41%), Positives = 144/246 (58%), Gaps = 5/246 (2%)
Query: 92 RREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKM 151
R +ALDWI+K ++YGF L+ L+VNY DRF++ +F W QL A+AC SLAAK+
Sbjct: 86 REKALDWIFKVKSHYGFNSLTALLAVNYFDRFITSRKFQTDKPWMSQLTALACLSLAAKV 145
Query: 152 EEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEK 211
EE++VP +D QV E ++VF+AKTIQRMEL++LS+L W+M +TP SF D+ + + S +
Sbjct: 146 EEIRVPFLLDFQVEEARYVFEAKTIQRMELLVLSTLDWRMHPVTPISFFDHIIRRYSFKS 205
Query: 212 YPDKSLIARSVQLILNIIKGIDFLEFRSSEIAAAVAIS----LKELPTQEVDKAITDFFI 267
+ ++R L+L+II FL F S +A A+ +S LK +
Sbjct: 206 HHQLEFLSRCESLLLSIIPDSRFLSFSPSVLATAIMVSVIRDLKMCDEAVYQSQLMTLLK 265
Query: 268 VDKERVLKCVELIRDLSLIKVGGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSH 327
VD E+V KC EL+ D S K N+ P SPIGV DA S S+E S S
Sbjct: 266 VDSEKVNKCYELVLDHSPSKKRMMNWMQ-QPASPIGVFDASFSSDSSNESWVVSASASVS 324
Query: 328 SSPNAK 333
SSP+++
Sbjct: 325 SSPSSE 330
>At5g67260 cyclin D3-like protein
Length = 367
Score = 161 bits (407), Expect = 6e-40
Identities = 112/327 (34%), Positives = 175/327 (53%), Gaps = 19/327 (5%)
Query: 12 LLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFL----CFVAQSEEIVKVMVE 67
L C E DD+ DD G D + ++V FL F+ +EI+ ++
Sbjct: 21 LYCEEETGFVEDDL--DDDG---DLDFLEKSDESVVKFQFLPLLDMFLWDDDEILS-LIS 74
Query: 68 KEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVF 127
KE + P + ++ G L +S R+EALDW+ + ++YGF L+ L+VNY DRF++
Sbjct: 75 KENETNPC--FGEQILDGFL-VSCRKEALDWVLRVKSHYGFTSLTAILAVNYFDRFMTSI 131
Query: 128 QFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSL 187
+ W QL+AVA SLAAK+EE++VP +DLQV E +++F+AKTIQRMEL+ILS+L
Sbjct: 132 KLQTDKPWMSQLVAVASLSLAAKVEEIQVPLLLDLQVEEARYLFEAKTIQRMELLILSTL 191
Query: 188 GWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFRSSEIAAAVA 247
W+M +TP SF D+ + + + + + +L++++I F+ + S +A A+
Sbjct: 192 QWRMHPVTPISFFDHIIRRFGSKWHQQLDFCRKCERLLISVIADTRFMRYFPSVLATAIM 251
Query: 248 I----SLKELPTQEVDKAITDFFIVDKERVLKCVELIRDLSLIKVGGNNFASFVPQSPIG 303
I LK E IT V++E+V +C EL+ + + K N SP G
Sbjct: 252 ILVFEELKPCDEVEYQSQITTLLKVNQEKVNECYELLLEHNPSKKRMMNLVD--QDSPSG 309
Query: 304 VLDAGCMSFKSDELTNGSCPNSSHSSP 330
VLD S S ++ + +SS SSP
Sbjct: 310 VLDFDDSSNSSWNVSTTASVSSSSSSP 336
>At4g34160 cyclin delta-3
Length = 376
Score = 156 bits (394), Expect = 2e-38
Identities = 99/292 (33%), Positives = 160/292 (53%), Gaps = 26/292 (8%)
Query: 57 QSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSV-RREALDWIWKAHAYYGFGPLSLCL 115
+ E++V + ++E+ L D D+ LS R+EA+ WI + +A+YGF L+ L
Sbjct: 59 EDEDLVTLFSKEEEQGLSCLD--------DVYLSTDRKEAVGWILRVNAHYGFSTLAAVL 110
Query: 116 SVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKT 175
++ YLD+F+ + R W +QL++VAC SLAAK+EE +VP +D QV E K+VF+AKT
Sbjct: 111 AITYLDKFICSYSLQRDKPWMLQLVSVACLSLAAKVEETQVPLLLDFQVEETKYVFEAKT 170
Query: 176 IQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFL 235
IQRMEL+ILS+L WKM +TP SF+D+ + ++ + + + +L+L++I F+
Sbjct: 171 IQRMELLILSTLEWKMHLITPISFVDHIIRRLGLKNNAHWDFLNKCHRLLLSVISDSRFV 230
Query: 236 EFRSSEIAAAVAISLKE----LPTQEVDKAITDFFIVDKERVLKCVELIRDLSLIKVG-- 289
+ S +AAA + + E + + KE+V C +LI L + ++G
Sbjct: 231 GYLPSVVAAATMMRIIEQVDPFDPLSYQTNLLGVLNLTKEKVKTCYDLILQLPVDRIGLQ 290
Query: 290 ---------GNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSPNA 332
++ +S SP V+DA F SDE +N S SS + P +
Sbjct: 291 IQIQSSKKRKSHDSSSSLNSPSCVIDAN--PFNSDESSNDSWSASSCNPPTS 340
>At4g03270 putative D-type cyclin
Length = 302
Score = 109 bits (273), Expect = 2e-24
Identities = 88/272 (32%), Positives = 144/272 (52%), Gaps = 19/272 (6%)
Query: 69 EKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQ 128
E H+P Y L+ LS R +A+ I + + P L+VNYLDRFLS
Sbjct: 35 EFQHMPSSHYFHSLKSSAFLLSNRNQAISSITQYSRKFD-DPSLTYLAVNYLDRFLSSED 93
Query: 129 FPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLG 188
P+ W ++L++++C SL+AKM + + S DL V E +F F A+ I+RME +IL +L
Sbjct: 94 MPQSKPWILKLISLSCVSLSAKMRKPDMSVS-DLPV-EGEF-FDAQMIERMENVILGALK 150
Query: 189 WKMRALTPCSFIDYFLAKISCEKYP----DKSLIARSVQLILNIIKGIDFLEFRSSEIAA 244
W+MR++TP SF+ +F++ ++ SL +++ L ++ I FLEF+ S IA
Sbjct: 151 WRMRSVTPFSFLAFFISLFELKEEDPLLLKHSLKSQTSDLTFSLQHDISFLEFKPSVIAG 210
Query: 245 AVAI--SLKELPTQE--VDKAITDFFIVDKERVLKCVELIRDLSLIKVGGNNFASFVPQS 300
A + S + P Q I V+K+ +++C + I++ +I VG N ++ ++
Sbjct: 211 AALLFASFELCPLQFPCFSNRINQCTYVNKDELMECYKAIQERDII-VGENEGST---ET 266
Query: 301 PIGVLD---AGCMSFKSDELTNGSCPNSSHSS 329
+ VLD + C S KS +T S P +S
Sbjct: 267 AVNVLDQQFSSCESDKSITITASSSPKRRKTS 298
>At4g37630 putative protein
Length = 321
Score = 100 bits (249), Expect = 1e-21
Identities = 65/161 (40%), Positives = 91/161 (56%), Gaps = 3/161 (1%)
Query: 90 SVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAA 149
S R A+DWI H + P S L N + R +S + R TW ++LL+VAC SLAA
Sbjct: 70 SDRLIAIDWILTVHKNKIWVPTSNSLHCNLILRSVSPQKIHRYETWAMRLLSVACLSLAA 129
Query: 150 KMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISC 209
KMEE VP + + FVF+ I++ EL+ILS+L WKM +TP + +YFLAKIS
Sbjct: 130 KMEERIVP-GLSQYPQDHDFVFKPDVIRKTELLILSTLDWKMNLITPFHYFNYFLAKISQ 188
Query: 210 EKYP-DKSLI-ARSVQLILNIIKGIDFLEFRSSEIAAAVAI 248
+ + K L+ RS +L + K I F E+R +AA +
Sbjct: 189 DNHSVSKDLVLLRSSDSLLALTKEISFTEYRQFVVAAVTTL 229
>At5g02110 putative protein
Length = 341
Score = 77.4 bits (189), Expect = 1e-14
Identities = 58/225 (25%), Positives = 109/225 (47%), Gaps = 7/225 (3%)
Query: 56 AQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCL 115
A EE + + +EKE D + R A W+ + + ++
Sbjct: 42 ATMEEAIAMDLEKELCFNNHGDKFVEFFVSKKLTDYRFHAFQWLIQTRSRLNLSYETVFS 101
Query: 116 SVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKT 175
+ N DRF+ + W V+L+AV S+A+K EV P +L++ +F T
Sbjct: 102 AANCFDRFVYMTCCDEWTNWMVELVAVTSLSIASKFNEVTTPLLEELEMEGLTHMFHVNT 161
Query: 176 IQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFL 235
+ +MEL+IL +L W++ A+T +F ++KI D ++ R +L++I + L
Sbjct: 162 VAQMELIILKALEWRVNAVTSYTFSQTLVSKIG--MVGDHMIMNRITNHLLDVICDLKML 219
Query: 236 EFRSSEIA-AAVAISLKELPTQEVDKAITDFFIVD-KERVLKCVE 278
++ S +A AA+ I +++ +E +I + F + KE+++KCV+
Sbjct: 220 QYPPSVVATAAIWILMEDKVCRE---SIMNLFEQNHKEKIVKCVD 261
>At1g44110 mitotic cyclin a2-type, putative
Length = 460
Score = 69.3 bits (168), Expect = 3e-12
Identities = 74/252 (29%), Positives = 116/252 (45%), Gaps = 23/252 (9%)
Query: 8 AESNLLCSENNST----CFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEEIVK 63
A SNL + N+ T C DV+ D + + VN+D+ D LC + I K
Sbjct: 145 ALSNLFITPNSETIDNYCSRDVLSDMKKMDKN---QIVNIDSNNGDPQLCATFACD-IYK 200
Query: 64 VMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRF 123
+ E P DY+ R++ D++ S+R +DW+ + Y P +L L+VNY+DR+
Sbjct: 201 HLRASEAKKRPDVDYMERVQK-DVNSSMRGILVDWLIEVSEEYRLVPETLYLTVNYIDRY 259
Query: 124 LSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMI 183
LS R +QLL VAC +AAK EE+ PQ + + + + ME +
Sbjct: 260 LSGNVISR---QKLQLLGVACMMIAAKYEEICAPQVEEFCYITDNTYLKDEVLD-MESDV 315
Query: 184 LSSLGWKMRALTPCSFIDYFL-AKISCEKYPDKSLIARSVQLILNIIKGIDFLEF----R 238
L+ L ++M A T F+ F+ A + P ++ + N I + LE+
Sbjct: 316 LNYLKFEMTAPTTKCFLRRFVRAAHGVHEAP-----LMQLECMANYIAELSLLEYTMLSH 370
Query: 239 SSEIAAAVAISL 250
S + AA AI L
Sbjct: 371 SPSLVAASAIFL 382
>At4g37490 cyclin cyc1
Length = 428
Score = 67.0 bits (162), Expect = 1e-11
Identities = 55/172 (31%), Positives = 85/172 (48%), Gaps = 8/172 (4%)
Query: 41 VNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIW 100
V++D+ ++ L V E+I E + PR DY+ D++ +R ++W+
Sbjct: 149 VDIDSADVENDLAAVEYVEDIYSFYKSVESEWRPR-DYMASQP--DINEKMRLILVEWLI 205
Query: 101 KAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSV 160
H + P + L+VN LDRFLSV PR +QL+ ++ ++AK EE+ PQ
Sbjct: 206 DVHVRFELNPETFYLTVNILDRFLSVKPVPRK---ELQLVGLSALLMSAKYEEIWPPQVE 262
Query: 161 DLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFL-AKISCEK 211
DL V + K I ME ILS+L W + T F+ F+ A I+ EK
Sbjct: 263 DL-VDIADHAYSHKQILVMEKTILSTLEWYLTVPTHYVFLARFIKASIADEK 313
>At5g06150 mitosis-specific cyclin 1b
Length = 445
Score = 61.6 bits (148), Expect = 6e-10
Identities = 45/164 (27%), Positives = 81/164 (48%), Gaps = 7/164 (4%)
Query: 41 VNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIW 100
+++D D+ L V +++ E EK+ P+ I+ +++ +R +DW+
Sbjct: 166 IDIDESDKDNHLAAVEYVDDMYSFYKEVEKESQPKMYMHIQT---EMNEKMRAILIDWLL 222
Query: 101 KAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSV 160
+ H + +L L+VN +DRFLSV P+ +QL+ ++ +A+K EE+ PQ
Sbjct: 223 EVHIKFELNLETLYLTVNIIDRFLSVKAVPKR---ELQLVGISALLIASKYEEIWPPQVN 279
Query: 161 DLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFL 204
DL V + ++ I ME IL +L W + T F+ F+
Sbjct: 280 DL-VYVTDNAYSSRQILVMEKAILGNLEWYLTVPTQYVFLVRFI 322
>At3g11520 cyclin box
Length = 414
Score = 60.1 bits (144), Expect = 2e-09
Identities = 39/119 (32%), Positives = 63/119 (52%), Gaps = 4/119 (3%)
Query: 86 DLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACF 145
++D +R +DW+ + H + P +L L+VN +DRFLS+ PR +QL+ V+
Sbjct: 185 EIDEKMRSILIDWLVEVHVKFDLSPETLYLTVNIIDRFLSLKTVPRR---ELQLVGVSAL 241
Query: 146 SLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFL 204
+A+K EE+ PQ DL V + ++ I ME IL +L W + T F+ F+
Sbjct: 242 LIASKYEEIWPPQVNDL-VYVTDNSYNSRQILVMEKTILGNLEWYLTVPTQYVFLVRFI 299
>At1g20610 hypothetical protein
Length = 429
Score = 60.1 bits (144), Expect = 2e-09
Identities = 48/160 (30%), Positives = 78/160 (48%), Gaps = 8/160 (5%)
Query: 86 DLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACF 145
DL+ +R +DW+ + H + +L L++N +DRFL+V Q R +QL+ V
Sbjct: 204 DLNERMRGILIDWLIEVHYKFELMEETLYLTINVIDRFLAVHQIVRK---KLQLVGVTAL 260
Query: 146 SLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLA 205
LA K EEV VP DL + K + + + ME ++ ++L + TP F+ FL
Sbjct: 261 LLACKYEEVSVPVVDDLILISDK-AYSRREVLDMEKLMANTLQFNFSLPTPYVFMKRFLK 319
Query: 206 KISCEKYPDKSLIARSVQLILNIIKGIDFLEFRSSEIAAA 245
DK L S +I + + LE+ S++AA+
Sbjct: 320 AAQ----SDKKLEILSFFMIELCLVEYEMLEYLPSKLAAS 355
>At1g47210 cyclin like protein
Length = 372
Score = 58.9 bits (141), Expect = 4e-09
Identities = 65/259 (25%), Positives = 126/259 (48%), Gaps = 20/259 (7%)
Query: 42 NLDNVGSDSFLC--FVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWI 99
++D+ D +C +VA E ++ + K K P DY+ +++ D+ S+R +DW+
Sbjct: 88 DIDSRSDDPQMCGPYVADIYEYLRQLEVKPKQR-PLPDYIEKVQK-DVTPSMRGVLVDWL 145
Query: 100 WKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQS 159
+ Y G +L L+V+++DRFLS+ + +QL+ V+ +A+K EE+ P
Sbjct: 146 VEVAEEYKLGSETLYLTVSHIDRFLSLKTVNK---QKLQLVGVSAMLIASKYEEIS-PPK 201
Query: 160 VDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIA 219
VD F + + +ME IL +L +++ T +F+ F +++ + + L
Sbjct: 202 VDDFCYITDNTFSKQDVVKMEADILLALQFELGRPTINTFMRRF-TRVAQDDFKVPHLQL 260
Query: 220 RSVQLILNIIKGIDF--LEFRSSEIAAAVA------ISLKELPTQEVDKAITDFFIVDKE 271
+ L+ + +D+ ++F S +AA+ I K+ P ++ + T + D +
Sbjct: 261 EPLCCYLSELSILDYKTVKFVPSLLAASAVFLARFIIRPKQHPWNQMLEEYTKYKAADLQ 320
Query: 272 RVLKCVELIRDLSLIKVGG 290
CV +I DL L + GG
Sbjct: 321 ---VCVGIIHDLYLSRRGG 336
>At1g47230 cyclin like protein
Length = 369
Score = 58.2 bits (139), Expect = 7e-09
Identities = 59/232 (25%), Positives = 107/232 (45%), Gaps = 17/232 (7%)
Query: 66 VEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLS 125
+E + H P DY+ +++ DL +R +DW+ + Y +L L+++Y+DRFLS
Sbjct: 106 MEGKPKHRPLPDYIEKVQS-DLTPHMRAVLVDWLVEVAEEYKLVSDTLYLTISYVDRFLS 164
Query: 126 VFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILS 185
V R +QL+ V+ +A+K EE+ P+ D F + + ME IL
Sbjct: 165 VKPINR---QKLQLVGVSAMLIASKYEEIGPPKVEDFCYITDN-TFTKQEVVSMEADILL 220
Query: 186 SLGWKMRALTPCSFIDYFLAKISCEKYPDKSL--------IARSVQLILNIIKGIDFLEF 237
+L +++ + T +F+ F +++ E + D L ++ L +K + L
Sbjct: 221 ALQFELGSPTIKTFLRRF-TRVAQEDFKDSQLQIEFLCCYLSELSMLDYTCVKYLPSLLS 279
Query: 238 RSSEIAAAVAISLKELPTQEVDKAITDFFIVDKERVLKCVELIRDLSLIKVG 289
S+ A I K+ P ++ + T + D + CV +I DL L + G
Sbjct: 280 ASAVFLARFIIRPKQHPWNQMLEEYTKYKAADLQ---VCVGIIHDLYLSRRG 328
>At2g26760 putative cyclin
Length = 387
Score = 57.4 bits (137), Expect = 1e-08
Identities = 49/181 (27%), Positives = 85/181 (46%), Gaps = 18/181 (9%)
Query: 41 VNLDNVGSDSFLCFVAQSEEIVKVM--VEKE---KDHLPREDYLIRLRGGDLDLSVRREA 95
+++D V +++ L V E+I K VE+E KD++ + +++ +R
Sbjct: 114 IDIDAVDANNELAAVEYVEDIFKFYRTVEEEGGIKDYIGSQP--------EINEKMRSIL 165
Query: 96 LDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVK 155
+DW+ H + P +L L++N +DRFLS+ R +QLL + +A K EE+
Sbjct: 166 IDWLVDVHRKFELMPETLYLTINLVDRFLSLTMVHRR---ELQLLGLGAMLIACKYEEIW 222
Query: 156 VPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFI-DYFLAKISCEKYPD 214
P+ D V + K + ME IL + W + TP F+ Y A + C+ +
Sbjct: 223 APEVNDF-VCISDNAYNRKQVLAMEKSILGQVEWYITVPTPYVFLARYVKAAVPCDAEME 281
Query: 215 K 215
K
Sbjct: 282 K 282
>At2g17620 putative cyclin 2
Length = 429
Score = 55.8 bits (133), Expect = 3e-08
Identities = 55/204 (26%), Positives = 94/204 (45%), Gaps = 10/204 (4%)
Query: 41 VNLDNVGSDSFLCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIW 100
V++D + S + L V +++ E+ DY+++ DL+ +R +DW+
Sbjct: 155 VDIDVLDSKNSLAAVEYVQDLYAFYRTMERFSCVPVDYMMQQI--DLNEKMRAILIDWLI 212
Query: 101 KAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSV 160
+ H + +L L+VN +DRFLS R +QL+ + LA K EEV VP
Sbjct: 213 EVHDKFDLINETLFLTVNLIDRFLSKQNVMRK---KLQLVGLVALLLACKYEEVSVPVVE 269
Query: 161 DLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEKYPDKSLIAR 220
DL + K + + ME +LS+L + + T F+ FL +K K +
Sbjct: 270 DLVLISDK-AYTRNDVLEMEKTMLSTLQFNISLPTQYPFLKRFLKAAQADK---KCEVLA 325
Query: 221 SVQLILNIIKGIDFLEFRSSEIAA 244
S + L +++ + L F S +AA
Sbjct: 326 SFLIELALVE-YEMLRFPPSLLAA 348
>At5g43080 cyclin A-type
Length = 355
Score = 54.7 bits (130), Expect = 7e-08
Identities = 65/264 (24%), Positives = 125/264 (46%), Gaps = 16/264 (6%)
Query: 34 PSWDHTNVNLDNVGSDSFLC--FVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSV 91
P+ + N ++D D +C +V E ++ + K + P DY+ +++ D+ ++
Sbjct: 65 PTIETLNSDIDTRSDDPQMCGPYVTSIFEYLRQLEVKSR---PLVDYIEKIQK-DVTSNM 120
Query: 92 RREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKM 151
R +DW+ + Y +L L+V+Y+DRFLS+ + +QLL V +A+K
Sbjct: 121 RGVLVDWLVEVAEEYKLLSDTLYLAVSYIDRFLSLKTVNK---QRLQLLGVTSMLIASKY 177
Query: 152 EEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSLGWKMRALTPCSFIDYFLAKISCEK 211
EE+ P +VD + + I +ME IL +L +++ T +F+ F +++ E
Sbjct: 178 EEI-TPPNVDDFCYITDNTYTKQEIVKMEADILLALQFELGNPTSNTFLRRF-TRVAQED 235
Query: 212 YPDKSLIARSVQLILNIIKGIDF--LEFRSSEIAAAVAISLKEL--PTQEVDKAITDFFI 267
+ L + L+ + +D+ ++F S +AA+ + + P Q + + +
Sbjct: 236 FEMSHLQMEFLCSYLSELSMLDYQSVKFLPSTVAASAVFLARFIIRPKQHPWNVMLEEYT 295
Query: 268 VDKERVLK-CVELIRDLSLIKVGG 290
K LK CV +I DL L + G
Sbjct: 296 RYKAGDLKECVAMIHDLYLSRKCG 319
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,853,361
Number of Sequences: 26719
Number of extensions: 334028
Number of successful extensions: 915
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 854
Number of HSP's gapped (non-prelim): 54
length of query: 346
length of database: 11,318,596
effective HSP length: 100
effective length of query: 246
effective length of database: 8,646,696
effective search space: 2127087216
effective search space used: 2127087216
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135848.10


