

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135799.9 - phase: 0
(754 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
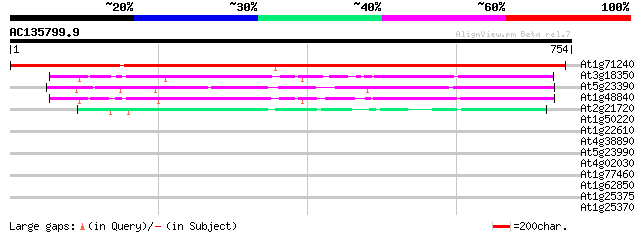
Score E
Sequences producing significant alignments: (bits) Value
At1g71240 hypothetical protein 874 0.0
At3g18350 unknown protein 277 1e-74
At5g23390 unknown protein 273 3e-73
At1g48840 unknown protein 258 1e-68
At2g21720 hypothetical protein 151 1e-36
At1g50220 hypothetical protein 33 0.77
At1g22610 Highly similar to phosphoribosylanthranilate transferase 32 1.3
At4g38890 unknown protein 31 2.9
At5g23990 FRO2 homolog 30 5.0
At4g02030 hypothetical protein 30 5.0
At1g77460 unknown protein 30 5.0
At1g62850 hypothetical protein 30 5.0
At1g25375 unknown protein 30 6.5
At1g25370 unknown protein 30 6.5
>At1g71240 hypothetical protein
Length = 823
Score = 874 bits (2257), Expect = 0.0
Identities = 451/754 (59%), Positives = 570/754 (74%), Gaps = 12/754 (1%)
Query: 1 MSRTQNFLNEVTSPLAKTAQS-RKPDPENDIGFQVMEDILMVEKTIDRKMPYGNLSLAAV 59
+S++Q L++VTSPL K +QS +K D E+ F+ +E++L VE+T+ P G LS A+
Sbjct: 75 VSKSQKILSDVTSPLKKKSQSLKKIDLEDQQDFEDLEELLTVEQTVRSDTPKGFLSFDAI 134
Query: 60 ICIEQFSRMSGLTGKKMKNIFETLVPETVYNDARNLVEYCCFRFLSRDNSDVHPSLQDPA 119
I IEQFSRM+G+TGKKM++IFET+V + DAR LVEYCCFRFLSRD+S+ HP L++PA
Sbjct: 135 ISIEQFSRMNGITGKKMQDIFETIVSPALSTDARYLVEYCCFRFLSRDSSEFHPCLKEPA 194
Query: 120 FQRLIFITMLAWENPYTYVLSSNAEKASLQVAWVTLQSKRVTEEAFVRIAPAVSGVVDRP 179
FQRLIFITMLAW NPY ++ + + + Q + + EEAF+RIAPA+SG+ DR
Sbjct: 195 FQRLIFITMLAWANPYCKERNARNDASGKP----SFQGRFIGEEAFIRIAPAISGLADRA 250
Query: 180 TVHNLFKVLAG--DKDGISMSTWLAYINEFVKVRRENRSYQIPEFPQIDEEKILCIGSNS 237
TVHNLFK LA D+ GIS+ WLAYI E VK+ +S+Q +FPQ+ E++LC+ +N
Sbjct: 251 TVHNLFKALATATDQKGISLEIWLAYIQELVKIHEGRKSHQTTDFPQLSSERLLCMAANR 310
Query: 238 KQPVLKWENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYDGLRVEKAKVGPLGSS 297
K PVLKWENN+AWPGKLTLTDKA+YFE + G+K +RLDL D VEKAKVGPLG S
Sbjct: 311 KGPVLKWENNVAWPGKLTLTDKALYFEPVDIKGSKGVLRLDLAGDKSTVEKAKVGPLGFS 370
Query: 298 LFDSAVSISSGSESNWWVLEFIDLGGDMRRDVWHALISEVIALHKFTHEYGPDEYGPN-- 355
LFDSAVS+SSG WVLEF+DLGG++RRDVWHA+ISEVIALH F E+GP E +
Sbjct: 371 LFDSAVSVSSGPGLATWVLEFVDLGGELRRDVWHAIISEVIALHTFLREFGPGEGDKSLY 430
Query: 356 -VFEARKGKQRATSSAINGIARLQALQHLRKLLDDPTKLVQFSYLQNAPNGDIVLQSLAV 414
VF A+KGK++A +SA N IARLQALQ++R L DDP KLVQFS+LQ GDIV Q+LAV
Sbjct: 431 QVFGAKKGKEKAIASASNCIARLQALQYMRNLPDDPIKLVQFSFLQQVAYGDIVCQTLAV 490
Query: 415 NYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGSVYLRKWMKSPSWGSSTSTSFW 474
N+WG L+T + R R S E ++ ++V D+DGSVYL++WM+SPSWGS+ S +FW
Sbjct: 491 NFWGGPLLTKVSDKRGDIA-RASRESYETFDNVSDLDGSVYLKRWMRSPSWGSTASMNFW 549
Query: 475 KNTSTK-GLVLSKNHVVADLSLTERAAKTSKQKSQVVEKTQATIDAATLKGIPSNIDLFK 533
KN+S + GLVLSK+ VADL+L ERA +T +QK +VVEKTQATIDAAT+KGIPSNIDLFK
Sbjct: 550 KNSSLRQGLVLSKHLAVADLTLVERATETCRQKYKVVEKTQATIDAATIKGIPSNIDLFK 609
Query: 534 ELIFPITLTAKNFEKLRHWEEPHLTVGFLGLAYTLIFRNLLSYIFPVMLMITAVGMLTIR 593
ELI P+++TA FEKLR WEEP++TV FL A T+IFRNLL Y+ PV L+ A GMLT++
Sbjct: 610 ELILPLSITATEFEKLRCWEEPYMTVSFLAFASTIIFRNLLQYVLPVSLIFLATGMLTLK 669
Query: 594 SLKEQGRLGRFFGGVMIRDQPPSNTIQKIIAVKDAMRDVENMTQKVNVSLLKIRSILLSG 653
L+ QGRLGR FG + IRDQP SNTIQKIIAVKDAM+++E+ QKVNV LLK+R+I+LSG
Sbjct: 670 GLRRQGRLGRLFGIISIRDQPSSNTIQKIIAVKDAMQNLESYLQKVNVVLLKLRTIVLSG 729
Query: 654 NPQITTEVAVLMLTWATILFIVPFKYILSFLLFDMFTRELEFRREMVERLTKLLRERWHA 713
+PQITTEVA+ ML+ AT+L IVPFKY+L+F+L+D FTRELEFR+EMV++ LRERW
Sbjct: 730 HPQITTEVALAMLSIATVLVIVPFKYVLAFVLYDQFTRELEFRKEMVKKFNAFLRERWEM 789
Query: 714 VPAAPVAVLPFENEESKSEVSLKELENKSKPPGN 747
VPAAPV VLPF NEES + K P G+
Sbjct: 790 VPAAPVIVLPFVNEESTPATQENKQLRKPTPRGD 823
>At3g18350 unknown protein
Length = 692
Score = 277 bits (709), Expect = 1e-74
Identities = 211/689 (30%), Positives = 350/689 (50%), Gaps = 59/689 (8%)
Query: 54 LSLAAVICIEQFSRMSGLTGKKMKNIFETLVPETVYNDA---RNLVEYCCFRFLSRDNSD 110
LS A + + + S++ G++ ++++ F+ E++ + RN +EYCCFR LS +
Sbjct: 49 LSPIANVVVRRCSKILGVSANELRDSFKQEAFESLKQPSLFPRNFLEYCCFRALSL-SVG 107
Query: 111 VHPSLQDPAFQRLIFITMLAWENPYTYVLSSNAEKASLQVAWVTLQSKRVTEEAFVRIAP 170
V L D F+RL F M+ WE P A +A L V + V+ EAF RIAP
Sbjct: 108 VTGHLADKKFRRLTFDMMVVWEVPAV------ASQALLSVE----EDATVSLEAFSRIAP 157
Query: 171 AVSGVVDRPTVHNLFKVLAGDKDG-ISMSTWLAYINEF---VKVRRENRSYQIPEFPQID 226
AV + D NLF++L G + S + Y++ +K R + +
Sbjct: 158 AVPIIADVIICDNLFQMLTSSTGGRLQFSVYDKYLHGLERAIKKMRTQSESSLLSGVRSK 217
Query: 227 EEKILCI-GSNSKQPVLKWENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYDGLR 285
EKIL I G+ + QPVL+ WPG+L LTD ++YFE ++ R L+ D +
Sbjct: 218 REKILEIDGTVTTQPVLEHVGISTWPGRLILTDHSLYFEALKVVSYDTPKRYHLSEDLKQ 277
Query: 286 VEKAKV-GPLGSSLFDSAVSISSGSESNWWVLEFIDLGGDMRRDVWHALISEVIALHKFT 344
+ K ++ GP G+ LFD AVS S S S V+EF +L G RRD W +I EV+ +H++
Sbjct: 278 IIKPELTGPWGTRLFDKAVSYQSISLSEPVVMEFPELKGHTRRDYWLTIIQEVLYVHRYI 337
Query: 345 HEYGPDEYGPNVFEARKGKQRATSSAINGIARLQALQHLRKLLDDPTK---LVQFSYLQN 401
++Y + A S A+ G+ R+QALQ L L + + L+ F+
Sbjct: 338 NKYKITGLA---------RDEALSKAVLGVMRVQALQELN--LTNAMRYENLLPFNLCDQ 386
Query: 402 APNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGSVYLRKWMK 461
P GD++L++LA +L +++ + + S++ + GSV+
Sbjct: 387 LPGGDLILETLAEMSTSREL-------HRSNKSKDTGTLHSSASDMVSQLGSVF------ 433
Query: 462 SPSWGSSTSTSFWKNTSTKGLVLSKNHVVADLSLTERAAKTSKQKSQVVEKTQATIDAAT 521
G S+ S T LV+ + VV D++ ERA K S++K + V Q TI+
Sbjct: 434 ----GGSSPRS---RRETSSLVVGEV-VVGDVNPLERAVKESRKKYEKVVLAQETINGVK 485
Query: 522 LKGIPSNIDLFKELIFPITLTAKNFEKLRHWEEPHLTVGFLGLAYTLIFRNLLSYIFPVM 581
+ GI +N+ + KEL+ PI T + +W++P + F L +I+R L Y+F +
Sbjct: 486 MGGIDTNLAVMKELMLPIMETWNLILSVVYWDDPTKSSVFCLLTTFIIWRGWLVYVFALA 545
Query: 582 LMITAVGMLTIRSLKEQGRLGRFFGGVMIRDQPPSNTIQKIIAVKDAMRDVENMTQKVNV 641
+ +A+ M+ R + + + + PP NT+++++AV++ + ++E Q N+
Sbjct: 546 SLFSAIFMVLTRCFSRE----KLMIELKVTAPPPMNTMEQLLAVQNGISELEQNIQDANI 601
Query: 642 SLLKIRSILLSGNPQITTEVAVLMLTWATILFIVPFKYILSFLLFDMFTRELEFRREMVE 701
LLK R++L S PQ + + A+ ++ AT++ VP +Y+LS + ++FTR RR E
Sbjct: 602 VLLKFRALLFSLFPQASQKFAIAIVVAATMMAFVPGRYLLSVVFVELFTRYSPPRRASTE 661
Query: 702 RLTKLLRERWHAVPAAPVAVLPFENEESK 730
RL + LRE W ++PAAPV +L +N + K
Sbjct: 662 RLIRRLREWWFSIPAAPVVLLHDKNNKKK 690
>At5g23390 unknown protein
Length = 730
Score = 273 bits (697), Expect = 3e-73
Identities = 208/720 (28%), Positives = 343/720 (46%), Gaps = 79/720 (10%)
Query: 50 PYGNLSLAAVICIEQFSRMSGLTGKKMKNIFETLVPETV---YNDARNLVEYCCFRFLSR 106
P LSL A + + S++ + + +++ F+ +PE+V ARN +E+C F+ L +
Sbjct: 53 PIPQLSLLANSVVSRCSKILNIQTEDLQHHFDVELPESVKQLLTYARNFLEFCSFQALHQ 112
Query: 107 DNSDVHPSLQDPAFQRLIFITMLAWENPYTYVLSSNAEKAS------------------- 147
L D F++L+F MLAWE P N + AS
Sbjct: 113 VMKKPD-YLSDQEFRQLMFDMMLAWETPSVTSEQENKDAASPSKQDSEDEDGWSLFYSSP 171
Query: 148 LQVAWVTLQSKRVTEEAFVRIAPAVSGVVDRPTVHNLFKVLAGDKDG----ISMSTWLAY 203
+A + K V +EAF RIAP + D TVHNLF L I +L
Sbjct: 172 TNMAMQVDEKKSVGQEAFARIAPVCPAIADAITVHNLFDALTSSSGHRLHYIVYDKYLRT 231
Query: 204 INEFVKVRRENRSYQIPEFPQIDEEKILCI-GSNSKQPVLKWENNMAWPGKLTLTDKAIY 262
+++ K + E +L + G+N PVLK AWPGKLTLT+ A+Y
Sbjct: 232 LDKIFKAAKSTLGPSAANLQLAKGEIVLDMDGANPVLPVLKHVGISAWPGKLTLTNCALY 291
Query: 263 FEGAGLLGNKRAMRLDLTYDGLRVEKAKV-GPLGSSLFDSAVSISSGSESNWWVLEFIDL 321
F+ G G ++ MR DLT D +V K ++ GPLG+ +FD A+ S + EF +
Sbjct: 292 FDSMG--GGEKPMRYDLTEDTKQVIKPELTGPLGARIFDKAIMYKSITVPEPVFFEFTEF 349
Query: 322 GGDMRRDVWHALISEVIALHKFTHEYGPDEYGPNVFEARKGKQRAT--SSAINGIARLQA 379
G+ RRD W + E++ + F Y KG QR+ + AI GI R +A
Sbjct: 350 KGNARRDYWLGICLEILRVQWFIRRYN-----------FKGIQRSEILARAILGIFRYRA 398
Query: 380 LQHLRKLLDDPTK-LVQFSYLQNAPNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSN 438
++ ++ K L+ F+ ++ P GD+VL++L S+
Sbjct: 399 IREAFQVFSSQYKTLLIFNLAESLPGGDMVLEAL------------------------SS 434
Query: 439 EIADSSNHVFDIDGSVYLRKWMKSPSWGS-STSTSFWKNTST-----KGLVLSKNHVVAD 492
++ + +V GSV KW + S S F N T + L + + V +
Sbjct: 435 RVSRITTNVASDIGSVQYMKWPSNLSPVSLKLLEHFGLNLETGTNMGEELTIVGDFCVGE 494
Query: 493 LSLTERAAKTSKQKSQVVEKTQATIDAATLKGIPSNIDLFKELIFPITLTAKNFEKLRHW 552
S E A K S + E QAT++ ++GI +N+ + KEL+ P + +L +W
Sbjct: 495 TSPLEIALKQSILDTDRAEAAQATVEQVKVEGIDTNVAVMKELLLPFIKLGLHINRLAYW 554
Query: 553 EEPHLTVGFLGLAYTLIFRNLLSYIFPVMLMITAVGMLTIRSLKEQGRLGRFFGGVMIRD 612
++P+ + F+ L +I + +I P +L++ A+ M+ ++Q G+ V ++
Sbjct: 555 QDPYKSTVFMILVSYMIISGWIGFILPSILLLVAIVMMW----RKQFNKGKEPKTVRVKA 610
Query: 613 QPPSNTIQKIIAVKDAMRDVENMTQKVNVSLLKIRSILLSGNPQITTEVAVLMLTWATIL 672
P N +++++ ++DA+ E++ Q VNV LLKIR+I L+ PQ T A+ ++ A IL
Sbjct: 611 PPSKNAVEQLLVLQDAISQFESLIQAVNVGLLKIRAITLAILPQATDTTAISLVVVAVIL 670
Query: 673 FIVPFKYILSFLLFDMFTRELEFRREMVERLTKLLRERWHAVPAAPVAVLPFENEESKSE 732
+VP KY+++ + FTRE+ +R+ +RL + +RE W VPAAPV ++ E+ + K +
Sbjct: 671 AVVPVKYLITVAFVEWFTREVGWRKASSDRLERRIREWWFRVPAAPVQLIRAEDSKKKKK 730
>At1g48840 unknown protein
Length = 691
Score = 258 bits (658), Expect = 1e-68
Identities = 203/692 (29%), Positives = 342/692 (49%), Gaps = 62/692 (8%)
Query: 54 LSLAAVICIEQFSRMSGLTGKKMKNIFETLVPETVYNDA---RNLVEYCCFRFLSRDNSD 110
LS A + I + S++ G+ ++++ F+ E+V + RN +EYCCFR L+ +
Sbjct: 49 LSPVANVVIRRCSKILGVAVSELQDSFKQEASESVKQPSMFPRNFLEYCCFRALAL-SVG 107
Query: 111 VHPSLQDPAFQRLIFITMLAWENPYTYVLSSNAEKASLQVAWVTLQSKRVTEEAFVRIAP 170
V L D +F+RL F M+AWE P S A + L V + V EAF RIAP
Sbjct: 108 VTGHLSDKSFRRLTFDMMVAWEVP------SAASQTLLSVD----EDPTVGLEAFSRIAP 157
Query: 171 AVSGVVDRPTVHNLFKVLAGDKDGISMS-----TWLAYINEFVKVRRENRSYQIPEFPQI 225
AV + D NLF +L + + + +L + +K + + +
Sbjct: 158 AVPIIADVIICENLFGILTSVSNSVRLQFYVYDKYLYGLERAIKKMKSQSESSLLSGVRS 217
Query: 226 DEEKILCI-GSNSKQPVLKWENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYDGL 284
EKIL + G+ + QPVL+ WPG+L LTD ++YFE ++ R L+ D
Sbjct: 218 KGEKILELDGTVTTQPVLEHIGISTWPGRLILTDHSLYFEAIKVVSFDTPKRYSLSDDLK 277
Query: 285 RVEKAKV-GPLGSSLFDSAVSISSGSESNWWVLEFIDLGGDMRRDVWHALISEVIALHKF 343
+V K ++ GP G+ LFD AVS S S V+EF +L G RRD W A+I EV+ +H++
Sbjct: 278 QVIKPELTGPWGTRLFDKAVSYKSISLPEPVVMEFPELKGHTRRDYWLAIILEVLYVHRY 337
Query: 344 THEYGPDEYGPNVFEARKGKQRATSSAINGIARLQALQHLRKLLDDPTK---LVQFSYLQ 400
++ + K A S A+ GI R+QA+Q + L +P + L+ F+
Sbjct: 338 IKKFKINSV---------AKDEAISKAVLGILRVQAIQEVG--LTNPVRYENLLPFNLCD 386
Query: 401 NAPNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGSVYLRKWM 460
P GD +L++LA +S+R + E S D+ + L
Sbjct: 387 QLPGGDRILETLA----------EMSSSRVLDRTAKAKEGTLHSISASDMVSQLGLVFGA 436
Query: 461 KSPSWGSSTSTSFWKNTSTKGLVLSKNHVVADLSLTERAAKTSKQKSQVVEKTQATIDAA 520
SP S LV+ + +V D++ E+A K S++ + V Q T++
Sbjct: 437 TSPK-------------SRSSLVVGE-VMVGDVNPLEKAVKQSRKNYEKVVLAQETVNGV 482
Query: 521 TLKGIPSNIDLFKELIFPITLTAKNFEKLRHWEEPHLTVGFLGLAYTLIFRNLLSYIFPV 580
+ GI +N+ + KEL+ P T L +WE+P + F L+ +I+R + Y+F +
Sbjct: 483 KVDGIDTNVAVMKELLLPATEIGNWLLSLVYWEDPLKSFVFCLLSTFIIYRGWIGYVFAI 542
Query: 581 MLMITAVGMLTIRSLKEQGRLGRFFGGVMIRDQPPSNTIQKIIAVKDAMRDVENMTQKVN 640
+ A M+ R + ++ + + PP NT+++++AV++A+ +E + Q N
Sbjct: 543 ATLFIAGFMVLTRYFSNREKV---MIELKVMAPPPMNTMEQLLAVQNAISQLEQLIQDAN 599
Query: 641 VSLLKIRSILLSGNPQITTEVAVLMLTWATILFIVPFKYILSFLLFDMFTRELEFRREMV 700
+ LLK R++LLS PQ + + AV ++ AT++ +VP+ ++ + ++FTR RR
Sbjct: 600 IVLLKFRALLLSLFPQASEKFAVAIVIAATMMALVPWNNLILVVFLELFTRYSPPRRAST 659
Query: 701 ERLTKLLRERWHAVPAAPVAVLPFENEESKSE 732
ERL + L+E W ++PAAPV + +++ K++
Sbjct: 660 ERLMRRLKEWWFSIPAAPVLLEQSKDDNKKTK 691
>At2g21720 hypothetical protein
Length = 703
Score = 151 bits (382), Expect = 1e-36
Identities = 154/661 (23%), Positives = 265/661 (39%), Gaps = 101/661 (15%)
Query: 92 ARNLVEYCCFRFLSRDNSDVHPSLQDPAFQRLIFITMLAWENP-------YTYVLSSNAE 144
++ VE+C + SR ++ ++D +F RL F MLAW+ P Y + +E
Sbjct: 107 SKKFVEFCNSKVTSRVCENILERIKDGSFTRLTFDMMLAWQQPDADDNESYKEAVGKESE 166
Query: 145 KASLQVAWVTLQSK-----------------RVTEEAFVRIAPAVSGVVDRPTVHNLFKV 187
+Q Q V E+AFV + + VD F+
Sbjct: 167 DKRIQATLSPEQDDISLFYSDMMPLLVDHEPSVGEDAFVYLGSIIPLPVDIINGRYTFET 226
Query: 188 L-AGDKDGISMSTWLAYINEFVKVRRENRSYQIPEFPQI-DEEKILCI-GSNSKQPVLKW 244
L A + + ++ E K + + P+ ++ D+E IL + G+ + Q V++
Sbjct: 227 LTAPTGHQLHFPAYDMFVKEIHKCMKHLQKQSTPKGIELADDEIILHVEGTMASQRVIRH 286
Query: 245 ENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYDGLRVEK-AKVGPLGSSLFDSAV 303
+WPG+LTLT+ A+YFE AG++ + A+++DL+ D + K GPLG+ LFD A+
Sbjct: 287 IKETSWPGRLTLTNYALYFEAAGIINYEDAIKIDLSKDNEKSTKPMSTGPLGAPLFDKAI 346
Query: 304 SISSGSESNWWVLEFIDLGGDMRRDVWHALISEVIALHKFTHEYGPDEYGPNVFEARKGK 363
S V+EF ++ RRD W L+ E+ +HKF ++ E+
Sbjct: 347 VYESPDFEEGIVIEFPEMTSSTRRDHWLMLVKEITLMHKFLRKFN--------VESPLQS 398
Query: 364 QRATSSAINGIARLQALQHLRKLL-DDPTKLVQFSYLQNAPNGDIVLQSLAVNYWGSQLV 422
S I GI RL A + + ++ DP + FS + P GD VL+ LA L
Sbjct: 399 WEIHSRTILGIIRLHAAREMLRISPPDPKNFLIFSLFEEVPKGDYVLEELA----EISLK 454
Query: 423 TGFTSTRHQPENRPSNEIADSSNHVFDIDG-SVYLRKWMKSPSWGSSTSTSFWKNTSTKG 481
G T + N D + +G + K +K
Sbjct: 455 IGTTRNPCSASSILRNMNMDQLGDMIKEEGEDICKEKVVK-------------------- 494
Query: 482 LVLSKNHVVADLSLTERAAKTSKQKSQVVEKTQATIDAATLKGIPSNIDLFKELIFPITL 541
V K ++A L E A S+++ +V+EK +AT +GI ++ + +L
Sbjct: 495 -VTDKEEMLASL---ESAVNQSREEGKVIEKARATTAELEEEGISESVAVLMKL------ 544
Query: 542 TAKNFEKLRHWEEPHLTVGFLGLAYTLIFRNLLSYIFPVMLMITAVGMLTIRSLKEQGRL 601
L +R + L+ M R+ +
Sbjct: 545 -------------------------NLCYREWVGKAIAACLIWVVAKMAQARNKMVHTKS 579
Query: 602 GRFFGGVMIRDQPPSNTIQKIIAVKDAMRDVENMTQKVNVSLLKIRSILLSGNPQITTEV 661
V + + + I++ + + + + Q VNV++LK+RS+ S + + V
Sbjct: 580 E---DAVTVSTESDQTVTESIVSAQYGLIRLHQLMQHVNVTILKLRSLYTSKASKHASMV 636
Query: 662 AVLMLTWATILFIVPFKYILSFLLFDMFTRELEFRREMV-ERLTKLLRERWHAVPAAPVA 720
LML A+ +VPFK + F + F M ++ + ++E W ++P PV
Sbjct: 637 MALMLVLASFFAVVPFKLFIIFGIVYCFVMTSSVGTYMSNDQSNRRMKEWWDSIPIVPVR 696
Query: 721 V 721
V
Sbjct: 697 V 697
>At1g50220 hypothetical protein
Length = 203
Score = 32.7 bits (73), Expect = 0.77
Identities = 21/91 (23%), Positives = 47/91 (51%), Gaps = 2/91 (2%)
Query: 1 MSRTQNFLNEVTSPLAKTAQSRKPDPENDIGFQVMEDILMVEKTIDRKMPYGNLSLAAVI 60
+SR +N +VTS A+ + + + + ++ +++ KT+ + GN++L
Sbjct: 53 ISRLKNVTLDVTS--ARFKELYITESKMNYARDELDGAMIISKTLTKSDIVGNVALPKAQ 110
Query: 61 CIEQFSRMSGLTGKKMKNIFETLVPETVYND 91
+ +RM+G+T + + N FE V + + +D
Sbjct: 111 VMSVLTRMNGVTDEGLDNGFEVQVHDIMEDD 141
>At1g22610 Highly similar to phosphoribosylanthranilate transferase
Length = 1029
Score = 32.0 bits (71), Expect = 1.3
Identities = 44/198 (22%), Positives = 78/198 (39%), Gaps = 25/198 (12%)
Query: 521 TLKGIPSNIDLFKELIFPITLTAKNFEKLRHWEEPHLTVGFLGLAYTLIFRNLLSY---I 577
+L+ +N L+ +TL K F + W P T ++F L+ Y I
Sbjct: 819 SLRRSKANFSRIMSLLSSVTLVCKWFNDICTWRNPITTC-----LVHVLFLILVCYPELI 873
Query: 578 FP-VMLMITAVGMLTIRSLKEQ-----GRLGRFFGGVMIR-----DQPPSNTIQKIIAVK 626
P V L + +GM R R+ + D P++ I+ ++
Sbjct: 874 LPTVFLYLFVIGMWNYRYRPRHPPHMDARVSQADNAHPDELDEEFDTFPTSRPADIVRMR 933
Query: 627 -DAMRDVENMTQKVNVSLL----KIRSILLSGNPQITTEVAVLMLTWATILFIVPFKYIL 681
D +R V Q V L +I+++L +P+ T V L WA +++ PF+ ++
Sbjct: 934 YDRLRSVGGRVQTVVGDLATQGERIQALLSWRDPRATALFIVFALIWAVFIYVTPFQ-VI 992
Query: 682 SFLLFDMFTRELEFRREM 699
+ ++ R FR M
Sbjct: 993 AIIIGLFMLRHPRFRSRM 1010
>At4g38890 unknown protein
Length = 700
Score = 30.8 bits (68), Expect = 2.9
Identities = 19/86 (22%), Positives = 38/86 (44%), Gaps = 3/86 (3%)
Query: 243 KWENNMAWPGKLTLTDKAIYFEGAGLLGNKRAMRLDLTYDGLRVEKAKVGPLGSSLFDSA 302
K + W K+T T+ + GLLG+ + + + EK + G G+ + A
Sbjct: 189 KETQRLLWKNKMTFTNADAKLKSLGLLGHAKKSN---AAEEITAEKTQNGMNGTQATEVA 245
Query: 303 VSISSGSESNWWVLEFIDLGGDMRRD 328
V + SE +++ +D+ G + +
Sbjct: 246 VDSAVSSEHTSEMIQDVDIPGPLETE 271
>At5g23990 FRO2 homolog
Length = 707
Score = 30.0 bits (66), Expect = 5.0
Identities = 29/120 (24%), Positives = 56/120 (46%), Gaps = 12/120 (10%)
Query: 109 SDVHPSLQDPAFQRLIFITMLAWENPYTYVLSSNAEKASLQVAWVTLQSKRVTEEAFVRI 168
S +H + F + I+ L W +P+T SSN EK +L V V + T++ + +
Sbjct: 339 SGLHYTPTSILFLHVPSISKLQW-HPFTITSSSNLEKDTLSV--VIRKQGSWTQKLYTHL 395
Query: 169 APAVSGV---VDRPTVHNLFKVLAGDK-----DGISMSTWLAYINEFVKVRRENRSYQIP 220
+ ++ + + P N F V D G ++ +++ I E + + +NRS ++P
Sbjct: 396 SSSIDSLEVSTEGPYGPNSFDVSRHDSLILVGGGSGVTPFISVIRELI-FQSQNRSTKLP 454
>At4g02030 hypothetical protein
Length = 694
Score = 30.0 bits (66), Expect = 5.0
Identities = 24/95 (25%), Positives = 42/95 (43%), Gaps = 1/95 (1%)
Query: 457 RKWMKSPSWGSSTSTSFWKNTSTKGLVLSKNHVVADLSLTE-RAAKTSKQKSQVVEKTQA 515
RK ++ S GS+T+TS NT + N A L E AK KQK ++ K +
Sbjct: 533 RKHKRTDSNGSNTTTSSRSNTLHNDKMARSNSQRARSQLFETHLAKLFKQKVEIFTKVEF 592
Query: 516 TIDAATLKGIPSNIDLFKELIFPITLTAKNFEKLR 550
T ++ + + +E + T F++++
Sbjct: 593 TQESVVTTTVKLCLKSLQEYVRLQTFNRSGFQQIQ 627
>At1g77460 unknown protein
Length = 2110
Score = 30.0 bits (66), Expect = 5.0
Identities = 23/83 (27%), Positives = 36/83 (42%), Gaps = 9/83 (10%)
Query: 388 DDPTKLVQF-----SYLQNAPNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIAD 442
DDPT +Q L A D+V + A N LV TS+R + + ++ +AD
Sbjct: 575 DDPTSKIQVIEVLGHVLSKASQEDLVHRGCAANKGLRSLVESLTSSREETKEHTASVLAD 634
Query: 443 SSNHVFDIDGSV----YLRKWMK 461
+ DI G + + W+K
Sbjct: 635 LFSSRQDICGHLATDDIINPWIK 657
>At1g62850 hypothetical protein
Length = 143
Score = 30.0 bits (66), Expect = 5.0
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query: 480 KGLVLSKNHVVADLSLTERAAKTSKQKSQV---VEKTQATIDAATLKGIPSNIDLFKELI 536
K L+ KN + D L + KT QK + +EK QA IDAA+ P + + K+++
Sbjct: 52 KILLTEKNRINKDGELVISSTKTRTQKGNIDDALEKLQAIIDAASYVPPPPSEEQKKKIV 111
>At1g25375 unknown protein
Length = 524
Score = 29.6 bits (65), Expect = 6.5
Identities = 19/79 (24%), Positives = 30/79 (37%), Gaps = 12/79 (15%)
Query: 396 FSYLQNAPNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGSVY 455
+ +L+ +PN I + VN W ++ G+ R E D + ++DI VY
Sbjct: 374 YKFLELSPNVVIPMHG-RVNLWPKHMLCGYLKNRRSREKSILKATVDGAQTLYDIVAKVY 432
Query: 456 LRKWMKSPSWGSSTSTSFW 474
SS FW
Sbjct: 433 -----------SSVDRKFW 440
>At1g25370 unknown protein
Length = 524
Score = 29.6 bits (65), Expect = 6.5
Identities = 19/79 (24%), Positives = 30/79 (37%), Gaps = 12/79 (15%)
Query: 396 FSYLQNAPNGDIVLQSLAVNYWGSQLVTGFTSTRHQPENRPSNEIADSSNHVFDIDGSVY 455
+ +L+ +PN I + VN W ++ G+ R E D + ++DI VY
Sbjct: 374 YKFLELSPNVVIPMHG-RVNLWPKHMLCGYLKNRRSREKSILKATVDGAQTLYDIVAKVY 432
Query: 456 LRKWMKSPSWGSSTSTSFW 474
SS FW
Sbjct: 433 -----------SSVDRKFW 440
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,199,375
Number of Sequences: 26719
Number of extensions: 678314
Number of successful extensions: 1836
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1792
Number of HSP's gapped (non-prelim): 21
length of query: 754
length of database: 11,318,596
effective HSP length: 107
effective length of query: 647
effective length of database: 8,459,663
effective search space: 5473401961
effective search space used: 5473401961
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC135799.9


