

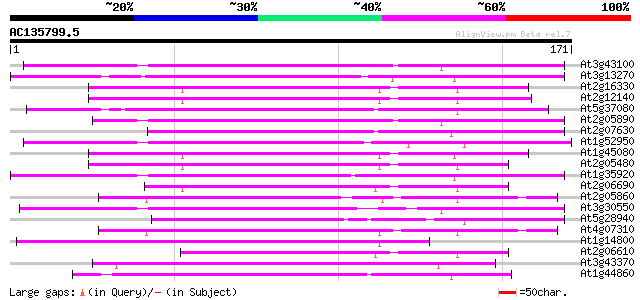
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135799.5 + phase: 0
(171 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g43100 putative protein 64 5e-11
At3g13270 hypothetical protein 62 2e-10
At2g16330 putative replication protein A1 60 8e-10
At2g12140 putative replication protein A1 58 3e-09
At5g37080 putative protein 57 6e-09
At2g05890 putative replication protein A1 56 1e-08
At2g07630 putative replication protein A1 55 1e-08
At1g52950 replication protein A1, putative 55 1e-08
At1g45080 hypothetical protein 55 1e-08
At2g05480 putative replication protein A1 50 6e-07
At1g35920 hypothetical protein 50 6e-07
At2g06690 putative replication protein A1 49 1e-06
At2g05860 putative replication protein A1 48 3e-06
At3g30550 putative replication protein 47 4e-06
At5g28940 46 9e-06
At4g07310 46 9e-06
At1g14800 hypothetical protein 46 9e-06
At2g06610 putative replication protein A1 45 2e-05
At3g43370 putative protein 45 3e-05
At1g44860 hypothetical protein 45 3e-05
>At3g43100 putative protein
Length = 269
Score = 63.5 bits (153), Expect = 5e-11
Identities = 39/166 (23%), Positives = 83/166 (49%), Gaps = 5/166 (3%)
Query: 5 YTPISAVSGGRKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRKDLVA 64
+TP+ ++ + ++ V++ H+W K SI M+LVDE G +++A R++ +
Sbjct: 9 FTPLKSLKPYKNAWRIQVKLLHVWRQYSVKAGESI---EMILVDEAGDKMYAAVRREQIK 65
Query: 65 KFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVFEFKP 124
KF + EG + ++ Y+++ K+K+ + +T + + +
Sbjct: 66 KFERCLTEGVWKIITTITLNPTSGKYRISDLKYKIGFVFKTTVSPCDSVSDAL-FLSLAK 124
Query: 125 FNEILS-STVEEVSTDVIGHVIERGDIRETEKDRRKSRVIDLTLED 169
F+ ILS S + DV+G V++R +I++ + + ++ ID L+D
Sbjct: 125 FDVILSGSANSNILHDVMGQVVDRSEIQDLNANNKPTKKIDFHLKD 170
>At3g13270 hypothetical protein
Length = 570
Score = 62.0 bits (149), Expect = 2e-10
Identities = 42/171 (24%), Positives = 87/171 (50%), Gaps = 7/171 (4%)
Query: 1 MSAKYTPISAVSGGRKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRK 60
M+A + ++ V + + ++ V+ H W R+ T + ++ DE G +IH + RK
Sbjct: 1 MAATFGYLTDVRPYKTSWRVQVKTLHAW--RQYTANTGET-LEVVFSDETGKKIHCSVRK 57
Query: 61 DLVAKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKVHLPAI-PMNV 119
DLV+K+ +M+ G +E +++ ++ TSH +K+S ++ ++ + P++ N
Sbjct: 58 DLVSKYANMLTVGEWVFIETFSLNYAGGSFRPTSHLYKMSFVNGTSV--IPSPSVSDSNY 115
Query: 120 FEFKPFNEILSSTVE-EVSTDVIGHVIERGDIRETEKDRRKSRVIDLTLED 169
F +I S + + DVIG ++ G++ E E + + + ID + D
Sbjct: 116 LTLATFEKIQSGELNPTMLVDVIGQIVMIGELEELEANNKPTTKIDFDIRD 166
>At2g16330 putative replication protein A1
Length = 233
Score = 59.7 bits (143), Expect = 8e-10
Identities = 41/138 (29%), Positives = 70/138 (50%), Gaps = 6/138 (4%)
Query: 25 AHIWLIREKKVPTSIIFMNMLLVDEKG--GRIHATTRKDLVAKFRSMVQEGGTYQLENAI 82
A + + +K V + ++ ++L D +G RI AT + L + + ++E +
Sbjct: 22 AFVVRVFQKCVGINEFYLGLILADNRGLRSRIEATINRRLASFYSDRIRENEWIVINTFS 81
Query: 83 VDFNESPYKVTSHKHKLSMMHNSTFTKVH-LPAIPMNVFEFKPFNEILSSTVEE-VSTDV 140
V + TSH +++ M + TKV LPAIP ++F PF+ I+ TV V DV
Sbjct: 82 VRLHNEGIHATSHDYRICFMDQTIVTKVSGLPAIP--YYDFTPFDYIIEKTVSTGVLVDV 139
Query: 141 IGHVIERGDIRETEKDRR 158
IG +IE G++ E + R+
Sbjct: 140 IGALIEIGNLTEDYRGRK 157
>At2g12140 putative replication protein A1
Length = 228
Score = 57.8 bits (138), Expect = 3e-09
Identities = 42/139 (30%), Positives = 69/139 (49%), Gaps = 6/139 (4%)
Query: 25 AHIWLIREKKVPTSIIFMNMLLVDEKG--GRIHATTRKDLVAKFRSMVQEGGTYQLENAI 82
A + + +K V + + ++L D +G RI AT + L + + ++E +
Sbjct: 22 AFVVRVFQKCVGINEFDLGLILADNRGMRSRIEATINRRLASFYSDRIRENEWIVINTFS 81
Query: 83 VDFNESPYKVTSHKHKLSMMHNSTFTKVH-LPAIPMNVFEFKPFNEILSSTVEE-VSTDV 140
V + TSH +++ M + TKV LPAIP ++F PF+ I+ TV V DV
Sbjct: 82 VRLHNEGIHATSHDYRICFMDQTIVTKVSGLPAIP--YYDFTPFDYIIERTVSTGVLVDV 139
Query: 141 IGHVIERGDIRETEKDRRK 159
IG +IE G++ E + R K
Sbjct: 140 IGVLIEVGNLTEDYRGRNK 158
>At5g37080 putative protein
Length = 566
Score = 56.6 bits (135), Expect = 6e-09
Identities = 37/160 (23%), Positives = 77/160 (48%), Gaps = 5/160 (3%)
Query: 6 TPISAVSGGRKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRKDLVAK 65
T +S V + N + V++ H+W R+ K S +++++ DE G ++H T RK + K
Sbjct: 36 TYLSDVKPFKTNWCVRVKILHLW--RQTK-DASRETLDVIICDENGSKMHITIRKHYIGK 92
Query: 66 FRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVFEFKPF 125
F ++ G ++N + YK++S ++++ HN+ K P + F
Sbjct: 93 FHRSIKVGDWKIIDNFTLSPLTGKYKISSLTYRMAFNHNTDVIKCD-PVSDSVFLDLADF 151
Query: 126 NEILSSTVEE-VSTDVIGHVIERGDIRETEKDRRKSRVID 164
+ + + +E V D++G V+ G + E + ++ +D
Sbjct: 152 EGVKTESYDENVLIDILGQVVRVGKVDEIMAQNKPNKKLD 191
>At2g05890 putative replication protein A1
Length = 456
Score = 55.8 bits (133), Expect = 1e-08
Identities = 37/145 (25%), Positives = 71/145 (48%), Gaps = 5/145 (3%)
Query: 26 HIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLENAIVDF 85
++W K SI M+LVD+ G +++A R++ + KF V EG + ++
Sbjct: 2 NVWRQYSVKAGESI---KMILVDKAGDKMYAAVRREQIKKFERCVTEGVWKIITTITLNP 58
Query: 86 NESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVFEFKPFNEILS-STVEEVSTDVIGHV 144
Y+++ K+K+ + +T + + + F+ ILS ST + DV+G V
Sbjct: 59 TSDQYRISDLKYKIGFVFKTTVSPCDTVSDSL-FLSLAKFDVILSGSTNSNILHDVMGQV 117
Query: 145 IERGDIRETEKDRRKSRVIDLTLED 169
I+R +I++ + + ++ ID L D
Sbjct: 118 IDRSEIQDLNANNKPTKKIDFHLRD 142
>At2g07630 putative replication protein A1
Length = 458
Score = 55.5 bits (132), Expect = 1e-08
Identities = 28/128 (21%), Positives = 64/128 (49%), Gaps = 2/128 (1%)
Query: 43 NMLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMM 102
+M+L D +G +IHA+ +++ + +F ++ G +EN + + +K T H++K+ M
Sbjct: 8 DMVLSDVRGKKIHASVKREHLNRFERLIVPGEWRAIENFGLTYATGQFKATDHRYKMGFM 67
Query: 103 HNSTFTKVHLPAIPMNVFEFKPFNEILSSTV-EEVSTDVIGHVIERGDIRETEKDRRKSR 161
+ ++ P FN++L+ + + DV+G ++ G++ + + ++
Sbjct: 68 AQTRVVRID-PLSDSYFLSLTAFNDVLNGGLNQNYLIDVVGQIVNVGEMETIDVHNQPTK 126
Query: 162 VIDLTLED 169
ID L D
Sbjct: 127 KIDFELRD 134
>At1g52950 replication protein A1, putative
Length = 566
Score = 55.5 bits (132), Expect = 1e-08
Identities = 41/169 (24%), Positives = 84/169 (49%), Gaps = 7/169 (4%)
Query: 5 YTPISAVSGGRKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRKDLVA 64
YT I + + + + V++ H W K S M+L DE G +I A +K+ +
Sbjct: 7 YTAIKNLKPFKTSWCIQVKILHAWNHYTKGSGMSY---EMMLADEDGNKIQAGIKKEHLL 63
Query: 65 KFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVF-EFK 123
K + V+ G +E V Y+ T+H +++++ ++ F+ + P I ++ +
Sbjct: 64 KLQRYVKIGHWTIIEEFSVTKASGLYRSTTHPYRINIQSSTRFS--NSPTISDEIWLDLV 121
Query: 124 PFNEILSSTVEEVS-TDVIGHVIERGDIRETEKDRRKSRVIDLTLEDLE 171
FN++LS T+++ +VIG ++ G+I+ + + ++ ID L D +
Sbjct: 122 NFNDVLSGTLDQNKLVNVIGQLVNVGEIQLLDVQGKPTKKIDFQLRDTD 170
>At1g45080 hypothetical protein
Length = 237
Score = 55.5 bits (132), Expect = 1e-08
Identities = 40/138 (28%), Positives = 68/138 (48%), Gaps = 6/138 (4%)
Query: 25 AHIWLIREKKVPTSIIFMNMLLVDEKG--GRIHATTRKDLVAKFRSMVQEGGTYQLENAI 82
A + + +K V + + ++L D +G RI T + L + + ++E +
Sbjct: 22 AFVVRVFQKCVGINEFDLGLILADNRGMRSRIETTINRCLASFYSDRIRENEWIVINTFS 81
Query: 83 VDFNESPYKVTSHKHKLSMMHNSTFTKVH-LPAIPMNVFEFKPFNEILSSTVE-EVSTDV 140
V + TSH +++ M + TKV LPAIP ++F PF+ I+ TV V DV
Sbjct: 82 VRLHNEGIHATSHDYRICFMDQTIVTKVSGLPAIP--YYDFTPFDYIIERTVSTRVLVDV 139
Query: 141 IGHVIERGDIRETEKDRR 158
IG +IE G++ E + R+
Sbjct: 140 IGALIEVGNLTEDYRGRK 157
>At2g05480 putative replication protein A1
Length = 237
Score = 50.1 bits (118), Expect = 6e-07
Identities = 39/132 (29%), Positives = 64/132 (47%), Gaps = 6/132 (4%)
Query: 25 AHIWLIREKKVPTSIIFMNMLLVDEKG--GRIHATTRKDLVAKFRSMVQEGGTYQLENAI 82
A + + +K V + M ++L D +G RI T + L + + ++E +
Sbjct: 22 AFVVRVFQKCVGINEFDMGLILADNRGMRSRIEGTINRRLASFYSDRIRENEWIVINTFS 81
Query: 83 VDFNESPYKVTSHKHKLSMMHNSTFTKVH-LPAIPMNVFEFKPFNEILSSTVEE-VSTDV 140
V + VTSH +++ M + TKV LPAIP ++F PF+ I+ TV V
Sbjct: 82 VRLHNEGIHVTSHDYRICFMDQTIVTKVSGLPAIP--YYDFTPFDYIIERTVSTGVLMLY 139
Query: 141 IGHVIERGDIRE 152
IG +IE G++ E
Sbjct: 140 IGALIEVGNLTE 151
>At1g35920 hypothetical protein
Length = 567
Score = 50.1 bits (118), Expect = 6e-07
Identities = 37/170 (21%), Positives = 77/170 (44%), Gaps = 5/170 (2%)
Query: 1 MSAKYTPISAVSGGRKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRK 60
M+A + + V + ++ V++ H W +I +++ DE G ++HAT +K
Sbjct: 1 MAATFAYLKDVRPYKNAWRVQVKILHSWKQYTSNTRETI---ELVISDEHGKKMHATVKK 57
Query: 61 DLVAKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVF 120
+LV+KF + G +E + + ++ T+H +K++ T N
Sbjct: 58 ELVSKFVHKLIVGEWVFIEIFGLTYASGQFRPTNHLYKMAFQVR-TEVMGCASVSDSNFL 116
Query: 121 EFKPFNEILSSTVE-EVSTDVIGHVIERGDIRETEKDRRKSRVIDLTLED 169
F++I S + + D IG +I G++ E + + +++ ID + D
Sbjct: 117 TLASFSKIQSGELNPHMLVDAIGQIITVGELEELKANNKRTTKIDFEIRD 166
>At2g06690 putative replication protein A1
Length = 253
Score = 49.3 bits (116), Expect = 1e-06
Identities = 34/115 (29%), Positives = 59/115 (50%), Gaps = 6/115 (5%)
Query: 42 MNMLLVDEKG--GRIHATTRKDLVAKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKL 99
+ ++L D +G RI AT + + + + ++E + V ++ TSH +++
Sbjct: 39 LGLILADNRGMRSRIEATINRRIASFYSDCIRENEWIIINTFSVRLHDEGIHTTSHDYRI 98
Query: 100 SMMHNSTFTKV-HLPAIPMNVFEFKPFNEILSSTVEE-VSTDVIGHVIERGDIRE 152
M + TKV L AIP ++F PF+ I+ TV V DVIG ++E G++ E
Sbjct: 99 CFMDQTIVTKVLGLAAIP--YYDFIPFDYIVEKTVSTGVLVDVIGALLEVGNLTE 151
>At2g05860 putative replication protein A1
Length = 238
Score = 47.8 bits (112), Expect = 3e-06
Identities = 41/152 (26%), Positives = 69/152 (44%), Gaps = 19/152 (12%)
Query: 28 WLIREKKVPTSII-------FMNMLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLEN 80
W IR + V T ++ M ++L DE G I AT + ++ + EG + N
Sbjct: 17 WCIRGRVVRTFLVSLVPSSKVMGLILADEHGMTIEATVGYKMSDHYKDFINEGEWVTITN 76
Query: 81 AIVDFNESPYKVTSHKHKLSMMHNSTFTKVHL----PAIPMNVFEFKPFNEILSSTVEE- 135
V N + T+H K+ S T V L PAIP + F+ I+ +++
Sbjct: 77 FGVVENSGSVRATTHSFKIGF---SVDTVVRLTSPVPAIPH--YRLASFSSIIDDEIDKS 131
Query: 136 VSTDVIGHVIERGDIRETEKDRRKSRVIDLTL 167
V D++G + + G++ T +++ V DLTL
Sbjct: 132 VLVDLVGAIYDVGELINTRP--KQNNVDDLTL 161
>At3g30550 putative replication protein
Length = 509
Score = 47.4 bits (111), Expect = 4e-06
Identities = 37/167 (22%), Positives = 79/167 (47%), Gaps = 13/167 (7%)
Query: 4 KYTPISAVSGGRKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRKDLV 63
++TP+ ++ + ++ V++ H+W K SI ++LVDE G +++A+ R++ +
Sbjct: 8 QFTPLKSLKPYKNAWRIQVKLLHVWRQYSVKAGESI---EIILVDEAGDKMYASVRREQI 64
Query: 64 AKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVFEFK 123
KF + EG + ++ Y + +S N + A+ +++ +
Sbjct: 65 KKFERCLTEGVWKIITTITLNPTSGQYHWFCFQTTVSPCDNVS------DALFLSLAK-- 116
Query: 124 PFNEILS-STVEEVSTDVIGHVIERGDIRETEKDRRKSRVIDLTLED 169
F+ ILS S + ++IG V+ R +I+ + ++ ID L D
Sbjct: 117 -FDVILSRSANSNILHNIIGQVVNRSEIQNLNASNKPTKKIDFHLRD 162
>At5g28940
Length = 485
Score = 46.2 bits (108), Expect = 9e-06
Identities = 33/130 (25%), Positives = 65/130 (49%), Gaps = 8/130 (6%)
Query: 44 MLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMH 103
M+L D++G +IHA+ +K LV K+ + +EN + ++VT+H+ K++ +
Sbjct: 1 MVLADQRGCKIHASVKKALVKKYERHIALDQWRIIENFFLGQVRGQFRVTNHQFKMAFV- 59
Query: 104 NSTFTKVHLPAIPMNVFEFKPFNEILSSTVEEVS----TDVIGHVIERGDIRETEKDRRK 159
NSTF P + + + P E S V E++ DV+G V+ G + + +
Sbjct: 60 NSTFVS-PCPTVTNDAYLSLPTFE--SIIVGELNLFYLVDVLGQVVNVGKVEDIVVNNES 116
Query: 160 SRVIDLTLED 169
++ ++ L +
Sbjct: 117 TKKLEFELRN 126
>At4g07310
Length = 242
Score = 46.2 bits (108), Expect = 9e-06
Identities = 37/149 (24%), Positives = 67/149 (44%), Gaps = 13/149 (8%)
Query: 28 WLIREKKVPTSII-------FMNMLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLEN 80
W IR + V T ++ M ++L +E G I AT + ++ + EG + N
Sbjct: 17 WCIRGRVVRTFLVSLVPSSKVMGLILAEEHGMTIEATVGYKMSDHYKDFINEGEWVTITN 76
Query: 81 AIVDFNESPYKVTSHKHKLSMMHNSTFTKVH-LPAIPMNVFEFKPFNEILSSTVEE-VST 138
V N + T+H K+ + LPAIP + F+ I+ +++ V
Sbjct: 77 FGVVENSGSVRATTHSFKIGFFVDIVVRLTSPLPAIPH--YRLASFSSIIDDEIDKSVLV 134
Query: 139 DVIGHVIERGDIRETEKDRRKSRVIDLTL 167
D++G + + G++ T +++ V DLTL
Sbjct: 135 DLVGAIYDVGELINTRP--KQNNVDDLTL 161
>At1g14800 hypothetical protein
Length = 384
Score = 46.2 bits (108), Expect = 9e-06
Identities = 34/128 (26%), Positives = 55/128 (42%), Gaps = 2/128 (1%)
Query: 3 AKYTPISAVSGGRKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRKDL 62
A Y A+ GR ++ R+ W R K + + +LL+DEK IHA L
Sbjct: 13 ANYVAFDALRLGRSTQQVVGRLLRFWDARNIKKDGQFMGIVLLLLDEKCSVIHAFIPAAL 72
Query: 63 VAKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKVH--LPAIPMNVF 120
+ FR +++EG + + V YK+T H L + +T +V P I F
Sbjct: 73 ASHFRQVLREGIIFNVSGFEVGRCTKLYKITDHPFLLRFLPATTIIEVSDVGPTIEREKF 132
Query: 121 EFKPFNEI 128
+ F+ +
Sbjct: 133 MLRNFDNL 140
>At2g06610 putative replication protein A1
Length = 189
Score = 45.1 bits (105), Expect = 2e-05
Identities = 31/102 (30%), Positives = 52/102 (50%), Gaps = 4/102 (3%)
Query: 53 RIHATTRKDLVAKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKV-H 111
RI AT + + + + ++E + V ++ TSH +++ M + TKV
Sbjct: 4 RIEATINRRIASFYSDCIRENEWIIINTFSVRLHDEGIHTTSHDYRICFMDQTIVTKVLG 63
Query: 112 LPAIPMNVFEFKPFNEILSSTVEE-VSTDVIGHVIERGDIRE 152
L AIP ++F PF+ I+ TV V DVIG ++E G++ E
Sbjct: 64 LAAIP--YYDFIPFDYIVEKTVSTGVLVDVIGALLEVGNLTE 103
>At3g43370 putative protein
Length = 235
Score = 44.7 bits (104), Expect = 3e-05
Identities = 27/126 (21%), Positives = 61/126 (47%), Gaps = 3/126 (2%)
Query: 26 HIWLIR--EKKVPTSIIFMNMLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLENAIV 83
+++++R +K + ++ + ++L D RI AT + L + E + + +V
Sbjct: 20 YVFVVRVFKKVISPNVFELGLILADYANNRIEATVDRRLAPFYEDRFVENEWKTITSFLV 79
Query: 84 DFNESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVFEFKPFNEIL-SSTVEEVSTDVIG 142
+ + T H++ + M+ + + + P+ F+F PF+ IL S + V DV+G
Sbjct: 80 RKSTDSVRATKHEYGILFMYRTVVVQAPPRSPPVPDFDFDPFDYILEKSAYKNVLVDVVG 139
Query: 143 HVIERG 148
+++ G
Sbjct: 140 ALVDVG 145
>At1g44860 hypothetical protein
Length = 244
Score = 44.7 bits (104), Expect = 3e-05
Identities = 35/135 (25%), Positives = 63/135 (45%), Gaps = 5/135 (3%)
Query: 20 MCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLE 79
+CV V I+ +K + T+ + ++L DE G +I AT A + +QE ++
Sbjct: 19 ICVIVLRIY---KKFLNTNAFELRLVLADEWGTQIEATIGPHFSAFYFDRIQENQWKKIS 75
Query: 80 NAIVDFNESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVFEFKPFNEILSSTV-EEVST 138
+V N P + T H+ ++ M ++ T P + +F PF+ I+ V +
Sbjct: 76 TFLVRPNCEPIRTTPHEFRILFMDHTIITS-STPLATGLLNKFTPFDYIVDDVVGKHTLV 134
Query: 139 DVIGHVIERGDIRET 153
DVIG ++ G + T
Sbjct: 135 DVIGALVNVGILTNT 149
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,641,814
Number of Sequences: 26719
Number of extensions: 142973
Number of successful extensions: 409
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 347
Number of HSP's gapped (non-prelim): 72
length of query: 171
length of database: 11,318,596
effective HSP length: 92
effective length of query: 79
effective length of database: 8,860,448
effective search space: 699975392
effective search space used: 699975392
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC135799.5


