

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135798.2 + phase: 0
(261 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
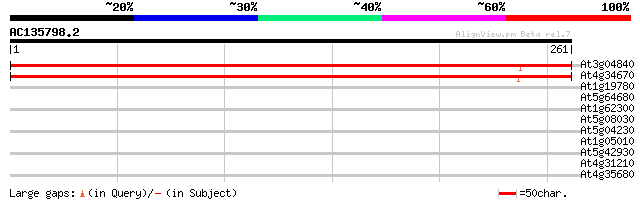
At3g04840 40S ribosomal protein S3A like protein (S phase speci... 456 e-129
At4g34670 S-phase-specific ribosomal protein like 455 e-128
At1g19780 cyclic nucleotide and calmodulin-regulated ion channel... 30 1.00
At5g64680 unknown protein 30 1.3
At1g62300 transcription factor WRKY6 29 2.2
At5g08030 glycerophosphodiester phosphodiesterase - like protein 28 5.0
At5g04230 phenylalanine ammonia-lyase PAL3 28 5.0
At1g05010 1-aminocyclopropane-1-carboxylate oxidase 28 5.0
At5g42930 unknown protein 28 6.5
At4g31210 DNA topoisomerase like- protein 28 6.5
At4g35680 putative protein 27 8.5
>At3g04840 40S ribosomal protein S3A like protein (S phase
specific)
Length = 262
Score = 456 bits (1172), Expect = e-129
Identities = 224/262 (85%), Positives = 244/262 (92%), Gaps = 1/262 (0%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKG+KGGKKKA DPF+KKDWYD+KAPS+F +NVGKTLVSRTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGRKGGKKKAVDPFSKKDWYDVKAPSIFTHRNVGKTLVSRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQGDEDNA+RKIRLRAEDVQG+N+L FWGMDFTTDKLRSLV+KWQTL
Sbjct: 61 LKHRVFEVSLADLQGDEDNAYRKIRLRAEDVQGRNVLCQFWGMDFTTDKLRSLVKKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTD+YTLR+FCI FTKRR+NQVKRTCYAQSSQIRQIRRKMR+IM+ +A+SCD
Sbjct: 121 IEAHVDVKTTDSYTLRLFCIAFTKRRANQVKRTCYAQSSQIRQIRRKMRDIMVREASSCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYS-EDV 239
LKDLV KFIPE IG+EIEKAT IYPLQNVFIRKVKILKAPKFDLGKLM+VHGDYS EDV
Sbjct: 181 LKDLVAKFIPEAIGREIEKATQGIYPLQNVFIRKVKILKAPKFDLGKLMDVHGDYSAEDV 240
Query: 240 GTKVERPADETMVEGTPEIVGA 261
G KV+RPADE VE EI+GA
Sbjct: 241 GVKVDRPADEMAVEEPTEIIGA 262
>At4g34670 S-phase-specific ribosomal protein like
Length = 262
Score = 455 bits (1170), Expect = e-128
Identities = 225/262 (85%), Positives = 242/262 (91%), Gaps = 1/262 (0%)
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKG+KGGKKKA DPF+KKDWYD+KAP F +NVGKTLVSRTQGTKIASEG
Sbjct: 1 MAVGKNKRISKGRKGGKKKAVDPFSKKDWYDVKAPGSFTNRNVGKTLVSRTQGTKIASEG 60
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQ DEDNA+RKIRLRAEDVQG+N+LT FWGMDFTTDKLRSLV+KWQTL
Sbjct: 61 LKHRVFEVSLADLQNDEDNAYRKIRLRAEDVQGRNVLTQFWGMDFTTDKLRSLVKKWQTL 120
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTD YTLRMFCI FTKRR+NQVKRTCYAQSSQIRQIRRKM EIM+ +A+SCD
Sbjct: 121 IEAHVDVKTTDGYTLRMFCIAFTKRRANQVKRTCYAQSSQIRQIRRKMSEIMVKEASSCD 180
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY-SEDV 239
LK+LV KFIPE IG+EIEKAT IYPLQNVFIRKVKILKAPKFDLGKLMEVHGDY +EDV
Sbjct: 181 LKELVAKFIPEAIGREIEKATQGIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYTAEDV 240
Query: 240 GTKVERPADETMVEGTPEIVGA 261
G KV+RPADETMVE EI+GA
Sbjct: 241 GVKVDRPADETMVEEPTEIIGA 262
>At1g19780 cyclic nucleotide and calmodulin-regulated ion channel,
putative
Length = 728
Score = 30.4 bits (67), Expect = 1.00
Identities = 32/144 (22%), Positives = 60/144 (41%), Gaps = 18/144 (12%)
Query: 75 GDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTT-DKLRSLVRKWQTLIEAHVDVKTTDNY 133
G F + L+ D G+ LLT W +D L S R + L E V+ +
Sbjct: 535 GGRSGFFNRGLLKEGDFCGEELLT--WALDPKAGSNLPSSTRTVKALTE--VEAFALEAE 590
Query: 134 TLRMFCIGFTKRRSNQVKRT-------------CYAQSSQIRQIRRKMREIMINQATSCD 180
L+ F + S QV++T C+ Q++ R +RRK+ E+ + +
Sbjct: 591 ELKFVASQFRRLHSRQVQQTFRFYSQQWRTWAACFIQAAWRRHLRRKIAELRRKEEEEEE 650
Query: 181 LKDLVRKFIPEMIGKEIEKATSSI 204
+ ++ + +G + ++ SS+
Sbjct: 651 MDYEDDEYYDDNMGGMVTRSDSSV 674
>At5g64680 unknown protein
Length = 203
Score = 30.0 bits (66), Expect = 1.3
Identities = 24/91 (26%), Positives = 43/91 (46%), Gaps = 12/91 (13%)
Query: 170 EIMINQATSCDLKDLVRK----FIPEMIGK--EIEKATSSIYPLQNVFIRKVKILKAPKF 223
+ +++ A D DL R I +G+ EIE+ T I P ++ + K K+
Sbjct: 10 DFIVSGAEQLDDTDLTRSDEFWLIQAPLGQFPEIEENTLKIEPDKDGLFGEFKDSNGAKY 69
Query: 224 DLGKLMEVHGDYSEDVGTKVERPADETMVEG 254
DL +S+D G ++ P++E+M+ G
Sbjct: 70 DLASF------HSQDAGAELIIPSEESMIVG 94
>At1g62300 transcription factor WRKY6
Length = 553
Score = 29.3 bits (64), Expect = 2.2
Identities = 46/202 (22%), Positives = 78/202 (37%), Gaps = 30/202 (14%)
Query: 52 QGTKIASEGLKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLR 111
Q T+I++ G + R F VSL+ ++ ED F + V G N +DF +DK
Sbjct: 46 QKTRISTNGSEFR-FPVSLSGIRDREDEDF------SSGVAGDNDREVPGEVDFFSDKKS 98
Query: 112 SLVRKWQTLIEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQI-RRKMRE 170
+ R+ GF ++ Q RT +R K E
Sbjct: 99 RVCREDDE---------------------GFRVKKEEQDDRTDVNTGLNLRTTGNTKSDE 137
Query: 171 IMINQATSCDLKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLME 230
MI+ S +++D K + E++K T L+ + + + + L LM+
Sbjct: 138 SMIDDGESSEMEDKRAKNELVKLQDELKKMTMDNQKLRELLTQVSNSYTSLQMHLVSLMQ 197
Query: 231 VHGDYSEDVGTKVERPADETMV 252
+ V E+P +ET+V
Sbjct: 198 QQQQQNNKVIEAAEKP-EETIV 218
>At5g08030 glycerophosphodiester phosphodiesterase - like protein
Length = 372
Score = 28.1 bits (61), Expect = 5.0
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query: 67 EVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRK 116
+V+L D D+ R R +VQG N+ T F+ +DFT +L++L K
Sbjct: 92 DVNLDDTTDVADHKEFADRKRTYEVQGMNM-TGFFTVDFTLKELKTLGAK 140
>At5g04230 phenylalanine ammonia-lyase PAL3
Length = 694
Score = 28.1 bits (61), Expect = 5.0
Identities = 11/24 (45%), Positives = 18/24 (74%)
Query: 61 LKHRVFEVSLADLQGDEDNAFRKI 84
L+H +F+ +LA+ +G+ D FRKI
Sbjct: 574 LRHVLFDKALAEPEGETDTVFRKI 597
>At1g05010 1-aminocyclopropane-1-carboxylate oxidase
Length = 323
Score = 28.1 bits (61), Expect = 5.0
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 1/34 (2%)
Query: 190 PEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKF 223
PE+IGKE EK YP + VF +K+ A KF
Sbjct: 264 PELIGKEAEKEKKENYP-RFVFEDYMKLYSAVKF 296
>At5g42930 unknown protein
Length = 237
Score = 27.7 bits (60), Expect = 6.5
Identities = 22/83 (26%), Positives = 36/83 (42%), Gaps = 4/83 (4%)
Query: 19 KAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGLKHRVFEVSLADLQGDED 78
+ DPF DW S ++VKNVGK + + EG EV+L + Q
Sbjct: 47 RGTDPFDADDWCTDLDLSWYEVKNVGKIHGGFMKALGLQKEGWPK---EVNLDETQNATT 103
Query: 79 -NAFRKIRLRAEDVQGKNLLTNF 100
A+ +R +++ +N + F
Sbjct: 104 LYAYYTVRRHLKEILDQNPTSKF 126
>At4g31210 DNA topoisomerase like- protein
Length = 1179
Score = 27.7 bits (60), Expect = 6.5
Identities = 14/43 (32%), Positives = 25/43 (57%), Gaps = 2/43 (4%)
Query: 89 EDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTLIEAHVDVKTTD 131
+D+Q +L+ +G +FT+D R +K + EAH ++ TD
Sbjct: 731 KDIQ--SLVAERYGKNFTSDSPRKYFKKVKNAQEAHEAIRPTD 771
>At4g35680 putative protein
Length = 503
Score = 27.3 bits (59), Expect = 8.5
Identities = 11/43 (25%), Positives = 27/43 (62%), Gaps = 2/43 (4%)
Query: 122 EAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQI 164
+A +D++ DN++ + +G T+R VKR ++Q + ++++
Sbjct: 398 KASLDIERPDNFSKEL--VGDTRREQRPVKRAKWSQEADLQKL 438
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,497,059
Number of Sequences: 26719
Number of extensions: 217151
Number of successful extensions: 615
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 608
Number of HSP's gapped (non-prelim): 12
length of query: 261
length of database: 11,318,596
effective HSP length: 97
effective length of query: 164
effective length of database: 8,726,853
effective search space: 1431203892
effective search space used: 1431203892
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135798.2


