

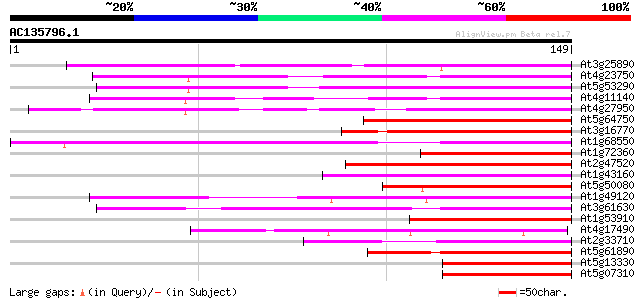
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135796.1 - phase: 0 /pseudo
(149 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g25890 unknown protein 91 3e-19
At4g23750 putative Ap2 domain protein 90 5e-19
At5g53290 unknown protein 82 1e-16
At4g11140 unknown protein with Ap2 domain 76 6e-15
At4g27950 unknown protein 75 2e-14
At5g64750 unknown protein 72 1e-13
At3g16770 AP2 domain containing protein RAP2.3 70 3e-13
At1g68550 putative AP2 domain transcription factor 68 2e-12
At1g72360 putative AP2 domain transcription factor 68 2e-12
At2g47520 putative AP2 domain transcription factor 67 3e-12
At1g43160 RAP2.6 (At1g43160) 67 4e-12
At5g50080 putative protein 67 5e-12
At1g49120 similar to AP2 domain containing protein RAP2.2 gb|AAC... 66 6e-12
At3g61630 unknown protein 66 8e-12
At1g53910 unknown protein 64 3e-11
At4g17490 ethylene responsive element binding factor-like protein 64 4e-11
At2g33710 putative AP2 domain transcription factor 64 4e-11
At5g61890 putative protein 63 5e-11
At5g13330 putative protein 63 7e-11
At5g07310 putative transcription factor 63 7e-11
>At3g25890 unknown protein
Length = 332
Score = 90.5 bits (223), Expect = 3e-19
Identities = 57/136 (41%), Positives = 73/136 (52%), Gaps = 6/136 (4%)
Query: 16 LKESKLMTRKLQLVFDDPDATDVSEDESQQSRTTTKRSFFEVTLPPRAPAALSLTSQNSF 75
L E RKL+ V +DP ATD S E ++ KR E+ LP A AA S++S+
Sbjct: 33 LSEDLKTMRKLRFVVNDPYATDYSSSEEEERSQRRKRYVCEIDLP-FAQAATQAESESSY 91
Query: 76 YVKPKTVTRRKTCLSVQPQTNKKVVQTSTSPTTGRRSC--AKYRGVRMRPWGKWAAEIRD 133
+ KT +S +KKV+++ SP GR S +K GVR R WGKWAAEIR
Sbjct: 92 CQESNNNGVSKTKISA---CSKKVLRSKASPVVGRSSTTVSKPVGVRQRKWGKWAAEIRH 148
Query: 134 PFRNVRIWLGTYNSAE 149
P VR WLGTY + E
Sbjct: 149 PITKVRTWLGTYETLE 164
>At4g23750 putative Ap2 domain protein
Length = 343
Score = 89.7 bits (221), Expect = 5e-19
Identities = 53/134 (39%), Positives = 74/134 (54%), Gaps = 19/134 (14%)
Query: 23 TRKLQLVFDDPDATDVSEDESQQS-------RTTTKRSFFEVTLPPRAPAALSLTSQNSF 75
TR L++ DPDATD S D+ ++ R K+ EV L A S S
Sbjct: 34 TRILRISVTDPDATDSSSDDEEEEHQRFVSKRRRVKKFVNEVYLDSGAVVTGSCGQMES- 92
Query: 76 YVKPKTVTRRKTCLSVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPF 135
+++ +V+ ++ V ++T+ TTG + K+RGVR RPWGKWAAEIRDP
Sbjct: 93 --------KKRQKRAVKSESTVSPVVSATTTTTGEK---KFRGVRQRPWGKWAAEIRDPL 141
Query: 136 RNVRIWLGTYNSAE 149
+ VR+WLGTYN+AE
Sbjct: 142 KRVRLWLGTYNTAE 155
>At5g53290 unknown protein
Length = 354
Score = 82.0 bits (201), Expect = 1e-16
Identities = 47/130 (36%), Positives = 67/130 (51%), Gaps = 12/130 (9%)
Query: 24 RKLQLVFDDPDATDVSEDESQQS----RTTTKRSFFEVTLPPRAPAALSLTSQNSFYVKP 79
+K+ + + DPDATD S DE ++ R KR E+T+ P ++ S
Sbjct: 37 KKVSICYTDPDATDSSSDEDEEDFLFPRRRVKRFVNEITVEPSCNNVVTGVSMKD----- 91
Query: 80 KTVTRRKTCLSVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNVR 139
+R + S + Q+ Q + + K+RGVR RPWGKWAAEIRDP + R
Sbjct: 92 ---RKRLSSSSDETQSPASSRQRPNNKVSVSGQIKKFRGVRQRPWGKWAAEIRDPEQRRR 148
Query: 140 IWLGTYNSAE 149
IWLGT+ +AE
Sbjct: 149 IWLGTFETAE 158
>At4g11140 unknown protein with Ap2 domain
Length = 287
Score = 76.3 bits (186), Expect = 6e-15
Identities = 52/136 (38%), Positives = 66/136 (48%), Gaps = 32/136 (23%)
Query: 22 MTRKLQLVFDDPDATDVSEDESQQ--------SRTTTKRSFFEVTLPPRAPAALSLTSQN 73
+ R L++ DP ATD S DE ++ R K+ EV L S+ S
Sbjct: 9 LPRILRISVTDPYATDSSSDEEEEVDFDALSTKRRRVKKYVKEVVLD-------SVVSDK 61
Query: 74 SFYVKPKTVTRRKTCLSVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRD 133
+K K K+VV TT R K+RGVR RPWGKWAAEIRD
Sbjct: 62 EKPMKKKR--------------KKRVVTVPVVVTTATR---KFRGVRQRPWGKWAAEIRD 104
Query: 134 PFRNVRIWLGTYNSAE 149
P R VR+WLGT+++AE
Sbjct: 105 PSRRVRVWLGTFDTAE 120
>At4g27950 unknown protein
Length = 335
Score = 74.7 bits (182), Expect = 2e-14
Identities = 51/146 (34%), Positives = 73/146 (49%), Gaps = 22/146 (15%)
Query: 6 KSRARQRPNPLKESKLMTRKLQLVFDDPDATDVSEDESQQ--SRTTTKRSFFEVTLPPRA 63
K + +R +++S R +++ D DATD S DE + R KR E+ + P
Sbjct: 26 KKSSMERKTSVRDS---ARLVRVSMTDRDATDSSSDEEEFLFPRRRVKRLINEIRVEP-- 80
Query: 64 PAALSLTSQNSFYVKPKTVTRRKTCLSVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRP 123
S +S P T+ + ++V K P+ ++ KYRGVR RP
Sbjct: 81 ----SSSSTGDVSASP---TKDRKRINVDSTVQK--------PSVSGQNQKKYRGVRQRP 125
Query: 124 WGKWAAEIRDPFRNVRIWLGTYNSAE 149
WGKWAAEIRDP + RIWLGT+ +AE
Sbjct: 126 WGKWAAEIRDPEQRRRIWLGTFATAE 151
>At5g64750 unknown protein
Length = 391
Score = 71.6 bits (174), Expect = 1e-13
Identities = 30/55 (54%), Positives = 39/55 (70%)
Query: 95 TNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
T + TS +G + +YRGVR RPWGKWAAEIRDPF+ R+WLGT+++AE
Sbjct: 164 TTTATASSETSSFSGDQPRRRYRGVRQRPWGKWAAEIRDPFKAARVWLGTFDNAE 218
>At3g16770 AP2 domain containing protein RAP2.3
Length = 248
Score = 70.5 bits (171), Expect = 3e-13
Identities = 32/61 (52%), Positives = 44/61 (71%), Gaps = 2/61 (3%)
Query: 89 LSVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSA 148
++V+ + KK + +T P R+ YRG+R RPWGKWAAEIRDP + VR+WLGT+N+A
Sbjct: 54 VNVKEEAVKK--EQATEPGKRRKRKNVYRGIRKRPWGKWAAEIRDPRKGVRVWLGTFNTA 111
Query: 149 E 149
E
Sbjct: 112 E 112
>At1g68550 putative AP2 domain transcription factor
Length = 324
Score = 68.2 bits (165), Expect = 2e-12
Identities = 43/150 (28%), Positives = 67/150 (44%), Gaps = 17/150 (11%)
Query: 1 MSGSRKSRARQRP-NPLKESKLMTRKLQLVFDDPDATDVSEDESQQSRTTTKRSFFEVTL 59
+S K RA++ + L + RK++++ +DP ATD S + ++ + R +
Sbjct: 13 VSSEIKKRAKRNTLSSLPQETQPLRKVRIIVNDPYATDDSSSDEEELKVPKPRKMKRIVR 72
Query: 60 PPRAPAALSLTSQNSFYVKPKTVTRRKTCLSVQPQTNKKVVQTSTSPTTGRRSCAKYRGV 119
P+ + + T T K +S P +K K GV
Sbjct: 73 EINFPSMEVSEQPSESSSQDSTKTDGKIAVSASPAVPRK----------------KPVGV 116
Query: 120 RMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
R R WGKWAAEIRDP + R WLGT+++ E
Sbjct: 117 RQRKWGKWAAEIRDPIKKTRTWLGTFDTLE 146
>At1g72360 putative AP2 domain transcription factor
Length = 262
Score = 67.8 bits (164), Expect = 2e-12
Identities = 24/40 (60%), Positives = 35/40 (87%)
Query: 110 RRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
++ ++Y+G+R RPWG+WAAEIRDP + VR+WLGT+N+AE
Sbjct: 70 KKQSSRYKGIRRRPWGRWAAEIRDPIKGVRVWLGTFNTAE 109
>At2g47520 putative AP2 domain transcription factor
Length = 171
Score = 67.4 bits (163), Expect = 3e-12
Identities = 30/60 (50%), Positives = 38/60 (63%)
Query: 90 SVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
SV + +K V S R YRG+R RPWGKWAAEIRDP + VR+WLGT+ +A+
Sbjct: 24 SVSSRKKRKPVSVSEERDGKRERKNLYRGIRQRPWGKWAAEIRDPSKGVRVWLGTFKTAD 83
>At1g43160 RAP2.6 (At1g43160)
Length = 192
Score = 67.0 bits (162), Expect = 4e-12
Identities = 31/66 (46%), Positives = 37/66 (55%)
Query: 84 RRKTCLSVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLG 143
R + L QP V + KYRGVR RPWGKWAAEIRDP + R+WLG
Sbjct: 29 REEFSLPPQPLITGSAVTKECESSMSLERPKKYRGVRQRPWGKWAAEIRDPHKATRVWLG 88
Query: 144 TYNSAE 149
T+ +AE
Sbjct: 89 TFETAE 94
>At5g50080 putative protein
Length = 220
Score = 66.6 bits (161), Expect = 5e-12
Identities = 30/51 (58%), Positives = 37/51 (71%), Gaps = 1/51 (1%)
Query: 100 VQTSTSPTT-GRRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
V +S +P R +YRGVR RPWGKWAAEIRDP R R+WLGT+++AE
Sbjct: 60 VDSSHNPIEEAREKKRRYRGVRQRPWGKWAAEIRDPHRAARVWLGTFDTAE 110
>At1g49120 similar to AP2 domain containing protein RAP2.2
gb|AAC49768.1
Length = 329
Score = 66.2 bits (160), Expect = 6e-12
Identities = 40/131 (30%), Positives = 64/131 (48%), Gaps = 26/131 (19%)
Query: 22 MTRKLQLVFDDPDATDVSEDESQQSRTTTKRSFFEVTLPPRAPAALSLTSQNSFYVKPKT 81
+ + + ++ +DPDATD S D+ S +R +KPK
Sbjct: 13 LIKTISVICNDPDATDSSSDDESISGNNPRRQ-----------------------IKPKP 49
Query: 82 VTR--RKTCLSVQPQTNKKVVQTSTSPTTG-RRSCAKYRGVRMRPWGKWAAEIRDPFRNV 138
R K C+ + + V ++ + G R++ + ++GVR RPWGK+AAEIR+PF
Sbjct: 50 PKRYVSKICVPTLIKRYENVSNSTGNKAAGNRKTSSGFKGVRRRPWGKFAAEIRNPFEKK 109
Query: 139 RIWLGTYNSAE 149
R WLGT+ + E
Sbjct: 110 RKWLGTFPTEE 120
>At3g61630 unknown protein
Length = 315
Score = 65.9 bits (159), Expect = 8e-12
Identities = 44/126 (34%), Positives = 57/126 (44%), Gaps = 16/126 (12%)
Query: 24 RKLQLVFDDPDATDVSEDESQQSRTTTKRSFFEVTLPPRAPAALSLTSQNSFYVKPKTVT 83
R +++ DP ATD S D+ + VT+ PR + T
Sbjct: 29 RLVRITVTDPFATDSSSDDDDNNN---------VTVVPRVKRYVKEIRFCQGESSSSTAA 79
Query: 84 RRKTCLSVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLG 143
R+ + + V TS P KYRGVR RPWGK+AAEIRDP RIWLG
Sbjct: 80 RKGKHKEEESVVVEDDVSTSVKPK-------KYRGVRQRPWGKFAAEIRDPSSRTRIWLG 132
Query: 144 TYNSAE 149
T+ +AE
Sbjct: 133 TFVTAE 138
>At1g53910 unknown protein
Length = 358
Score = 63.9 bits (154), Expect = 3e-11
Identities = 26/43 (60%), Positives = 32/43 (73%)
Query: 107 TTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
+ R+ +YRG+R RPWGKWAAEIRDP RIWLGT+ +AE
Sbjct: 116 SANRKRKNQYRGIRQRPWGKWAAEIRDPREGARIWLGTFKTAE 158
>At4g17490 ethylene responsive element binding factor-like protein
Length = 282
Score = 63.5 bits (153), Expect = 4e-11
Identities = 41/112 (36%), Positives = 53/112 (46%), Gaps = 14/112 (12%)
Query: 49 TTKRSFFEVTLPPRAPAALSLTSQNSFYVKPKTVT---RRKTCLSVQPQTNKKVVQTSTS 105
T K F+ + P+ + +F P VT RK L + P K +Q +T
Sbjct: 61 TPKPEIFDFDVKSEIPSESN--DSFTFQSNPPRVTVQSNRKPPLKIAPPNRTKWIQFATG 118
Query: 106 --------PTTGRRSCAKYRGVRMRPWGKWAAEIRDPF-RNVRIWLGTYNSA 148
P YRGVRMRPWGK+AAEIRDP R R+WLGT+ +A
Sbjct: 119 NPKPELPVPVVAAEEKRHYRGVRMRPWGKFAAEIRDPTRRGTRVWLGTFETA 170
>At2g33710 putative AP2 domain transcription factor
Length = 218
Score = 63.5 bits (153), Expect = 4e-11
Identities = 29/71 (40%), Positives = 43/71 (59%), Gaps = 14/71 (19%)
Query: 79 PKTVTRRKTCLSVQPQTNKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNV 138
P ++ + + L++Q ++N + YRGVR RPWGKWAAEIRDP +
Sbjct: 47 PSSIDDQSSSLTLQEKSNSRQ--------------RNYRGVRQRPWGKWAAEIRDPNKAA 92
Query: 139 RIWLGTYNSAE 149
R+WLGT+++AE
Sbjct: 93 RVWLGTFDTAE 103
>At5g61890 putative protein
Length = 248
Score = 63.2 bits (152), Expect = 5e-11
Identities = 30/54 (55%), Positives = 36/54 (66%), Gaps = 2/54 (3%)
Query: 96 NKKVVQTSTSPTTGRRSCAKYRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
N++V T RR YRGVR RPWGKWAAEIRDP + R+WLGT+ +AE
Sbjct: 72 NQQVAPTHQDQGDLRRR--HYRGVRQRPWGKWAAEIRDPKKAARVWLGTFETAE 123
>At5g13330 putative protein
Length = 212
Score = 62.8 bits (151), Expect = 7e-11
Identities = 25/34 (73%), Positives = 29/34 (84%)
Query: 116 YRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
YRGVR RPWGKWAAEIRDP + R+WLGT+ +AE
Sbjct: 39 YRGVRQRPWGKWAAEIRDPKKAARVWLGTFETAE 72
>At5g07310 putative transcription factor
Length = 263
Score = 62.8 bits (151), Expect = 7e-11
Identities = 25/34 (73%), Positives = 29/34 (84%)
Query: 116 YRGVRMRPWGKWAAEIRDPFRNVRIWLGTYNSAE 149
YRGVR RPWGKWAAEIRDP + R+WLGT+ +AE
Sbjct: 92 YRGVRQRPWGKWAAEIRDPQKAARVWLGTFETAE 125
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.126 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,270,945
Number of Sequences: 26719
Number of extensions: 118855
Number of successful extensions: 583
Number of sequences better than 10.0: 159
Number of HSP's better than 10.0 without gapping: 131
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 408
Number of HSP's gapped (non-prelim): 164
length of query: 149
length of database: 11,318,596
effective HSP length: 90
effective length of query: 59
effective length of database: 8,913,886
effective search space: 525919274
effective search space used: 525919274
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC135796.1


