

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.6 - phase: 0
(267 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
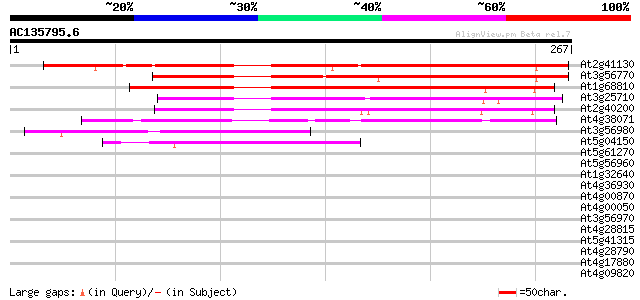
Score E
Sequences producing significant alignments: (bits) Value
At2g41130 putative bHLH transcription factor (bHLH106) 233 6e-62
At3g56770 putative HLH DNA binding protein 196 8e-51
At1g68810 putative bHLH transcription factor (bHLH030) 154 5e-38
At3g25710 putative bHLH transcription factor (bHLH032) 139 1e-33
At2g40200 putative bHLH transcription factor (bHLH051) 106 1e-23
At4g38071 putative bHLH transcription factor (bHLH131) 72 2e-13
At3g56980 putative bHLH transcription factor (bHLH039) 44 7e-05
At5g04150 myc - like protein 41 8e-04
At5g61270 bHLH transcription factor like protein 39 0.003
At5g56960 putative protein 39 0.004
At1g32640 transcription factor like protein, BHLH6 39 0.004
At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATU... 37 0.011
At4g00870 putative bHLH transcription factor (bHLH014) 37 0.011
At4g00050 bHLH like transcription factor (bHLH016) 37 0.011
At3g56970 putative bHLH transcription factor (bHLH038) 37 0.014
At4g28815 AtbHLH127 36 0.024
At5g41315 bHLH transcription factor-like protein 35 0.054
At4g28790 bHLH transcription factor like protein (bHLH023) 35 0.054
At4g17880 putative transcription factor BHLH4 34 0.071
At4g09820 putative protein 34 0.071
>At2g41130 putative bHLH transcription factor (bHLH106)
Length = 253
Score = 233 bits (595), Expect = 6e-62
Identities = 137/263 (52%), Positives = 176/263 (66%), Gaps = 33/263 (12%)
Query: 17 LFQFLVANNPSFFEYSTPSSTMM-QQSFCSSSDNNNNYYHPFEVSEITDTPSQQDRALAA 75
L+ FL N Y M QS C SS + ++YY P +S I +T +Q DRALAA
Sbjct: 10 LYSFLAGNEVGGGGYCVSGDYMTTMQSLCGSSSSTSSYY-PLAISGIGETMAQ-DRALAA 67
Query: 76 LKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVE 135
L+NHKEAE+RRRERINSHL+ LR +L CNSKT K + LAKVV+
Sbjct: 68 LRNHKEAERRRRERINSHLNKLRNVLSCNSKT-----------------DKATLLAKVVQ 110
Query: 136 RVKELRQQTCQITQVET--VPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLIPDL 193
RV+EL+QQT + + + +P E D +V+ G D S +G +IFKASLCCEDRSDL+PDL
Sbjct: 111 RVRELKQQTLETSDSDQTLLPSETDEISVLHFG-DYSNDGHIIFKASLCCEDRSDLLPDL 169
Query: 194 IEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQNSLRSLLDRSS---- 249
+EILKSL++KTL+AEM T+GGRTR+VLVVAA+KE + +ES+HFLQN+L+SLL+RSS
Sbjct: 170 MEILKSLNMKTLRAEMVTIGGRTRSVLVVAADKEMHGVESVHFLQNALKSLLERSSKSLM 229
Query: 250 ------GCNDRSKRRRGLDRRMM 266
G +RSKRRR LD +M
Sbjct: 230 ERSSGGGGGERSKRRRALDHIIM 252
>At3g56770 putative HLH DNA binding protein
Length = 230
Score = 196 bits (499), Expect = 8e-51
Identities = 110/209 (52%), Positives = 145/209 (68%), Gaps = 29/209 (13%)
Query: 69 QDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGS 128
+D+ALA+L+NHKEAE++RR RINSHL+ LR LL CNSKT K +
Sbjct: 39 EDKALASLRNHKEAERKRRARINSHLNKLRKLLSCNSKT-----------------DKST 81
Query: 129 RLAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGR-LIFKASLCCEDRS 187
LAKVV+RVKEL+QQT +IT ET+P E D +V++ G+ R +IFK S CCEDR
Sbjct: 82 LLAKVVQRVKELKQQTLEITD-ETIPSETDEISVLNIEDCSRGDDRRIIFKVSFCCEDRP 140
Query: 188 DLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAEKEHNSIESIHFLQNSLRSLLDR 247
+L+ DL+E LKSL ++TL A+M T+GGRTRNVLVVAA+KEH+ ++S++FLQN+L+SLL+R
Sbjct: 141 ELLKDLMETLKSLQMETLFADMTTVGGRTRNVLVVAADKEHHGVQSVNFLQNALKSLLER 200
Query: 248 SS----------GCNDRSKRRRGLDRRMM 266
SS G +R KRRR LD +M
Sbjct: 201 SSKSVMVGHGGGGGEERLKRRRALDHIIM 229
>At1g68810 putative bHLH transcription factor (bHLH030)
Length = 368
Score = 154 bits (389), Expect = 5e-38
Identities = 87/210 (41%), Positives = 131/210 (61%), Gaps = 25/210 (11%)
Query: 58 EVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSS 117
E+ ++T +ALAA K+H EAE+RRRERIN+HL LR++LP +KT
Sbjct: 157 ELGKMTAQEIMDAKALAASKSHSEAERRRRERINNHLAKLRSILPNTTKT---------- 206
Query: 118 RTCERFKSKGSRLAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIF 177
K S LA+V++ VKEL+++T I++ VP E+D TV + +G+GR +
Sbjct: 207 -------DKASLLAEVIQHVKELKRETSVISETNLVPTESDELTVAFTEEEETGDGRFVI 259
Query: 178 KASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAAE----KEHNSIES 233
KASLCCEDRSDL+PD+I+ LK++ LKTLKAE+ T+GGR +NVL V E +E
Sbjct: 260 KASLCCEDRSDLLPDMIKTLKAMRLKTLKAEITTVGGRVKNVLFVTGEESSGEEVEEEYC 319
Query: 234 IHFLQNSLRSLLDRS----SGCNDRSKRRR 259
I ++ +L++++++S S + +KR+R
Sbjct: 320 IGTIEEALKAVMEKSNVEESSSSGNAKRQR 349
>At3g25710 putative bHLH transcription factor (bHLH032)
Length = 344
Score = 139 bits (351), Expect = 1e-33
Identities = 83/216 (38%), Positives = 126/216 (57%), Gaps = 42/216 (19%)
Query: 71 RALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRL 130
+ALAA K+H EAE+RRRERIN+HL LR++LP +KT K S L
Sbjct: 128 KALAASKSHSEAERRRRERINTHLAKLRSILPNTTKT-----------------DKASLL 170
Query: 131 AKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLI 190
A+V++ +KEL++QT QIT VP E D TV S+ +D EG L+ +AS CC+DR+DL+
Sbjct: 171 AEVIQHMKELKRQTSQITDTYQVPTECDDLTVDSSYNDE--EGNLVIRASFCCQDRTDLM 228
Query: 191 PDLIEILKSLHLKTLKAEMATLGGRTRNVLVVAA----EKEHNSI--------------- 231
D+I LKSL L+TLKAE+AT+GGR +N+L ++ E++H+S
Sbjct: 229 HDVINALKSLRLRTLKAEIATVGGRVKNILFLSREYDDEEDHDSYRRNFDGDDVEDYDEE 288
Query: 232 ----ESIHFLQNSLRSLLDRSSGCNDRSKRRRGLDR 263
+ ++ +L++++++ ND S L++
Sbjct: 289 RMMNNRVSSIEEALKAVIEKCVHNNDESNDNNNLEK 324
>At2g40200 putative bHLH transcription factor (bHLH051)
Length = 254
Score = 106 bits (265), Expect = 1e-23
Identities = 75/203 (36%), Positives = 110/203 (53%), Gaps = 30/203 (14%)
Query: 70 DRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSR 129
++A + ++H+ AEKRRR+RINSHL LR L+P + K K +
Sbjct: 58 EKAESLSRSHRLAEKRRRDRINSHLTALRKLVPNSDKL-----------------DKAAL 100
Query: 130 LAKVVERVKELRQQTCQITQVETVPCEADGXTVISAG-SDM-SGEGRLIFKASLCCEDRS 187
LA V+E+VKEL+Q+ + + +P EAD TV SD S +IFKAS CCED+
Sbjct: 101 LATVIEQVKELKQKAAESPIFQDLPTEADEVTVQPETISDFESNTNTIIFKASFCCEDQP 160
Query: 188 DLIPDLIEILKSLHLKTLKAEMATLGGRTRNVLVVA---AEKEHNSIESIHFLQNSLRSL 244
+ I ++I +L L L+T++AE+ ++GGR R ++ + N S L+ SL S
Sbjct: 161 EAISEIIRVLTKLQLETIQAEIISVGGRMRINFILKDSNCNETTNIAASAKALKQSLCSA 220
Query: 245 LDR--------SSGCNDRSKRRR 259
L+R SS C RSKR+R
Sbjct: 221 LNRITSSSTTTSSVCRIRSKRQR 243
>At4g38071 putative bHLH transcription factor (bHLH131)
Length = 256
Score = 72.4 bits (176), Expect = 2e-13
Identities = 60/226 (26%), Positives = 100/226 (43%), Gaps = 31/226 (13%)
Query: 35 SSTMMQQSFCSSSDNNNNYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHL 94
S+TM + F ++ ++ + F V T S+ + A K H +AE+RRR RINS
Sbjct: 55 STTMGRSFFAGAATSSKLFSRGFSV---TKPKSKTESKEVAAKKHSDAERRRRLRINSQF 111
Query: 95 DHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQTCQITQVETVP 154
LRT+LP K + K S L + V EL++ + + T P
Sbjct: 112 ATLRTILPNLVK-----------------QDKASVLGETVRYFNELKK---MVQDIPTTP 151
Query: 155 CEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLIPDLIEILKSLHLKTLKAEMATLGG 214
D + D R + + C DR L+ ++ E +K++ K ++AE+ T+GG
Sbjct: 152 SLEDNLRL-----DHCNNNRDLARVVFSCSDREGLMSEVAESMKAVKAKAVRAEIMTVGG 206
Query: 215 RTRNVLVVAAEKEHNSIESIHFLQNSLRSLLDRSSGCNDRSKRRRG 260
RT+ L V N E + L+ SL+ +++ S ++ G
Sbjct: 207 RTKCALFVQGV---NGNEGLVKLKKSLKLVVNGKSSSEAKNNNNGG 249
>At3g56980 putative bHLH transcription factor (bHLH039)
Length = 258
Score = 44.3 bits (103), Expect = 7e-05
Identities = 32/140 (22%), Positives = 62/140 (43%), Gaps = 9/140 (6%)
Query: 8 NIGMENNSELFQFLVA----NNPSFFEYSTPSSTMMQQSFCSSSDNNNNYYHPFEVSEIT 63
N G + E + +A NN F ++ P T + + N+ E +EI
Sbjct: 11 NFGWPSTGEYDSYYLAGDILNNGGFLDFPVPEETYGAVTAVTQHQNSFGVSVSSEGNEID 70
Query: 64 DTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERF 123
+ P + NH +E+ RR +INS LR+ LP + ++++ + SR+ +
Sbjct: 71 NNP-----VVVKKLNHNASERDRRRKINSLFSSLRSCLPASGQSKKLSIPATVSRSLKYI 125
Query: 124 KSKGSRLAKVVERVKELRQQ 143
++ K++++ +EL Q
Sbjct: 126 PELQEQVKKLIKKKEELLVQ 145
>At5g04150 myc - like protein
Length = 240
Score = 40.8 bits (94), Expect = 8e-04
Identities = 30/126 (23%), Positives = 58/126 (45%), Gaps = 16/126 (12%)
Query: 45 SSSDNNNNYYHPFEVSEITDTPSQQDRALAALK---NHKEAEKRRRERINSHLDHLRTLL 101
+++ NNNNY ++DR L+ NH +E+ RR ++N+ LR LL
Sbjct: 46 NTNSNNNNY-------------QEEDRGAVVLEKKLNHNASERDRRRKLNALYSSLRALL 92
Query: 102 PCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQTCQITQVETVPCEADGXT 161
P + + R+ +R + + L ++ R +EL ++ + T E + +A +
Sbjct: 93 PLSDQKRKLSIPMTVARVVKYIPEQKQELQRLSRRKEELLKRISRKTHQEQLRNKAMMDS 152
Query: 162 VISAGS 167
+ S+ S
Sbjct: 153 IDSSSS 158
>At5g61270 bHLH transcription factor like protein
Length = 366
Score = 38.9 bits (89), Expect = 0.003
Identities = 30/112 (26%), Positives = 49/112 (42%), Gaps = 26/112 (23%)
Query: 41 QSFCSSSDNNNNYYHPFEVSEITDTPSQQDRALAALKN---------HKEAEKRRRERIN 91
+S ++ + +Y+ ++ T+ Q+ R A N H E+E+RRR+RIN
Sbjct: 124 RSLKTARTGDRDYFRSGSETQDTEGDEQETRGEAGRSNGRRGRAAAIHNESERRRRDRIN 183
Query: 92 SHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQ 143
+ L+ LLP SK K S L V+E +K+L+ Q
Sbjct: 184 QRMRTLQKLLPTASKA-----------------DKVSILDDVIEHLKQLQAQ 218
>At5g56960 putative protein
Length = 466
Score = 38.5 bits (88), Expect = 0.004
Identities = 39/155 (25%), Positives = 76/155 (48%), Gaps = 15/155 (9%)
Query: 79 HKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVK 138
H +E++RRE++N LR+LLP +K + +S S E+ S ++K++ER +
Sbjct: 290 HMISERKRREKLNESFQALRSLLPPGTKKDK---ASVLSIAREQLSSLQGEISKLLERNR 346
Query: 139 ELRQQTCQITQVETVPCEADGXTV------ISAGSDMSGEGRLIFKASLCCEDRSDLIPD 192
E+ + ++E + V S + + + R++ + + D DL+
Sbjct: 347 EVEAKLAGEREIENDLRPEERFNVRIRHIPESTSRERTLDLRVVLRGDIIRVD--DLMIR 404
Query: 193 LIEILKSLH---LKTLKAE-MATLGGRTRNVLVVA 223
L+E LK ++ L +++A +A G T VLV++
Sbjct: 405 LLEFLKQINNVSLVSIEARTLARAEGDTSIVLVIS 439
>At1g32640 transcription factor like protein, BHLH6
Length = 623
Score = 38.5 bits (88), Expect = 0.004
Identities = 29/94 (30%), Positives = 42/94 (43%), Gaps = 11/94 (11%)
Query: 78 NHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERV 137
NH EAE++RRE++N LR ++P SK + + KSK V
Sbjct: 452 NHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAIAYINELKSK----------V 501
Query: 138 KELRQQTCQI-TQVETVPCEADGXTVISAGSDMS 170
+ + QI Q+E V E G ++G DMS
Sbjct: 502 VKTESEKLQIKNQLEEVKLELAGRKASASGGDMS 535
>At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATULA
(SPT)
Length = 373
Score = 37.0 bits (84), Expect = 0.011
Identities = 26/105 (24%), Positives = 44/105 (41%), Gaps = 17/105 (16%)
Query: 44 CSSSDNNNNYYHPFEVSEITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPC 103
C S + S+ + + A + H +EKRRR RIN + L++L+P
Sbjct: 167 CESEEGGEAVVDEAPSSKSGPSSRSSSKRCRAAEVHNLSEKRRRSRINEKMKALQSLIPN 226
Query: 104 NSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVKELRQQTCQIT 148
++KT K S L + +E +K+L+ Q +T
Sbjct: 227 SNKT-----------------DKASMLDEAIEYLKQLQLQVQMLT 254
>At4g00870 putative bHLH transcription factor (bHLH014)
Length = 423
Score = 37.0 bits (84), Expect = 0.011
Identities = 23/84 (27%), Positives = 39/84 (46%)
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVV 134
A+ +H EAEK+RRE++N LR ++P S+ + S + E KSK L +
Sbjct: 246 AVLSHVEAEKQRREKLNHRFYALRAIVPKVSRMDKASLLSDAVSYIESLKSKIDDLETEI 305
Query: 135 ERVKELRQQTCQITQVETVPCEAD 158
+++K + T P +
Sbjct: 306 KKMKMTETDKLDNSSSNTSPSSVE 329
>At4g00050 bHLH like transcription factor (bHLH016)
Length = 399
Score = 37.0 bits (84), Expect = 0.011
Identities = 20/72 (27%), Positives = 39/72 (53%), Gaps = 17/72 (23%)
Query: 79 HKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERVK 138
H ++E++RR++IN + L+ L+P +SKT K S L +V+E +K
Sbjct: 218 HNQSERKRRDKINQRMKTLQKLVPNSSKT-----------------DKASMLDEVIEYLK 260
Query: 139 ELRQQTCQITQV 150
+L+ Q ++++
Sbjct: 261 QLQAQVSMMSRM 272
>At3g56970 putative bHLH transcription factor (bHLH038)
Length = 253
Score = 36.6 bits (83), Expect = 0.014
Identities = 28/132 (21%), Positives = 61/132 (46%), Gaps = 17/132 (12%)
Query: 26 PSFFEYSTPSSTMMQQSFCSSSDNNNN-------YYHPFEVSE--------ITDTPSQQD 70
PSFF ST +S+ + DN NN +EV+ ++ ++ D
Sbjct: 6 PSFFTNFGWPSTNQYESYYGAGDNLNNGTFLELTVPQTYEVTHHQNSLGVSVSSEGNEID 65
Query: 71 RALAALK--NHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGS 128
+K NH +E+ RR++IN+ LR+ LP + ++++ S++ +
Sbjct: 66 NNPVVVKKLNHNASERDRRKKINTLFSSLRSCLPASDQSKKLSIPETVSKSLKYIPELQQ 125
Query: 129 RLAKVVERVKEL 140
++ +++++ +E+
Sbjct: 126 QVKRLIQKKEEI 137
>At4g28815 AtbHLH127
Length = 307
Score = 35.8 bits (81), Expect = 0.024
Identities = 26/73 (35%), Positives = 33/73 (44%), Gaps = 19/73 (26%)
Query: 75 ALKNHKEAEKRRRERINSHLDHLRTLLP-CNSKTRQGFTSSKSSRTCERFKSKGSRLAKV 133
A + H AE+RRRE+IN + L+ L+P CN T K S L V
Sbjct: 151 AAEMHNLAERRRREKINERMKTLQQLIPRCNKST------------------KVSMLEDV 192
Query: 134 VERVKELRQQTCQ 146
+E VK L Q Q
Sbjct: 193 IEYVKSLEMQINQ 205
>At5g41315 bHLH transcription factor-like protein
Length = 637
Score = 34.7 bits (78), Expect = 0.054
Identities = 25/113 (22%), Positives = 47/113 (41%), Gaps = 6/113 (5%)
Query: 64 DTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERF 123
D+P +D NH EK+RRE++N LR ++P +K + + +
Sbjct: 431 DSPEARDET----GNHAVLEKKRREKLNERFMTLRKIIPSINKIDKVSILDDTIEYLQEL 486
Query: 124 KSKGSRLAKVVERVKELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLI 176
+ + L E + T +T PC+A T + ++ +G G+ +
Sbjct: 487 ERRVQELESCRESTDTETRGT--MTMKRKKPCDAGERTSANCANNETGNGKKV 537
>At4g28790 bHLH transcription factor like protein (bHLH023)
Length = 413
Score = 34.7 bits (78), Expect = 0.054
Identities = 28/83 (33%), Positives = 42/83 (49%), Gaps = 19/83 (22%)
Query: 61 EITDTPSQQDRALAALKNHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTC 120
E D+ S + R+ AA+ HK +E+RRR++IN + L+ LLP +KT
Sbjct: 266 EARDSTSSK-RSRAAIM-HKLSERRRRQKINEMMKALQELLPRCTKT------------- 310
Query: 121 ERFKSKGSRLAKVVERVKELRQQ 143
+ S L V+E VK L+ Q
Sbjct: 311 ----DRSSMLDDVIEYVKSLQSQ 329
>At4g17880 putative transcription factor BHLH4
Length = 589
Score = 34.3 bits (77), Expect = 0.071
Identities = 22/66 (33%), Positives = 33/66 (49%), Gaps = 3/66 (4%)
Query: 78 NHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERV 137
NH EAE++RRE++N LR ++P SK + + KSK L K
Sbjct: 416 NHVEAERQRREKLNQRFYSLRAVVPNVSKMDKASLLGDAISYISELKSK---LQKAESDK 472
Query: 138 KELRQQ 143
+EL++Q
Sbjct: 473 EELQKQ 478
>At4g09820 putative protein
Length = 379
Score = 34.3 bits (77), Expect = 0.071
Identities = 27/127 (21%), Positives = 51/127 (39%), Gaps = 15/127 (11%)
Query: 78 NHKEAEKRRRERINSHLDHLRTLLPCNSKTRQGFTSSKSSRTCERFKSKGSRLAKVVERV 137
+H AE+RRRE++N LR+++P +K + + + + L
Sbjct: 224 SHVVAERRRREKLNEKFITLRSMVPFVTKMDKVSILGDTIAYVNHLRKRVHELENTHHEQ 283
Query: 138 KELRQQTCQITQVETVPCEADGXTVISAGSDMSGEGRLIFKASLCCEDRSDLIPDLIEIL 197
+ R +TC+ E V V+ + CE R L+ D++++L
Sbjct: 284 QHKRTRTCKRKTSEEVEVSIIENDVL---------------LEMRCEYRDGLLLDILQVL 328
Query: 198 KSLHLKT 204
L ++T
Sbjct: 329 HELGIET 335
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,356,468
Number of Sequences: 26719
Number of extensions: 206357
Number of successful extensions: 1022
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 927
Number of HSP's gapped (non-prelim): 104
length of query: 267
length of database: 11,318,596
effective HSP length: 98
effective length of query: 169
effective length of database: 8,700,134
effective search space: 1470322646
effective search space used: 1470322646
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135795.6


