

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.3 + phase: 0
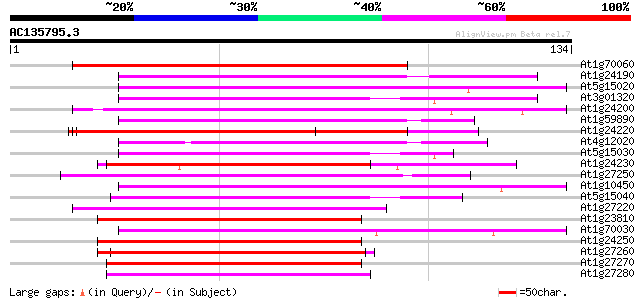
(134 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g70060 hypothetical protein 76 5e-15
At1g24190 hypothetical protein 74 2e-14
At5g15020 transcriptional regulatory - like protein 71 2e-13
At3g01320 unknown protein 70 3e-13
At1g24200 hypothetical protein 68 1e-12
At1g59890 hypothetical protein 64 2e-11
At1g24220 hypothetical protein 63 5e-11
At4g12020 putative disease resistance protein 62 1e-10
At5g15030 putative protein 60 3e-10
At1g24230 hypothetical protein 60 3e-10
At1g27250 hypothetical protein 59 1e-09
At1g10450 unknown protein 59 1e-09
At5g15040 putative protein 55 8e-09
At1g27220 putative protein 53 4e-08
At1g23810 hypothetical protein 53 4e-08
At1g70030 unknown protein 50 3e-07
At1g24250 hypothetical protein 50 4e-07
At1g27260 hypothetical protein 49 8e-07
At1g27270 hypothetical protein 48 1e-06
At1g27280 hypothetical protein 48 2e-06
>At1g70060 hypothetical protein
Length = 1386
Score = 76.3 bits (186), Expect = 5e-15
Identities = 38/80 (47%), Positives = 51/80 (63%)
Query: 16 GYAHQNQQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFK 75
G + + + DALA++K V+ +F+DK DKYDEFL VM DFK R+D G R+ ELFK
Sbjct: 3 GGSSAQKLTTNDALAYLKAVKDKFQDKRDKYDEFLEVMKDFKAQRVDTTGVILRVKELFK 62
Query: 76 EHRHLIMRFNSLTLKGSHFT 95
+R LI+ FN+ KG T
Sbjct: 63 GNRELILGFNTFLPKGFEIT 82
Score = 29.3 bits (64), Expect = 0.64
Identities = 13/59 (22%), Positives = 29/59 (49%)
Query: 26 KDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
++A++FV ++ F+ Y FL+++N ++ + LF++H L+ F
Sbjct: 100 EEAISFVNKIKTRFQGDDRVYKSFLDILNMYRKENKSITEVYHEVAILFRDHHDLLGEF 158
>At1g24190 hypothetical protein
Length = 1263
Score = 74.3 bits (181), Expect = 2e-14
Identities = 40/100 (40%), Positives = 59/100 (59%), Gaps = 5/100 (5%)
Query: 27 DALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRFNS 86
DALA++K V+ +F+D+ KYDEFL VM +FK+ R+D G R+ ELFK H+ LI+ FN+
Sbjct: 14 DALAYLKAVKDKFQDQRGKYDEFLEVMKNFKSQRVDTAGVITRVKELFKGHQELILGFNT 73
Query: 87 LTLKGSHFTKHGYETEEEEDMEILDKLEKLESALQLILEI 126
KG T + E+ L K + E A+ + +I
Sbjct: 74 FLPKGFEIT-----LQPEDGQPPLKKRVEFEEAISFVNKI 108
Score = 30.0 bits (66), Expect = 0.38
Identities = 13/59 (22%), Positives = 30/59 (50%)
Query: 26 KDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
++A++FV ++ F+ Y FL+++N ++ + LF++H L++ F
Sbjct: 99 EEAISFVNKIKTRFQGDDRVYKSFLDILNMYRRDSKSITEVYQEVAILFRDHSDLLVEF 157
>At5g15020 transcriptional regulatory - like protein
Length = 1343
Score = 70.9 bits (172), Expect = 2e-13
Identities = 42/117 (35%), Positives = 61/117 (51%), Gaps = 10/117 (8%)
Query: 27 DALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRFNS 86
DAL ++K V+ F+D+ DKYD FL VM DFK + D G +R+ ELFK H +LI FN+
Sbjct: 52 DALTYLKEVKEMFQDQRDKYDMFLEVMKDFKAQKTDTSGVISRVKELFKGHNNLIFGFNT 111
Query: 87 LTLKGSHFTKHGYETEEEEDME----------ILDKLEKLESALQLILEILVIKKKE 133
KG T E ++ +E I + + E + LEIL + +K+
Sbjct: 112 FLPKGFEITLDDVEAPSKKTVEFEEAISFVNKIKTRFQHNELVYKSFLEILNMYRKD 168
Score = 31.2 bits (69), Expect = 0.17
Identities = 14/59 (23%), Positives = 29/59 (48%)
Query: 26 KDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
++A++FV ++ F+ Y FL ++N ++ D + LF++H L+ F
Sbjct: 135 EEAISFVNKIKTRFQHNELVYKSFLEILNMYRKDNKDITEVYNEVSTLFEDHSDLLEEF 193
>At3g01320 unknown protein
Length = 1324
Score = 70.5 bits (171), Expect = 3e-13
Identities = 42/101 (41%), Positives = 58/101 (56%), Gaps = 8/101 (7%)
Query: 27 DALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRFNS 86
DAL++++ V+ F+D+ +KYD FL VM DFK R D G AR+ ELFK H +LI FN+
Sbjct: 57 DALSYLREVKEMFQDQREKYDRFLEVMKDFKAQRTDTGGVIARVKELFKGHNNLIYGFNT 116
Query: 87 LTLKGSHFTKHGYE-TEEEEDMEILDKLEKLESALQLILEI 126
F GYE T EED + K + E A+ + +I
Sbjct: 117 -------FLPKGYEITLIEEDDALPKKTVEFEQAINFVNKI 150
Score = 27.3 bits (59), Expect = 2.4
Identities = 13/59 (22%), Positives = 29/59 (49%)
Query: 26 KDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
+ A+ FV +++ F+ Y FL ++N ++ + + + LF+ H L+ +F
Sbjct: 141 EQAINFVNKIKMRFKHDEHVYKSFLEILNMYRKENKEIKEVYNEVSILFQGHLDLLEQF 199
>At1g24200 hypothetical protein
Length = 196
Score = 68.2 bits (165), Expect = 1e-12
Identities = 41/127 (32%), Positives = 72/127 (56%), Gaps = 11/127 (8%)
Query: 16 GYAHQNQQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFK 75
G AH+ + DAL +++ V+ +F+ + +KYDEFL +M D+KT RID G R+ EL K
Sbjct: 5 GGAHK--PTTNDALKYLRAVKAKFQGQREKYDEFLQIMIDYKTQRIDISGVIIRMKELLK 62
Query: 76 EHRHLIMRFNSLTLKGSHFTKHGYETEEE--EDMEILDKLEKLESALQ-------LILEI 126
E + L++ FN+ G T H ++++ E E + + K+++ Q +L+I
Sbjct: 63 EQQGLLLGFNAFLPNGYMITHHEQPSQKKPVELGEAISFINKIKTRFQGDDRVYKSVLDI 122
Query: 127 LVIKKKE 133
L + +K+
Sbjct: 123 LNMYRKD 129
>At1g59890 hypothetical protein
Length = 1125
Score = 63.9 bits (154), Expect = 2e-11
Identities = 33/85 (38%), Positives = 51/85 (59%), Gaps = 3/85 (3%)
Query: 27 DALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRFNS 86
DAL ++K V+ F+D +KY+ FL VM DFK R+D G AR+ +LFK + L++ FN+
Sbjct: 45 DALTYLKAVKDMFQDNKEKYETFLGVMKDFKAQRVDTNGVIARVKDLFKGYDDLLLGFNT 104
Query: 87 LTLKGSHFTKHGYETEEEEDMEILD 111
KG T + E+E+ + +D
Sbjct: 105 FLPKGYKIT---LQPEDEKPKKPVD 126
Score = 26.9 bits (58), Expect = 3.2
Identities = 13/57 (22%), Positives = 26/57 (44%)
Query: 28 ALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
A+ FV ++ F Y +FL+++N ++ + LF++H L+ F
Sbjct: 130 AIEFVNRIKARFGGDDRAYKKFLDILNMYRKETKSINEVYQEVTLLFQDHEDLLGEF 186
>At1g24220 hypothetical protein
Length = 744
Score = 62.8 bits (151), Expect = 5e-11
Identities = 28/81 (34%), Positives = 52/81 (63%)
Query: 15 YGYAHQNQQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELF 74
+G + ++++ DA +++ V+ F D+ KY EFL ++ND+K R+DA+ AR+DEL
Sbjct: 164 FGKSVPLEKTLDDARSYIDSVKEAFHDEPAKYAEFLKLLNDYKARRLDADSVIARVDELT 223
Query: 75 KEHRHLIMRFNSLTLKGSHFT 95
K+HR+L++ ++ L + T
Sbjct: 224 KDHRNLLLGLRAILLPAAKST 244
Score = 41.6 bits (96), Expect = 1e-04
Identities = 18/58 (31%), Positives = 37/58 (63%)
Query: 16 GYAHQNQQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDEL 73
G + + +M DA+++++ V+ EF D+ K+DEF +N+ + R++ + AR++EL
Sbjct: 3 GRSVSQEPTMADAVSYIESVKEEFHDEPAKFDEFRMRLNEVRDDRVEKDRITARINEL 60
Score = 40.0 bits (92), Expect = 4e-04
Identities = 22/96 (22%), Positives = 43/96 (43%)
Query: 17 YAHQNQQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKE 76
Y + ++++ + ++ V+ F D+ K+ EFL +MND +I+ AR+ E+ K
Sbjct: 492 YRLEIASTIRETVTYIADVKEAFLDEPAKFHEFLRLMNDVCDHKIEEANGSARMAEIIKG 551
Query: 77 HRHLIMRFNSLTLKGSHFTKHGYETEEEEDMEILDK 112
H L++ + K + + E I K
Sbjct: 552 HPRLLLVLSVFFPKSKQYEEAARSISSEASRTIPPK 587
>At4g12020 putative disease resistance protein
Length = 1895
Score = 61.6 bits (148), Expect = 1e-10
Identities = 36/88 (40%), Positives = 52/88 (58%), Gaps = 4/88 (4%)
Query: 27 DALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRFNS 86
DAL ++K V+ +FED +KYD FL V+ND K +D G ARL +LFK H L++ FN+
Sbjct: 308 DALNYLKAVKDKFEDS-EKYDTFLEVLNDCKHQGVDTSGVIARLKDLFKGHDDLLLGFNT 366
Query: 87 LTLKGSHFTKHGYETEEEEDMEILDKLE 114
K T E++ ++ LDK+E
Sbjct: 367 YLSKEYQIT---ILPEDDFPIDFLDKVE 391
>At5g15030 putative protein
Length = 271
Score = 60.1 bits (144), Expect = 3e-10
Identities = 34/81 (41%), Positives = 47/81 (57%), Gaps = 8/81 (9%)
Query: 27 DALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRFNS 86
DALA++K ++ F D+ KY FL +M+DFK R D AR+ +L K H HLI+ FN
Sbjct: 196 DALAYLKEIKDVFHDQKYKYHLFLEIMSDFKAQRTDTSVVIARVKDLLKGHNHLILVFNK 255
Query: 87 LTLKGSHFTKHGYE-TEEEED 106
F HG+E T ++ED
Sbjct: 256 -------FLPHGFEITLDDED 269
Score = 30.0 bits (66), Expect = 0.38
Identities = 23/79 (29%), Positives = 38/79 (47%), Gaps = 10/79 (12%)
Query: 24 SMKDALAFVKGVEVEF--EDKGDKYDEFLNVMNDFKTLRIDAEGAKAR--------LDEL 73
++ DA A+++ V+ F D+ DKY F V+ DFK RI A+ + + L +L
Sbjct: 83 TISDARAYLQQVKNTFIDHDERDKYAMFRKVLFDFKAQRIVADRSDSARFCSHLKDLSDL 142
Query: 74 FKEHRHLIMRFNSLTLKGS 92
F+ + R + GS
Sbjct: 143 FQIFSERMKRIGDKEIDGS 161
>At1g24230 hypothetical protein
Length = 245
Score = 60.1 bits (144), Expect = 3e-10
Identities = 29/101 (28%), Positives = 58/101 (56%), Gaps = 1/101 (0%)
Query: 22 QQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLI 81
+++ D +++ ++ F D+ KY +FL ++ND+ R+DA A AR+ EL K+HR+L+
Sbjct: 81 KETFHDVRSYIYSLKESFRDEPAKYAQFLEILNDYSARRVDAPSAVARMTELMKDHRNLV 140
Query: 82 MRFNSLTLKG-SHFTKHGYETEEEEDMEILDKLEKLESALQ 121
+ F+ L G + T E + + + + + + KL++ Q
Sbjct: 141 LGFSVLLSTGDTKTTPLEAEPDNNKRIRVANFISKLKARFQ 181
Score = 45.8 bits (107), Expect = 7e-06
Identities = 22/64 (34%), Positives = 40/64 (62%), Gaps = 1/64 (1%)
Query: 24 SMKDALAFVKGVEVEF-EDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIM 82
++ +A +++ V+ F D+ KY EFL++M D + R+D R+ EL K+H +L++
Sbjct: 11 TIDEATSYINAVKEAFGADQPAKYREFLDIMLDLRANRVDLATVVPRMRELLKDHVNLLL 70
Query: 83 RFNS 86
RFN+
Sbjct: 71 RFNA 74
>At1g27250 hypothetical protein
Length = 137
Score = 58.5 bits (140), Expect = 1e-09
Identities = 32/98 (32%), Positives = 51/98 (51%), Gaps = 2/98 (2%)
Query: 13 KEYGYAHQNQQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDE 72
K G + + + DA A+++ V +F + KYD+F+ VM +FK RID +G +++
Sbjct: 7 KHVGEGSKPRATKDDAYAYLRAVRAKFHNDSKKYDDFVAVMTNFKARRIDRDGCIKEVEQ 66
Query: 73 LFKEHRHLIMRFNSLTLKGSHFTKHGYETEEEEDMEIL 110
L K HR LI FN+ K Y ED+E++
Sbjct: 67 LLKGHRDLISGFNAFLPKCLEI--KNYYFGAGEDLEVV 102
>At1g10450 unknown protein
Length = 1173
Score = 58.5 bits (140), Expect = 1e-09
Identities = 37/117 (31%), Positives = 59/117 (49%), Gaps = 10/117 (8%)
Query: 27 DALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRFNS 86
DAL ++K V+ F D +KY+ FL +M +FK ID G R+ LFK +R L++ FN+
Sbjct: 84 DALTYLKAVKDIFHDNKEKYESFLELMKEFKAQTIDTNGVIERIKVLFKGYRDLLLGFNT 143
Query: 87 LTLKGSHFTKHGYETEEEEDMEILDKLEKL----------ESALQLILEILVIKKKE 133
KG T E + + ++ D + + E A + L+IL + +KE
Sbjct: 144 FLPKGYKITLLPEEEKPKIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRKE 200
Score = 37.7 bits (86), Expect = 0.002
Identities = 18/59 (30%), Positives = 29/59 (48%)
Query: 26 KDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
KDA+ FV ++ F D Y FL+++N ++ + + LFK H L+M F
Sbjct: 167 KDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRKEKKSISEVYEEVTMLFKGHEDLLMEF 225
>At5g15040 putative protein
Length = 87
Score = 55.5 bits (132), Expect = 8e-09
Identities = 32/84 (38%), Positives = 46/84 (54%), Gaps = 7/84 (8%)
Query: 25 MKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
M + A+ V+ F D+ +KYD F N++ D K RI A A+L ELFKEH LI+ F
Sbjct: 1 MAECRAYFMEVKDTFHDQIEKYDMFKNILLDLKARRIGRHTAFAQLKELFKEHNELIIGF 60
Query: 85 NSLTLKGSHFTKHGYETEEEEDME 108
N+ F GY+ ++D+E
Sbjct: 61 NT-------FLPTGYKIALDDDVE 77
>At1g27220 putative protein
Length = 90
Score = 53.1 bits (126), Expect = 4e-08
Identities = 26/75 (34%), Positives = 42/75 (55%)
Query: 16 GYAHQNQQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFK 75
G + + ++ DA A+++ V+ F + DKYD+F+ +M +FK +ID + EL K
Sbjct: 3 GEGSKPKATVDDAYAYIRTVKSTFHNDPDKYDDFMAIMKNFKARKIDRNTCIEEVKELLK 62
Query: 76 EHRHLIMRFNSLTLK 90
HR LI FN+ K
Sbjct: 63 GHRDLISGFNAFLPK 77
>At1g23810 hypothetical protein
Length = 241
Score = 53.1 bits (126), Expect = 4e-08
Identities = 23/63 (36%), Positives = 41/63 (64%)
Query: 22 QQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLI 81
+ ++ DA +++ V+ F D+ KY+E L ++NDFK R++A AR++EL K+H +L+
Sbjct: 104 EPTIDDATSYLIAVKEAFHDEPAKYEEMLKLLNDFKARRVNAASVIARVEELMKDHSNLL 163
Query: 82 MRF 84
F
Sbjct: 164 FGF 166
Score = 37.0 bits (84), Expect = 0.003
Identities = 20/63 (31%), Positives = 34/63 (53%), Gaps = 1/63 (1%)
Query: 24 SMKDALAFVKGVEVEFED-KGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIM 82
S++D A+V V+V E+ + KY EFL + ++ R+ AR+ +L K+H L +
Sbjct: 11 SLEDGKAYVNAVKVALEEAEPAKYQEFLRLFHEVIARRMGMATFSARMQDLLKDHPSLCL 70
Query: 83 RFN 85
N
Sbjct: 71 GLN 73
>At1g70030 unknown protein
Length = 160
Score = 50.4 bits (119), Expect = 3e-07
Identities = 33/117 (28%), Positives = 56/117 (47%), Gaps = 10/117 (8%)
Query: 27 DALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRFNS 86
DAL F+ V+ +++D + YD FL +M DF+ R +++ ELFK L++ FN+
Sbjct: 11 DALDFLHLVKTKYQDNREIYDSFLTIMKDFRGQRAKTCDVISKVKELFKGQPELLLGFNT 70
Query: 87 -------LTLKGSHFTKHGYETEEEEDMEILDKLE---KLESALQLILEILVIKKKE 133
+TL T + +E E ++K++ + LE+L KKE
Sbjct: 71 FLPTGFEITLSDDELTSNSKFAHFDEAYEFVNKVKTRFQNNDVFNSFLEVLKTHKKE 127
>At1g24250 hypothetical protein
Length = 252
Score = 50.1 bits (118), Expect = 4e-07
Identities = 23/63 (36%), Positives = 39/63 (61%)
Query: 22 QQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLI 81
+ ++ DA +++ V+ F D+ KY E L ++ DFK R+DA AR++EL K+H +L+
Sbjct: 115 EPTIDDATSYLIAVKEAFHDEPAKYGEMLKLLKDFKARRVDAACVIARVEELMKDHLNLL 174
Query: 82 MRF 84
F
Sbjct: 175 FGF 177
Score = 33.9 bits (76), Expect = 0.026
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 1/65 (1%)
Query: 24 SMKDALAFVKGVEVEFED-KGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIM 82
S++D A+V VEV ++ + ++ F+ + F RI AR+ +L K+H L +
Sbjct: 11 SLEDVKAYVNAVEVALQEMEPARFGMFVRLFRGFTAPRIGMPTFSARMQDLLKDHPSLCL 70
Query: 83 RFNSL 87
N L
Sbjct: 71 GLNVL 75
>At1g27260 hypothetical protein
Length = 222
Score = 48.9 bits (115), Expect = 8e-07
Identities = 21/64 (32%), Positives = 41/64 (63%)
Query: 22 QQSMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLI 81
+ +++DA +++ V+ F D+ KY+E L ++ND + R+DA A ++EL K+H+ L+
Sbjct: 88 ETTIEDATSYLNSVKRAFHDEPAKYEELLKLLNDIEARRVDAASFIASVEELMKDHQTLL 147
Query: 82 MRFN 85
F+
Sbjct: 148 NGFS 151
Score = 44.3 bits (103), Expect = 2e-05
Identities = 23/63 (36%), Positives = 35/63 (55%)
Query: 25 MKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
M A A++ V F D+ KY +FL+++ D + RID L EL K+H L++ F
Sbjct: 1 MNKAFAYIIAVRDRFRDEPAKYRQFLSLLRDRRARRIDKATFFVGLVELIKDHLDLLLGF 60
Query: 85 NSL 87
N+L
Sbjct: 61 NAL 63
>At1g27270 hypothetical protein
Length = 241
Score = 48.1 bits (113), Expect = 1e-06
Identities = 22/61 (36%), Positives = 37/61 (60%)
Query: 24 SMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMR 83
+M DA +++ V+ F D+ KY E ++ D K RI+A AR++EL K+H +L++
Sbjct: 110 TMDDATSYLNAVKEAFHDEPAKYMEITKLLTDLKARRINAASVIARMEELLKDHLNLLLG 169
Query: 84 F 84
F
Sbjct: 170 F 170
Score = 38.1 bits (87), Expect = 0.001
Identities = 21/73 (28%), Positives = 37/73 (49%), Gaps = 1/73 (1%)
Query: 16 GYAHQNQQSMKDALAFVKGVEVEFED-KGDKYDEFLNVMNDFKTLRIDAEGAKARLDELF 74
G + + + +D +A+ V+V +D + +KY EF+ + D+ R E A L EL
Sbjct: 3 GKSKSPEFNFEDGMAYFDAVKVALQDTEPEKYQEFVRIFIDYTANRFGFETLSASLQELL 62
Query: 75 KEHRHLIMRFNSL 87
++H +L N L
Sbjct: 63 RDHANLCHGSNVL 75
>At1g27280 hypothetical protein
Length = 225
Score = 47.8 bits (112), Expect = 2e-06
Identities = 21/63 (33%), Positives = 38/63 (59%)
Query: 24 SMKDALAFVKGVEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMR 83
++ DA++++ V+ F D+ KY EF + D + ID G R++EL K H++L++R
Sbjct: 78 TIDDAVSYINTVKEAFHDEPAKYYEFFQLFYDIRYRLIDVAGGITRVEELLKAHKNLLVR 137
Query: 84 FNS 86
N+
Sbjct: 138 LNA 140
Score = 32.7 bits (73), Expect = 0.058
Identities = 14/50 (28%), Positives = 30/50 (60%)
Query: 35 VEVEFEDKGDKYDEFLNVMNDFKTLRIDAEGAKARLDELFKEHRHLIMRF 84
+E + D+ +K++ L+++ D+ R D A + EL ++H +L++RF
Sbjct: 1 MEEVYHDEPEKFNHILHLIRDYFNHRDDRARITACMGELMRDHLNLLVRF 50
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,761,346
Number of Sequences: 26719
Number of extensions: 108811
Number of successful extensions: 506
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 444
Number of HSP's gapped (non-prelim): 86
length of query: 134
length of database: 11,318,596
effective HSP length: 88
effective length of query: 46
effective length of database: 8,967,324
effective search space: 412496904
effective search space used: 412496904
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC135795.3


