

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135604.6 - phase: 0
(314 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
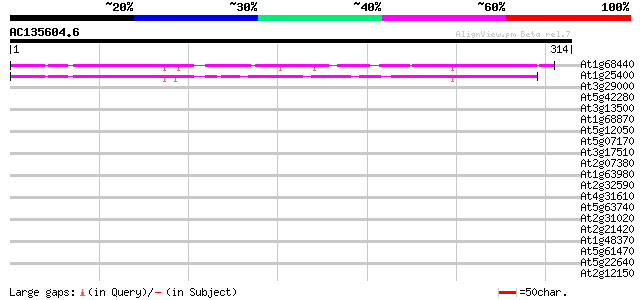
Score E
Sequences producing significant alignments: (bits) Value
At1g68440 unknown protein 157 9e-39
At1g25400 unknown protein 133 1e-31
At3g29000 unknown protein 36 0.024
At5g42280 putative protein 35 0.069
At3g13500 hypothetical protein 34 0.12
At1g68870 hypothetical protein 34 0.12
At5g12050 putative serine rich protein 33 0.15
At5g07170 putative protein 33 0.15
At3g17510 CBL-interacting protein kinase 1 (CIPK1) 33 0.15
At2g07380 Mutator-like transposase 33 0.15
At1g63980 unknown protein 33 0.15
At2g32590 hypothetical protein 33 0.20
At4g31610 reproductive meristem protein 1 (REM1) 33 0.26
At5g63740 unknown protein 32 0.34
At2g31020 putative oxysterol-binding protein 32 0.34
At2g21420 Mutator-like transposase 32 0.34
At1g48370 unknown protein 32 0.34
At5g61470 putative protein 32 0.45
At5g22640 unknown protein 32 0.45
At2g12150 pseudogene 32 0.45
>At1g68440 unknown protein
Length = 306
Score = 157 bits (396), Expect = 9e-39
Identities = 114/325 (35%), Positives = 179/325 (55%), Gaps = 41/325 (12%)
Query: 1 MQVPLIDRLNDFQAGFNSLQQNPSFPSQITATSFNGIQSVSIAFNFCKWGAVILALVATF 60
M+VP+I+R+ DF+ G NS+ +PSF S+ A S GI + A+ F KWGA+I+A +A F
Sbjct: 1 MEVPVINRIRDFEVGINSIN-DPSFLSRSVAVS--GIGKLHQAYGFWKWGALIIAFLAYF 57
Query: 61 TSLINKVTIFIIHLRKKASSLPSIT----FDNDDDFS------SDDEDNENDDIVSLSSS 110
T+ ++K+ ++ LRK S+ S T +D+D D S SDDE +E D+ +
Sbjct: 58 TNFVSKLNSLVVRLRKIDVSVSSPTLFDDYDSDSDVSCSSTVSSDDEKDEEDE------A 111
Query: 111 SEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVFQTQNS-----GHRRQHSIGDFFSLL 165
+ ++D S+ + FR+ G+ DY ++ Q N G R S GD FS
Sbjct: 112 DDEDEDVDSIFNRRRVNGGFRVRGS--DYYDDDDDQGDNGNCTWMGRRYSGSFGDLFSWP 169
Query: 166 ELAN--GDNVVKLWDSIGFGLGLDFDEYEDGVISSTNPHAPLTSAAASPDVIVSAGEGAH 223
+L VVKLWD L +D D++E+ V + + ++ +SP + +G
Sbjct: 170 DLGGIGSSGVVKLWDH----LDIDGDDHENVVATFLKNY----NSTSSPFFWAAEKKGVD 221
Query: 224 GNLAVEIWDTRLRRRKPSVVAEW---GPTVGNTLRVESGGVQKVYVRDNGRQRLTVGDMR 280
+ V+ D R R P+++AEW G +GN + V++GGV+KVYVRD+ + VGD+R
Sbjct: 222 A-VKVKACDPRAGFRMPALLAEWRQPGRLLGNIIGVDTGGVEKVYVRDDVSGEIAVGDLR 280
Query: 281 KVSFPLGNVTESDADNTWWDADAVI 305
K + L ++TE +A+ TWWDAD +I
Sbjct: 281 KFNGVLTDLTECEAE-TWWDADVLI 304
>At1g25400 unknown protein
Length = 288
Score = 133 bits (335), Expect = 1e-31
Identities = 105/306 (34%), Positives = 156/306 (50%), Gaps = 39/306 (12%)
Query: 1 MQVPLIDRLNDFQAGFNSLQQNPSFPSQITATSFNGIQSVSIAFNFCKWGAVIL-ALVAT 59
M+VP+I+R+ DF G NS+ +PS+ S+ A S G+ + A++F KWGA++L A A+
Sbjct: 1 MEVPIINRIGDFDMGINSIN-DPSYLSRALAVS--GVGKLHQAYSFWKWGALLLLAFFAS 57
Query: 60 FTSLINKVTIFIIHLRKKASSLPSIT----FDNDDD--FSSDDEDNENDDIVSLSSSSEF 113
FTSL ++ + LR SLPS T +D+D D FSSD D E D+
Sbjct: 58 FTSLTTRIKTLVFRLRNVNVSLPSQTLLCNYDSDSDWSFSSDSSDEEKDE------DDNK 111
Query: 114 EDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVFQTQNSGHRRQHSIGDFFSLLELANGDNV 173
EDD S++ S + F G +D D + + R S GD L V
Sbjct: 112 EDD--SVNGDSRVQRF----GYYHDDDDKGISGSVPWLRRCSGSFGDLLDL----GSSGV 161
Query: 174 VKLWDSIGF-GLGLDFDEYEDGVISSTNPHAPLTSAAASPDVIVSAGEGAHGNLAVEIWD 232
VKLWD++ F G G + S ++ L+SA V+++A + L V WD
Sbjct: 162 VKLWDNLDFNGEGSPVASF----FSKCGSYSLLSSA-----VLLAAEKKGSDGLEVSAWD 212
Query: 233 TRLRRRKPSVVAEW---GPTVGNTLRVESGGVQKVYVRDNGRQRLTVGDMRKVSFPLGNV 289
R+ P+++AEW G +G +RV+ G V K+YV D+ +TVGDMR V+ L +
Sbjct: 213 ARVGFGVPALLAEWKQPGRLLGKIIRVDVGDVDKIYVGDDVEGEITVGDMRMVNGALTEL 272
Query: 290 TESDAD 295
TES+ +
Sbjct: 273 TESEVE 278
>At3g29000 unknown protein
Length = 194
Score = 36.2 bits (82), Expect = 0.024
Identities = 25/118 (21%), Positives = 52/118 (43%), Gaps = 5/118 (4%)
Query: 45 NFCKWGAVILALVATFTSLINKVTIFIIHLRKKASSLPSITFDNDDDFSSDDEDNENDDI 104
+FC+W + ++ F L+ + + ++T DDD DD+D+++DD
Sbjct: 22 SFCRWVSSTRIFLSRFVPLLQHHQR-VFDKKNNKDQQETLTKQEDDDDDDDDDDDDDDDD 80
Query: 105 VSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVFQTQNSGHRRQHSIGDFF 162
+ +S E+ E M S F + +L + +V+++F+ + + D F
Sbjct: 81 IDISR----EEAEMVMRSLGLFYNDDQLQEQYSAKEVSSLFEEKEASLEEVKQAFDVF 134
>At5g42280 putative protein
Length = 694
Score = 34.7 bits (78), Expect = 0.069
Identities = 14/31 (45%), Positives = 20/31 (64%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDE 117
DNDDD DD+D++NDD V + +DD+
Sbjct: 161 DNDDDDDDDDDDDDNDDDVVDDDDDDDDDDD 191
Score = 31.2 bits (69), Expect = 0.76
Identities = 11/31 (35%), Positives = 20/31 (64%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDE 117
++DDD DD+D+ +DD+V + +DD+
Sbjct: 162 NDDDDDDDDDDDDNDDDVVDDDDDDDDDDDD 192
>At3g13500 hypothetical protein
Length = 110
Score = 33.9 bits (76), Expect = 0.12
Identities = 17/50 (34%), Positives = 30/50 (60%), Gaps = 6/50 (12%)
Query: 86 FDNDDDF-----SSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFF 130
+D+DDD+ DD+D ++DD+V+ +S + D+ SSS F D++
Sbjct: 56 YDSDDDYDYVDDDDDDDDVDDDDVVAFDHNSNGDFDDHG-SSSLDFDDYY 104
>At1g68870 hypothetical protein
Length = 147
Score = 33.9 bits (76), Expect = 0.12
Identities = 22/63 (34%), Positives = 32/63 (49%), Gaps = 7/63 (11%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVFQ 146
D+DDD D +NE+DD ++ +SS +P S+ + NSN VNN Q
Sbjct: 59 DDDDDDDDDSSNNESDDSMTSDASSWPSTHQPPRSTKN-----HAAAKNSNAKQVNN--Q 111
Query: 147 TQN 149
T+N
Sbjct: 112 TEN 114
>At5g12050 putative serine rich protein
Length = 362
Score = 33.5 bits (75), Expect = 0.15
Identities = 14/27 (51%), Positives = 20/27 (73%)
Query: 90 DDFSSDDEDNENDDIVSLSSSSEFEDD 116
+D+ DDED+++DD+ S SSS FE D
Sbjct: 309 EDYEDDDEDDDDDDVASDSSSDLFELD 335
>At5g07170 putative protein
Length = 542
Score = 33.5 bits (75), Expect = 0.15
Identities = 14/31 (45%), Positives = 21/31 (67%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDE 117
D+DDD DD+D+++DD S SE E++E
Sbjct: 124 DDDDDDDDDDDDDDDDDDDDESKDSEVEEEE 154
Score = 29.3 bits (64), Expect = 2.9
Identities = 12/37 (32%), Positives = 19/37 (50%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSS 123
D DDD DD+D+++DD + +DD+ S
Sbjct: 112 DTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDESKDS 148
Score = 28.9 bits (63), Expect = 3.8
Identities = 12/42 (28%), Positives = 22/42 (51%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKD 128
D+ DD DD+D+++DD + +DD+ S S ++
Sbjct: 111 DDTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDESKDSEVEE 152
Score = 27.7 bits (60), Expect = 8.4
Identities = 15/59 (25%), Positives = 26/59 (43%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVF 145
D+DDD DD+D++++ S E +DD D F + +D++ F
Sbjct: 129 DDDDDDDDDDDDDDDESKDSEVEEEEGDDDLRMRKIDPETMDIFAEDFSLEKFDIDFEF 187
>At3g17510 CBL-interacting protein kinase 1 (CIPK1)
Length = 444
Score = 33.5 bits (75), Expect = 0.15
Identities = 23/78 (29%), Positives = 39/78 (49%), Gaps = 8/78 (10%)
Query: 77 KASSLPSITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNS 136
K +PSI D+D++ E + +DD S+ E +E S S + + F+L G S
Sbjct: 276 KLEYIPSIPDDDDEE-----EVDTDDDAFSIQ---ELGSEEGKGSDSPTIINAFQLIGMS 327
Query: 137 NDYDVNNVFQTQNSGHRR 154
+ D++ F+ +N RR
Sbjct: 328 SFLDLSGFFEQENVSERR 345
>At2g07380 Mutator-like transposase
Length = 408
Score = 33.5 bits (75), Expect = 0.15
Identities = 18/55 (32%), Positives = 27/55 (48%)
Query: 84 ITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSND 138
+ +D +D + DDE E++ V +S E D+E SS F F G S+D
Sbjct: 85 VCYDYQEDEAGDDEAGEDESGVDEASDKEASDEEEDHEFSSRFDFFDNSDGASSD 139
>At1g63980 unknown protein
Length = 391
Score = 33.5 bits (75), Expect = 0.15
Identities = 15/41 (36%), Positives = 22/41 (53%)
Query: 85 TFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSS 125
+FDN DD DD+D++ +D SE +DD+ SS
Sbjct: 262 SFDNSDDDDDDDDDDDEEDEEEDEDESEADDDDKDSVIESS 302
Score = 30.4 bits (67), Expect = 1.3
Identities = 17/48 (35%), Positives = 25/48 (51%), Gaps = 4/48 (8%)
Query: 77 KASSLPSITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSS 124
K +S + D+DDD D+ED E D+ S +DD+ S+ SS
Sbjct: 259 KKTSFDNSDDDDDDDDDDDEEDEEEDE----DESEADDDDKDSVIESS 302
>At2g32590 hypothetical protein
Length = 704
Score = 33.1 bits (74), Expect = 0.20
Identities = 45/197 (22%), Positives = 80/197 (39%), Gaps = 30/197 (15%)
Query: 15 GFNSLQQNPSFPSQITATSFNGIQSVSIAFNFCKWGAVILA-----------LVATFTSL 63
G L + P ++ +++ +S +I +F K G +LA L F+ +
Sbjct: 238 GCQVLFDSQEIPGKLVSSANKHDKSETIDLSFVK-GLALLADMPRSTIVHVLLHYAFSDV 296
Query: 64 INK----VTIFIIHLRKKASSLPSIT-----FDND-----DDFSSDDEDNENDDIVSLSS 109
+ K V ++++RKK +PS+ FD + D FS + E+ DI +
Sbjct: 297 VVKFVECVEQMVLNMRKKDEIVPSLRAIINQFDEENQRPSDTFSCGQQTTESFDISHGND 356
Query: 110 SSEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVFQTQNSGHRRQHSIGDFFSLLELAN 169
+S +DDE +F F G S D D N + H + SL ++ +
Sbjct: 357 ASYADDDE----GYENFGTSFDYEGQSGDVDENFGPNEAEPIYSNFHEEVEPASLQDMDS 412
Query: 170 GDNVVKLWDSIGFGLGL 186
D + + D + LG+
Sbjct: 413 DDRLENVDDYLFLSLGI 429
>At4g31610 reproductive meristem protein 1 (REM1)
Length = 517
Score = 32.7 bits (73), Expect = 0.26
Identities = 29/95 (30%), Positives = 44/95 (45%), Gaps = 16/95 (16%)
Query: 76 KKASSLPSITFD--------NDDDFSSDDEDNENDDIV---SLSSSSEFEDDEPSMSSSS 124
+ ASS IT D +DDD+ DDED+++DD ++ + SE D SSS
Sbjct: 98 RNASSSQVITEDTYIDVDDVDDDDYGQDDEDDDDDDDEGEDNIENISEKTDKRQEADSSS 157
Query: 125 SFKDFF--RLTGNS---NDYDVNNVFQTQNSGHRR 154
F R+T S + D++ F GH++
Sbjct: 158 DHSGFITARVTRYSLLHDRLDLSRNFTLLFGGHQK 192
>At5g63740 unknown protein
Length = 226
Score = 32.3 bits (72), Expect = 0.34
Identities = 22/86 (25%), Positives = 37/86 (42%), Gaps = 15/86 (17%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSNDYDVNNVFQ 146
D+DDD +DD D++ DD E +DD+ + D +YD + +
Sbjct: 96 DDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDENDEECDD---------EYDSHRLIS 146
Query: 147 TQNSGHR------RQHSIGDFFSLLE 166
T H+ R++ +F+SL E
Sbjct: 147 TPYCTHKFCKTCWREYLETNFYSLEE 172
>At2g31020 putative oxysterol-binding protein
Length = 760
Score = 32.3 bits (72), Expect = 0.34
Identities = 34/126 (26%), Positives = 57/126 (44%), Gaps = 16/126 (12%)
Query: 28 QITATSFNGIQSVSIAFNFCKWGAVILALVATFTSLINKVTIFIIHLRKKAS---SLPSI 84
QIT + F+ IQS + +W +I L T ++ + +++A S +I
Sbjct: 246 QITRSEFSAIQSQLLLLKQKQW-LLIDTLRQLETEKVDLENTVVDETQRQAGNGDSEETI 304
Query: 85 TFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSNDYD-VNN 143
+ +DD+ D+ + E D SLSS SS S FR + S+D D + N
Sbjct: 305 SESDDDNEQFDEAEEEMDTCDSLSS-----------SSFKSIGSVFRTSSFSSDDDGLTN 353
Query: 144 VFQTQN 149
F+++N
Sbjct: 354 GFESEN 359
>At2g21420 Mutator-like transposase
Length = 468
Score = 32.3 bits (72), Expect = 0.34
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 3/52 (5%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKDFFRLTGNSND 138
D+DDD DD+D+++DD + EDDE S D + G+ ND
Sbjct: 391 DDDDDDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYIDSDDDD---VDGDDND 439
Score = 28.9 bits (63), Expect = 3.8
Identities = 15/47 (31%), Positives = 25/47 (52%), Gaps = 4/47 (8%)
Query: 87 DNDDDFSSDDEDNENDDIVSLSSSSEFED----DEPSMSSSSSFKDF 129
D+DDD DD+D +++D + S + D D+ S +F+DF
Sbjct: 406 DDDDDDDDDDDDEDDEDDGYIDSDDDDVDGDDNDDGSELDGINFEDF 452
>At1g48370 unknown protein
Length = 724
Score = 32.3 bits (72), Expect = 0.34
Identities = 22/76 (28%), Positives = 35/76 (45%), Gaps = 24/76 (31%)
Query: 25 FPSQITATSFNGIQSVSI-----------AFNFCKWGAVILALVATFTSLINKVTIFIIH 73
FPS + ++S NG+Q+ + +NFCK L+ TF+ LI+++
Sbjct: 332 FPSNVDSSSMNGLQAYKVFIAVATILGDGLYNFCK------VLIRTFSGLISQI------ 379
Query: 74 LRKKASSLPSITFDND 89
R KA S S+ D
Sbjct: 380 -RGKAGSRSSLAHKED 394
>At5g61470 putative protein
Length = 304
Score = 32.0 bits (71), Expect = 0.45
Identities = 14/28 (50%), Positives = 21/28 (75%), Gaps = 2/28 (7%)
Query: 87 DNDDDF--SSDDEDNENDDIVSLSSSSE 112
D+DD++ +DEDNE+DD+ SL+S E
Sbjct: 168 DSDDEYVVEDEDEDNEDDDVKSLTSDVE 195
>At5g22640 unknown protein
Length = 871
Score = 32.0 bits (71), Expect = 0.45
Identities = 17/43 (39%), Positives = 25/43 (57%), Gaps = 3/43 (6%)
Query: 87 DNDDDFSSDDEDNENDDIV---SLSSSSEFEDDEPSMSSSSSF 126
D DDD DD+D+++DD + S S+ + + P SSS SF
Sbjct: 686 DEDDDDGDDDDDDDDDDDLGPSSFGSADKGRRNSPFSSSSLSF 728
>At2g12150 pseudogene
Length = 942
Score = 32.0 bits (71), Expect = 0.45
Identities = 21/83 (25%), Positives = 32/83 (38%), Gaps = 6/83 (7%)
Query: 84 ITFDNDDDFSSDDEDNENDDIVSLSSSSEFEDDEPSMSSSSSFKD--FFRLTGNSNDYDV 141
I D +DD DED D+ + DD+ S S D G+S +YD
Sbjct: 134 INVDEEDDIDEGDEDEHEDEDNEEDEDMDVHDDDTDGESRGSTLDPTVDFSEGSSEEYDY 193
Query: 142 NN----VFQTQNSGHRRQHSIGD 160
N + + +GH + + D
Sbjct: 194 NKWGDMIVEQYGNGHVEEGVVND 216
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,469,239
Number of Sequences: 26719
Number of extensions: 336810
Number of successful extensions: 3241
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 62
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 2487
Number of HSP's gapped (non-prelim): 394
length of query: 314
length of database: 11,318,596
effective HSP length: 99
effective length of query: 215
effective length of database: 8,673,415
effective search space: 1864784225
effective search space used: 1864784225
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135604.6


