

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135566.8 - phase: 0
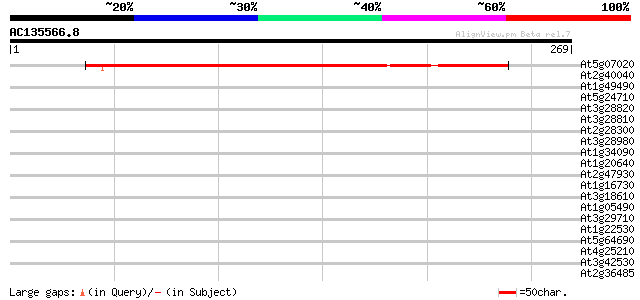
(269 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g07020 unknown protein 245 2e-65
At2g40040 unknown protein 39 0.004
At1g49490 hypothetical protein 37 0.008
At5g24710 unknown protein 36 0.025
At3g28820 unknown protein 36 0.025
At3g28810 hypothetical protein 36 0.025
At2g28300 unknown protein 36 0.025
At3g28980 unknown protein 34 0.072
At1g34090 putative protein 34 0.072
At1g20640 unknown protein 34 0.094
At2g47930 predicted GPI-anchored protein 33 0.12
At1g16730 hypothetical protein 33 0.12
At3g18610 unknown protein 33 0.16
At1g05490 hypothetical protein 33 0.16
At3g29710 hypothetical protein 33 0.21
At1g22530 unknown protein 33 0.21
At5g64690 unknown protein 32 0.27
At4g25210 unknown protein 32 0.27
At3g42530 putative protein 32 0.36
At2g36485 unknown protein 32 0.36
>At5g07020 unknown protein
Length = 235
Score = 245 bits (625), Expect = 2e-65
Identities = 129/204 (63%), Positives = 157/204 (76%), Gaps = 5/204 (2%)
Query: 37 IVRASSE-SDCNDEECAPDKEVGKVSMEWLAEEKTKVVGTYPPRRKQGWTGYVEKDTAGQ 95
+VRASS+ +DCN EECAP+KEVG VSMEWLA EKTKVVGT+PPR+ +GWTGYVEKDTAGQ
Sbjct: 36 VVRASSDDADCNAEECAPEKEVGTVSMEWLAGEKTKVVGTFPPRKPRGWTGYVEKDTAGQ 95
Query: 96 TNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPP 155
TN+YS+EPAVYVAESAISSGTAG+S+DGA+NTAAI AG+ALI++AAASSILLQVGK++P
Sbjct: 96 TNVYSIEPAVYVAESAISSGTAGSSADGAENTAAIVAGIALIAVAAASSILLQVGKDAPT 155
Query: 156 QVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSES 215
+ + V+YSGPSLSYYIN+FK EI Q PS + TE E+ +P E S Q E+
Sbjct: 156 RPKAVDYSGPSLSYYINKFKPSEIVQ-PSTPSVTEAPPVAELETSLP---ETPSVAQQET 211
Query: 216 IAPAKTELSSSIQTESVATDASIS 239
P + + SV T +S S
Sbjct: 212 SLPETMASEAQPEASSVPTTSSTS 235
>At2g40040 unknown protein
Length = 839
Score = 38.5 bits (88), Expect = 0.004
Identities = 23/68 (33%), Positives = 39/68 (56%), Gaps = 1/68 (1%)
Query: 197 TESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVES 256
T+S P+QT S Q+++ +P++T+ S Q++S + S S S ++ SQ Q +S
Sbjct: 752 TQSQSPSQTRAQSPSQAQAQSPSQTQSQSQSQSQSQSQSQSQSQSQSQSQSQ-SQSQSQS 810
Query: 257 QIQTETSS 264
QT+T S
Sbjct: 811 PSQTQTQS 818
Score = 38.1 bits (87), Expect = 0.005
Identities = 27/113 (23%), Positives = 57/113 (49%), Gaps = 5/113 (4%)
Query: 153 SPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVS-----TETELSSSVPTESDVPTQTEL 207
+PP ++ + S+ ++F+TQ +Q+PS + ++ + S T+S +Q++
Sbjct: 727 TPPGEEQSQPPNQSIGNGGDDFQTQTQSQSPSQTRAQSPSQAQAQSPSQTQSQSQSQSQS 786
Query: 208 SSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQT 260
S QS+S + ++++ S Q++S + + S + A+ Q SQ QT
Sbjct: 787 QSQSQSQSQSQSQSQSQSQSQSQSPSQTQTQSPSQTQAQAQSPSSQSPSQTQT 839
>At1g49490 hypothetical protein
Length = 847
Score = 37.4 bits (85), Expect = 0.008
Identities = 26/85 (30%), Positives = 42/85 (48%), Gaps = 9/85 (10%)
Query: 180 TQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASIS 239
TQAP+ S+ET + VPT S Q+++ S VQ+ + + T S Q + ++ S
Sbjct: 656 TQAPTPSSET---TQVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQVPTPSSSESYQ 712
Query: 240 VPPESSPAEVSQVQVESQIQTETSS 264
P +S VQ + +Q T+S
Sbjct: 713 AP------NLSPVQAPTPVQAPTTS 731
Score = 35.0 bits (79), Expect = 0.042
Identities = 27/74 (36%), Positives = 41/74 (54%), Gaps = 6/74 (8%)
Query: 177 QEITQAPSVSTE---TELSSSVPTESDVPTQTELSSSVQSESI-APAKTELS-SSIQTES 231
Q +QAP+ E +S P +S P+ +SS QSE + AP T ++ SS+ + S
Sbjct: 746 QSPSQAPTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEAPEPTPVNPSSVPSSS 805
Query: 232 VATDASISVPPESS 245
+TD SI PPE++
Sbjct: 806 PSTDTSIP-PPENN 818
Score = 32.0 bits (71), Expect = 0.36
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 1/73 (1%)
Query: 176 TQEITQAPSVSTETELS-SSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVAT 234
+ E +Q P+ S+E+ S S PT P +S +S P+ +SS Q+E V
Sbjct: 731 SSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQSEEVEA 790
Query: 235 DASISVPPESSPA 247
V P S P+
Sbjct: 791 PEPTPVNPSSVPS 803
Score = 31.6 bits (70), Expect = 0.47
Identities = 39/150 (26%), Positives = 60/150 (40%), Gaps = 18/150 (12%)
Query: 116 TAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFK 175
T + SD +Q + + A + S + SS QV P Y P+LS +
Sbjct: 670 TPSSESDQSQILSPVQAPTPVQS-STPSSEPTQV----PTPSSSESYQAPNLS----PVQ 720
Query: 176 TQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATD 235
QAP+ S+ET S VPT S Q+ + A T S +Q+ + +++
Sbjct: 721 APTPVQAPTTSSET---SQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSE 777
Query: 236 ASISVPPESSPAEVSQVQVESQIQTETSSV 265
P SSP + +V+ SSV
Sbjct: 778 ------PVSSPEQSEEVEAPEPTPVNPSSV 801
>At5g24710 unknown protein
Length = 1003
Score = 35.8 bits (81), Expect = 0.025
Identities = 37/143 (25%), Positives = 60/143 (41%), Gaps = 16/143 (11%)
Query: 103 PAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEY 162
P A + + TA + A TAA+A A + AA + + PP V+K E
Sbjct: 860 PETAAAAESPAPETAAVAESPAPGTAAVAEAPASETAAAPVDGPVTETVSEPPPVEKEET 919
Query: 163 SGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESI--APAK 220
S E PS + TE ++S S T E ++ E I AP +
Sbjct: 920 S-------------LEEKSDPSSTPNTETATSTENTSQTTTTPESVTTAPPEPITTAPPE 966
Query: 221 TELSSSIQ-TESVATDASISVPP 242
T +++++ TE+ AT+ ++ P
Sbjct: 967 TVTTTAVKPTENAATERRVTNYP 989
>At3g28820 unknown protein
Length = 434
Score = 35.8 bits (81), Expect = 0.025
Identities = 23/88 (26%), Positives = 42/88 (47%)
Query: 177 QEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDA 236
+E + S+E SS +ES ++E S+ +S++ A ++E SSS +T A
Sbjct: 196 EESSVGKEASSENSKSSGKESESSAKGESETSAKGESKTSAKGESETSSSKSAGGSSTSA 255
Query: 237 SISVPPESSPAEVSQVQVESQIQTETSS 264
+ S + V+ QVE + + S+
Sbjct: 256 TKEESSASQSSGVTVTQVEEETSKDVST 283
>At3g28810 hypothetical protein
Length = 434
Score = 35.8 bits (81), Expect = 0.025
Identities = 23/88 (26%), Positives = 42/88 (47%)
Query: 177 QEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDA 236
+E + S+E SS +ES ++E S+ +S++ A ++E SSS +T A
Sbjct: 196 EESSVGKEASSENSKSSGKESESSAKGESETSAKGESKTSAKGESETSSSKSAGGSSTSA 255
Query: 237 SISVPPESSPAEVSQVQVESQIQTETSS 264
+ S + V+ QVE + + S+
Sbjct: 256 TKEESSASQSSGVTVTQVEEETSKDVST 283
>At2g28300 unknown protein
Length = 2218
Score = 35.8 bits (81), Expect = 0.025
Identities = 26/140 (18%), Positives = 58/140 (40%), Gaps = 11/140 (7%)
Query: 139 IAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTE 198
+ A L+ G +S PQV + P +++ +T++ V T SS+P
Sbjct: 1862 VKTAGCELVSTGCSSEPQVHLPPSAEPDGDIHVHLKETEKSESMVVVGEGTAFPSSLPVT 1921
Query: 199 SDVPTQTELSSSVQSESIAPAKTELSSSIQTESV-----------ATDASISVPPESSPA 247
+ +++L+ + S + + Q E+ +++ + +PP + P
Sbjct: 1922 EEGNAESQLADTEPFTSPTVVEKNIKDQEQVETTGCGLVDDSTGCSSEPQVQLPPSAEPM 1981
Query: 248 EVSQVQVESQIQTETSSVDI 267
E + + +E ++ET +I
Sbjct: 1982 EGTHMHLEETKKSETVVTEI 2001
Score = 33.1 bits (74), Expect = 0.16
Identities = 45/178 (25%), Positives = 74/178 (41%), Gaps = 30/178 (16%)
Query: 100 SVEPAVYV---AESAISSGTAGTSS---DGAQNTAAIAAGLALISIAAASSILLQVGKNS 153
S EP V + AE ++ GT SS G N++ + G + A + L V +S
Sbjct: 1716 STEPQVQLPPSAEPVVAEGTEFPSSLLMTGVDNSSHLMTG-----VDNAKTHLADVVPSS 1770
Query: 154 PPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQS 213
P + K E V+T V TE Q +L S +
Sbjct: 1771 SPTTME---------------KNIEAQDQDQVTTGGCGLVDVLTECSSEPQLQLPPS--A 1813
Query: 214 ESIAPAKTELSSSIQTESVATDASI-SVPPESSPAEVSQ-VQVESQIQTETSSVDIVS 269
E + TEL++ TE D+ + ++ P SSP+ V + ++ + Q Q +T+ ++VS
Sbjct: 1814 EPVISEGTELATLPLTEEENADSQLANIEPSSSPSVVEKNIEAQDQDQVKTAGCELVS 1871
>At3g28980 unknown protein
Length = 438
Score = 34.3 bits (77), Expect = 0.072
Identities = 25/89 (28%), Positives = 42/89 (47%), Gaps = 5/89 (5%)
Query: 176 TQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATD 235
T + + S S + SSV ES+ + E +S ++ES AK E +S + ES
Sbjct: 174 TNTMVSSTSTSASSSEESSVGKESETSAKGESETSAKNESKTSAKGESETSAKGES---- 229
Query: 236 ASISVPPESSPAEVSQVQVESQIQTETSS 264
S ES + + + ++ ++ETSS
Sbjct: 230 -KTSAKGESETSAKGESETSAKGESETSS 257
Score = 32.3 bits (72), Expect = 0.27
Identities = 28/92 (30%), Positives = 41/92 (44%), Gaps = 8/92 (8%)
Query: 180 TQAPSVSTETELS------SSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVA 233
++ SV E+E S +S ES + E +S + ES AK E +S + ES
Sbjct: 188 SEESSVGKESETSAKGESETSAKNESKTSAKGESETSAKGESKTSAKGESETSAKGESET 247
Query: 234 TDASISVPPESSPAEVSQVQ--VESQIQTETS 263
+ S S AE SQ +Q++ ETS
Sbjct: 248 SAKGESETSSSKTAEASQSSSLTVTQVEEETS 279
>At1g34090 putative protein
Length = 761
Score = 34.3 bits (77), Expect = 0.072
Identities = 50/199 (25%), Positives = 78/199 (39%), Gaps = 29/199 (14%)
Query: 64 WLAEEKTKVVGTYPPRRKQGWTGYVEKDTAGQTNIYSVE--PAVYVAESAISSGTAGTSS 121
WL GT P +R Q + EK + T +++ + P + V+ A+ +G S
Sbjct: 4 WLQSLDLPTKGTRPLQRDQDYDSNTEKVS---TTLFTPDDFPPLPVSSRAVVIPESGEQS 60
Query: 122 DGAQNTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQ 181
+ G SI S+ +SPP + + S K++ T
Sbjct: 61 ------SECTTGFDPASIGNNST-------SSPPGTPTATQASSTSSE-----KSKTPTS 102
Query: 182 APSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVP 241
PS T + SS TES T+T +SS V ++ A + + T S +T SV
Sbjct: 103 IPSTPTTPQRSS---TES---TETPMSSPVITQPSPSASSIPTMHAATSSASTSQQSSVA 156
Query: 242 PESSPAEVSQVQVESQIQT 260
S +V Q+Q S T
Sbjct: 157 SNKSTTDVVQIQEASPKST 175
Score = 30.4 bits (67), Expect = 1.0
Identities = 24/88 (27%), Positives = 43/88 (48%), Gaps = 5/88 (5%)
Query: 183 PSVSTETELSSSVPTESDVPTQTELSSSV-QSESIAPAKTELSSSIQTESVATDASI-SV 240
P T T+ SS+ +S PT + + Q S +T +SS + T+ + +SI ++
Sbjct: 81 PGTPTATQASSTSSEKSKTPTSIPSTPTTPQRSSTESTETPMSSPVITQPSPSASSIPTM 140
Query: 241 PPESSPAEVSQVQVESQIQTETSSVDIV 268
+S A SQ +S + + S+ D+V
Sbjct: 141 HAATSSASTSQ---QSSVASNKSTTDVV 165
>At1g20640 unknown protein
Length = 844
Score = 33.9 bits (76), Expect = 0.094
Identities = 28/111 (25%), Positives = 46/111 (41%)
Query: 157 VQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELSSSVQSESI 216
VQ V P S+Y N +Q PS +T P + + S S S
Sbjct: 620 VQGVSGPLPIGSFYANFPNLVSQSQEPSQQAKTTPPPPPPVQLAKSPVSSYSHSSNSSQC 679
Query: 217 APAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVDI 267
++T+L+S T+ +TD ++ SS E+ ++ I T +S +I
Sbjct: 680 CSSETQLNSGATTDPPSTDVGGALKKTSSEIELQSSSLDETILTLSSLENI 730
>At2g47930 predicted GPI-anchored protein
Length = 136
Score = 33.5 bits (75), Expect = 0.12
Identities = 30/100 (30%), Positives = 42/100 (42%), Gaps = 11/100 (11%)
Query: 172 NEFKTQEITQAPSVSTE--TELSSSVPTES-------DVPTQTELSSSVQSESIAPAKTE 222
+EF+ I+ APS E + S+S P S P +E+S S SI P
Sbjct: 21 SEFQLSTISAAPSFLPEAPSSFSASTPAMSPDTSPLFPTPGSSEMSPSPSESSIMPTIPS 80
Query: 223 LSSSIQTESVATDASISVPPESS--PAEVSQVQVESQIQT 260
S ++V D + V P S PA S V SQ+ +
Sbjct: 81 SLSPPNPDAVTPDPLLEVSPVGSPLPASSSVCLVSSQLSS 120
>At1g16730 hypothetical protein
Length = 202
Score = 33.5 bits (75), Expect = 0.12
Identities = 32/138 (23%), Positives = 64/138 (46%), Gaps = 5/138 (3%)
Query: 91 DTAGQTNIYSVEPAVYVAESAISSGTAGTSSDGAQNTAAIAAGLALISIAAASSILLQVG 150
+ GQ N+YS + A+ +A + S ++ +G + I GL ++ A +L ++
Sbjct: 41 ENVGQGNLYSPDHAIKIATNPDSVDSSSEEEEGEKQRKTIMLGLKNLA-AVKDKVLEKLS 99
Query: 151 KNSPPQVQKVEYSGPSLSYYINEF--KTQEITQAPSVSTETELSSSVPTESDVPTQTELS 208
S P + +E + L I +F Q +T+ +T+L+ VP+ S T+ ++
Sbjct: 100 AASVPS-ESLENAKQFLEGAIKDFAGAAQGMTKDALHRIKTQLAVIVPSVSPAVTE-KIV 157
Query: 209 SSVQSESIAPAKTELSSS 226
+ E+I + + S S
Sbjct: 158 DDAEKEAIDGEEEDESKS 175
>At3g18610 unknown protein
Length = 636
Score = 33.1 bits (74), Expect = 0.16
Identities = 32/110 (29%), Positives = 50/110 (45%), Gaps = 10/110 (9%)
Query: 158 QKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPTQTELS-SSVQSESI 216
+KVE S S E KT+E ++ + SS E D + E++ + + E I
Sbjct: 60 KKVESSSSDASDSDEEEKTKE------TPSKLKDESSSEEEDDSSSDEEIAPAKKRPEPI 113
Query: 217 APAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETSSVD 266
AK E SSS + +T + P + PA + + +VES + SS D
Sbjct: 114 KKAKVESSSS---DDDSTSDEETAPVKKQPAVLEKAKVESSSSDDDSSSD 160
Score = 30.0 bits (66), Expect = 1.4
Identities = 22/84 (26%), Positives = 39/84 (46%), Gaps = 5/84 (5%)
Query: 183 PSVSTETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVPP 242
P + E SSS D T E ++ V+ + K ++ SS + ++D +VP
Sbjct: 112 PIKKAKVESSSS----DDDSTSDEETAPVKKQPAVLEKAKVESSSSDDDSSSDEE-TVPV 166
Query: 243 ESSPAEVSQVQVESQIQTETSSVD 266
+ PA + + ++ES + SS D
Sbjct: 167 KKQPAVLEKAKIESSSSDDDSSSD 190
Score = 27.7 bits (60), Expect = 6.7
Identities = 42/199 (21%), Positives = 75/199 (37%), Gaps = 31/199 (15%)
Query: 67 EEKTKVVGTYPPRRKQGWTGYVEKDTAGQTNIYSVEPAVY-VAESAISSGTAGTSSDGAQ 125
EEKTK P + K + E D++ I + + ++ + S ++ S +
Sbjct: 75 EEKTKET---PSKLKDESSSEEEDDSSSDEEIAPAKKRPEPIKKAKVESSSSDDDSTSDE 131
Query: 126 NTAAIAAGLALISIAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSV 185
TA + A++ A KVE S +E +T + + P+V
Sbjct: 132 ETAPVKKQPAVLEKA------------------KVESSSSDDDSSSDE-ETVPVKKQPAV 172
Query: 186 STETELSSSVPTESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESS 245
+ ++ SS + + + Q+ + AK E SSS D S E +
Sbjct: 173 LEKAKIESSSSDDDSSSDEETVPMKKQTAVLEKAKAESSSS--------DDGSSSDEEPT 224
Query: 246 PAEVSQVQVESQIQTETSS 264
PA+ + V+ E+SS
Sbjct: 225 PAKKEPIVVKKDSSDESSS 243
>At1g05490 hypothetical protein
Length = 731
Score = 33.1 bits (74), Expect = 0.16
Identities = 26/126 (20%), Positives = 57/126 (44%), Gaps = 8/126 (6%)
Query: 144 SILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTESDVPT 203
S+ + GK + + E P +N + I++ +TE + VP E P+
Sbjct: 435 SVSINSGKTTGAP-SRPEVENPETGKELNTPEKPSISRPEIFTTEKAIDVQVPEE---PS 490
Query: 204 QTELSSSVQSESIA----PAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQ 259
+ E+ SS +++ + P++ E+ SS + + + S+P + + +VQ +++
Sbjct: 491 RPEIYSSEKAKEVQAPEMPSRPEVFSSEKAKEIQVPEMPSIPEIQNSEKAKEVQANNRMG 550
Query: 260 TETSSV 265
T +V
Sbjct: 551 LTTPAV 556
Score = 27.7 bits (60), Expect = 6.7
Identities = 20/72 (27%), Positives = 34/72 (46%), Gaps = 1/72 (1%)
Query: 198 ESDVPTQTELSSSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQ 257
E VP + E+ S +SE+ A + + ES SIS E S +E + + E+Q
Sbjct: 204 EKSVPGEIEILSDSESETEARRRASAKKKLFEESSRIVESIS-DGEDSSSETDEEEEENQ 262
Query: 258 IQTETSSVDIVS 269
+ ++ D V+
Sbjct: 263 DSEDNNTKDNVT 274
>At3g29710 hypothetical protein
Length = 669
Score = 32.7 bits (73), Expect = 0.21
Identities = 24/90 (26%), Positives = 44/90 (48%), Gaps = 4/90 (4%)
Query: 178 EITQAPSVSTETELSSSVPTESDVPTQ---TELSSSVQSESIAPAKTELSSSIQTES-VA 233
E ++ E+E VP E PTQ T ++++ + AP T +++ TES +
Sbjct: 49 EESEGSGAVEESEDKEKVPAEEQSPTQTTATAMATNAAPTTAAPTTTAPTTAPTTESPML 108
Query: 234 TDASISVPPESSPAEVSQVQVESQIQTETS 263
D++ + PAE Q +++ +TET+
Sbjct: 109 DDSTFYDALKHIPAEEIQENMQTDEETETN 138
>At1g22530 unknown protein
Length = 683
Score = 32.7 bits (73), Expect = 0.21
Identities = 22/84 (26%), Positives = 36/84 (42%), Gaps = 1/84 (1%)
Query: 139 IAAASSILLQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPSVSTETELSSSVPTE 198
+AA + I+ + K P + P+ + E K +EI A V+TET++ V
Sbjct: 175 LAAPAPIVAETKKEETPVAPAPVETKPAAPV-VAETKKEEILPAAPVTTETKVEEKVVPV 233
Query: 199 SDVPTQTELSSSVQSESIAPAKTE 222
P + + + E AP TE
Sbjct: 234 ETTPAAPVTTETKEEEKAAPVTTE 257
>At5g64690 unknown protein
Length = 344
Score = 32.3 bits (72), Expect = 0.27
Identities = 27/95 (28%), Positives = 43/95 (44%), Gaps = 8/95 (8%)
Query: 175 KTQEITQAPSVSTETELSSSVPTESDVPTQTELSS-----SVQSESIAPAKTELSSSIQT 229
K E ++ P VS V ESDVP E + V+++ + A++EL +
Sbjct: 189 KVAEESKVPEVSAPELSEIKVTKESDVPEVLEDKNIPEVVEVKTDEVKVAESELLKVSEV 248
Query: 230 ESVATDASISVPP--ESSPAEVSQVQVESQIQTET 262
E+ D I VP E+ E S V+V + + +T
Sbjct: 249 EA-TEDLEIKVPKVFEAKTPETSNVKVTGEAEVKT 282
>At4g25210 unknown protein
Length = 368
Score = 32.3 bits (72), Expect = 0.27
Identities = 32/117 (27%), Positives = 55/117 (46%), Gaps = 27/117 (23%)
Query: 175 KTQEITQAPSVSTE----------TELSSSVP----------TESDVPTQTELSSSVQSE 214
K +E+ ++P VS+E +E S+ VP ++S+ +++E SS + E
Sbjct: 5 KAEEVVESPPVSSEEEESGSSGEESESSAEVPKKVESSQKPESDSEGESESESSSGPEPE 64
Query: 215 SIAPAKTELSSSIQTESV-ATDASISVPPESSPA-----EVSQVQVESQIQTETSSV 265
S PAKT + T+ + T S + PESS A E + ++ Q ++T V
Sbjct: 65 S-EPAKTIKLKPVGTKPIPETSGSAATVPESSTAKRPLKEAAPEAIKKQKTSDTEHV 120
>At3g42530 putative protein
Length = 889
Score = 32.0 bits (71), Expect = 0.36
Identities = 32/128 (25%), Positives = 56/128 (43%), Gaps = 18/128 (14%)
Query: 147 LQVGKNSPPQVQKVEYSGPSLSYYINEFKTQEITQAPS---VSTE--TELSSSVPTESDV 201
+QV +S P V+KV P +I++ ++ + PS +S E TE S E DV
Sbjct: 546 MQVDPSSNPSVEKVL---PLNQDHISDDASERVPAIPSGLDLSKEHNTEEQESNANEEDV 602
Query: 202 PTQTELS--SSVQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQ 259
+Q ++ S E + P + + + DAS VP S ++ + ++Q
Sbjct: 603 DSQMKVDPRSDPSVEKLLP--------LHQDHIIADASERVPATHSGLDLPKEHNSEELQ 654
Query: 260 TETSSVDI 267
T + D+
Sbjct: 655 TNANETDV 662
>At2g36485 unknown protein
Length = 158
Score = 32.0 bits (71), Expect = 0.36
Identities = 16/53 (30%), Positives = 27/53 (50%)
Query: 211 VQSESIAPAKTELSSSIQTESVATDASISVPPESSPAEVSQVQVESQIQTETS 263
+ ESI P + + ++ T +++VPP SS VS + ES I ++ S
Sbjct: 22 LSEESIRPVNSNARQFLSQRTLGTATAVTVPPASSRFRVSGRETESSIVSDPS 74
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.303 0.118 0.308
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,192,104
Number of Sequences: 26719
Number of extensions: 199839
Number of successful extensions: 771
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 53
Number of HSP's that attempted gapping in prelim test: 718
Number of HSP's gapped (non-prelim): 94
length of query: 269
length of database: 11,318,596
effective HSP length: 98
effective length of query: 171
effective length of database: 8,700,134
effective search space: 1487722914
effective search space used: 1487722914
T: 11
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135566.8


