

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135565.15 + phase: 0
(557 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
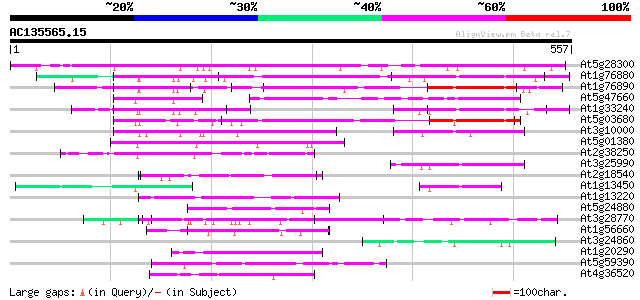
Score E
Sequences producing significant alignments: (bits) Value
At5g28300 GTL1 - like protein 399 e-111
At1g76880 unknown protein 226 2e-59
At1g76890 unknown protein 181 7e-46
At5g47660 unknown protein 126 4e-29
At1g33240 DNA-binding factor, putative 115 6e-26
At5g03680 GT2 -like protein 112 5e-25
At3g10000 hypothetical protein 77 3e-14
At5g01380 transcription factor GT-3a 67 2e-11
At2g38250 putative GT-1-like transcription factor 62 1e-09
At3g25990 putative DNA-binding protein, GT-1 59 5e-09
At2g18540 putative vicilin storage protein (globulin-like) 53 4e-07
At1g13450 hypothetical protein 51 1e-06
At1g13220 putative nuclear matrix constituent protein 48 1e-05
At5g24880 glutamic acid-rich protein 46 6e-05
At3g28770 hypothetical protein 46 6e-05
At1g56660 hypothetical protein 45 8e-05
At3g24860 unknown protein 44 3e-04
At1g20290 hypothetical protein 43 4e-04
At5g59390 transcriptional regulator - like protein 42 7e-04
At4g36520 trichohyalin like protein 42 9e-04
>At5g28300 GTL1 - like protein
Length = 619
Score = 399 bits (1024), Expect = e-111
Identities = 263/643 (40%), Positives = 355/643 (54%), Gaps = 124/643 (19%)
Query: 1 MYDG-VPDQFHQFITPRTSSSSLPLH-------LPFPLSTPNNTFPPFDPYNQQNHPSQH 52
M+DG VP+Q H+FI LP H LPFP+S F +N + P
Sbjct: 1 MFDGGVPEQIHRFIASPPPPPPLPPHQPAAERSLPFPVS--------FSSFNTNHQPQ-- 50
Query: 53 HQLPLQVQPNLLHPLHPHKDDEDKEQNSTPSMNNFQIDRDQRQILPQLID--PWTNDEVL 110
H L L + ++H H H + K+ +T Q D D Q PW +DEVL
Sbjct: 51 HMLSLDSR-KIIHHHHHHHHHDIKDGGATTGEWIGQTDHDDSDNHHQHHHHHPWCSDEVL 109
Query: 111 ALLKIRSSMESWFPDFTWEHVSRKLAEVGYKRSAEKCKEKFEEESR-FFNNINHNQNSF- 168
ALL+ RS++E+WFP+FTWEH SRKLAEVG+KRS ++CKEKFEEE R +FN+ N+N N+
Sbjct: 110 ALLRFRSTVENWFPEFTWEHTSRKLAEVGFKRSPQECKEKFEEEERRYFNSNNNNNNNTN 169
Query: 169 -----------GKNFRFVTELEEVYQGG----------GGENNK--NLVEAE-------K 198
G N+R +E+EE Y G G NK NLVE + +
Sbjct: 170 DHQHIGNYNNKGNNYRIFSEVEEFYHHGHDNEHVSSEVGDNQNKRTNLVEGKGNVGETVQ 229
Query: 199 QNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTND---------------EKKRKR- 242
+DK+ ++ + + ++++ EV + G D EKKRK+
Sbjct: 230 DLMAEDKLRDQDQGQVEEASMENQRNSIEVGKVGNVEDDAKSSSSSSLMMIMKEKKRKKR 289
Query: 243 -SGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNK 301
+RF V KGFCE +V+ M+ QQEEMH KL+EDMVK++EEK +REEAWKKQE+E++NK
Sbjct: 290 KKEKERFGVLKGFCEGLVRNMIAQQEEMHKKLLEDMVKKEEEKIAREEAWKKQEIERVNK 349
Query: 302 ELELMAHEQAIAGDRQAHIIQFLNKF------------STSANSSSLTSMSTQLQAYLAT 349
E+E+ A EQA+A DR +II+F++KF S S +SSSL TQ + T
Sbjct: 350 EVEIRAQEQAMASDRNTNIIKFISKFTDHDLDVVQNPTSPSQDSSSLALRKTQGRRKFQT 409
Query: 350 LTSNSSSSTLHSQNPNPE---TLKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPISSYSL 406
SS+L Q P T+ K+L+P +T +P+N NP
Sbjct: 410 ------SSSLLPQTLTPHNLLTIDKSLEPF------------STKTLKPKNQNPKPP--- 448
Query: 407 ISSGERDDIGRRWPKDEVLALINLRCN----NNNEEKEGNS----NNKAPLWERISQGML 458
S ++ D+G+RWPKDEVLALIN+R + N+++ K+ NS + PLWERIS+ ML
Sbjct: 449 -KSDDKSDLGKRWPKDEVLALINIRRSISNMNDDDHKDENSLSTSSKAVPLWERISKKML 507
Query: 459 ELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQ---GKLVLQ 515
E+GYKRSAKRCKEKWENINKYFRKTKD N+KR LDSRTCPYFH LT LY+Q G
Sbjct: 508 EIGYKRSAKRCKEKWENINKYFRKTKDVNKKRPLDSRTCPYFHQLTALYSQPPTGTTATT 567
Query: 516 SDQKQESNNVNV-PEENVVQEK-----AKQDENQDGAGESSQV 552
+ + +++ PEEN V + + DGAG+ S V
Sbjct: 568 ATTATSARDLDTRPEENRVGSQDPDISVPMHVDGDGAGDKSNV 610
>At1g76880 unknown protein
Length = 603
Score = 226 bits (577), Expect = 2e-59
Identities = 161/506 (31%), Positives = 234/506 (45%), Gaps = 84/506 (16%)
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGYKRSAEKCKEKFEEESRFF 158
W E LALLKIRS M F D + WE VSRK+AE GY R+A+KCKEKFE ++
Sbjct: 62 WPRQETLALLKIRSDMGIAFRDASVKGPLWEEVSRKMAEHGYIRNAKKCKEKFENVYKYH 121
Query: 159 NNINHNQN--SFGKNFRFVTELEEV-------------------YQGGGGENNKNL---- 193
+ S GK +RF +LE + Q NN N
Sbjct: 122 KRTKEGRTGKSEGKTYRFFDQLEALESQSTTSLHHHQQQTPLRPQQNNNNNNNNNNNSSI 181
Query: 194 ------VEAEKQNEVQDKMDPHEE----------------DSRMDDVLVSKKSEEEVVEK 231
V + P+ + D+ S + +
Sbjct: 182 FSTPPPVTTVMPTLPSSSIPPYTQQINVPSFPNISGDFLSDNSTSSSSSYSTSSDMEMGG 241
Query: 232 GTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAW 291
GT KKRKR +K F E ++K+++D+QEE+ K +E + KR+ E+ REE+W
Sbjct: 242 GTATTRKKRKRK-------WKVFFERLMKQVVDKQEELQRKFLEAVEKREHERLVREESW 294
Query: 292 KKQEMEKMNKELELMAHEQAIAGDRQAHIIQFLNKFSTSA-NSSSLTSMSTQLQAYLATL 350
+ QE+ ++N+E E++A E++++ + A ++ FL K S N Q++ +
Sbjct: 295 RVQEIARINREHEILAQERSMSAAKDAAVMAFLQKLSEKQPNQPQPQPQPQQVRPSMQLN 354
Query: 351 TSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPISSYSLISSG 410
+N S P P P+P+ + + + TT + N + S SS
Sbjct: 355 NNNQQQPPQRSPPPQPPA------PLPQPIQAVVSTLDTTKTDNGGDQNMTPAASASSS- 407
Query: 411 ERDDIGRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCK 470
RWPK E+ ALI LR N +++ +E K PLWE IS GM LG+ R++KRCK
Sbjct: 408 -------RWPKVEIEALIKLRTNLDSKYQENGP--KGPLWEEISAGMRRLGFNRNSKRCK 458
Query: 471 EKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSDQKQESNNVNVPEE 530
EKWENINKYF+K K++N+KR DS+TCPYFH L LY + K SNN N+
Sbjct: 459 EKWENINKYFKKVKESNKKRPEDSKTCPYFHQLDALYRE-------RNKFHSNN-NIAAS 510
Query: 531 NVVQEKAKQDENQDGAGESSQVGPPS 556
+ K D + + Q PP+
Sbjct: 511 SSSSGLVKPDNSVPLMVQPEQQWPPA 536
Score = 80.5 bits (197), Expect = 2e-15
Identities = 53/171 (30%), Positives = 86/171 (49%), Gaps = 22/171 (12%)
Query: 380 PSSTLPSSSTTLVAQP---RNNNPISSYSLISSG-----------ERDDIGRRWPKDEVL 425
P++T +++ T P NNN ++ ++ +R G RWP+ E L
Sbjct: 9 PTTTAAATTVTTATAPPPQSNNNDSAATEAAAAAVGAFEVSEEMHDRGFGGNRWPRQETL 68
Query: 426 ALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKD 485
AL+ +R + ++ + K PLWE +S+ M E GY R+AK+CKEK+EN+ KY ++TK+
Sbjct: 69 ALLKIRSDMGIAFRDASV--KGPLWEEVSRKMAEHGYIRNAKKCKEKFENVYKYHKRTKE 126
Query: 486 ANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSDQKQ-----ESNNVNVPEEN 531
+S + +T +F L L +Q L Q+Q + NN N N
Sbjct: 127 GRTGKS-EGKTYRFFDQLEALESQSTTSLHHHQQQTPLRPQQNNNNNNNNN 176
Score = 52.8 bits (125), Expect = 5e-07
Identities = 43/196 (21%), Positives = 80/196 (39%), Gaps = 28/196 (14%)
Query: 27 PFPLSTPNNTFPPFD-PYNQQNHPSQHHQLPLQVQP------NLLHPLHPHKDDEDKEQN 79
P P P P N Q P Q P P ++ L K D +QN
Sbjct: 338 PQPQPQPQQVRPSMQLNNNNQQQPPQRSPPPQPPAPLPQPIQAVVSTLDTTKTDNGGDQN 397
Query: 80 STPSMNNFQIDRDQRQILPQLIDPWTNDEVLALLKIRSSMESWFPDF-----TWEHVSRK 134
TP+ + W E+ AL+K+R++++S + + WE +S
Sbjct: 398 MTPAASASS-------------SRWPKVEIEALIKLRTNLDSKYQENGPKGPLWEEISAG 444
Query: 135 LAEVGYKRSAEKCKEKFEEESRFFNNI---NHNQNSFGKNFRFVTELEEVYQGGGGENNK 191
+ +G+ R++++CKEK+E +++F + N + K + +L+ +Y+ ++
Sbjct: 445 MRRLGFNRNSKRCKEKWENINKYFKKVKESNKKRPEDSKTCPYFHQLDALYRERNKFHSN 504
Query: 192 NLVEAEKQNEVQDKMD 207
N + A + K D
Sbjct: 505 NNIAASSSSSGLVKPD 520
>At1g76890 unknown protein
Length = 575
Score = 181 bits (460), Expect = 7e-46
Identities = 124/358 (34%), Positives = 177/358 (48%), Gaps = 74/358 (20%)
Query: 221 SKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKR 280
S S+EE + KKRK +KG + K++M++QE+M + +E + R
Sbjct: 239 STASDEEEDHHQVKSSRKKRK--------YWKGLFTKLTKELMEKQEKMQKRFLETLEYR 290
Query: 281 DEEKFSREEAWKKQEMEKMNKELELMAHEQAIAGDRQAHIIQFLNKFSTSA--NSSSLTS 338
++E+ SREEAW+ QE+ ++N+E E + HE++ A + A II FL+K S
Sbjct: 291 EKERISREEAWRVQEIGRINREHETLIHERSNAAAKDAAIISFLHKISGGQPQQPQQHNH 350
Query: 339 MSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIP-ENPSSTLPSSSTTLVAQPRN 397
+Q + Y + S T S+ P L T++ +N S PSSS
Sbjct: 351 KPSQRKQY-----QSDHSITFESKEPRAVLLDTTIKMGNYDNNHSVSPSSS--------- 396
Query: 398 NNPISSYSLISSGERDDIGRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGM 457
RWPK EV ALI +R N E + K PLWE IS GM
Sbjct: 397 --------------------RWPKTEVEALIRIR--KNLEANYQENGTKGPLWEEISAGM 434
Query: 458 LELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYN--------- 508
LGY RSAKRCKEKWENINKYF+K K++N+KR LDS+TCPYFH L LYN
Sbjct: 435 RRLGYNRSAKRCKEKWENINKYFKKVKESNKKRPLDSKTCPYFHQLEALYNERNKSGAMP 494
Query: 509 ---------QGKLVL--------QSDQKQESNNVNVPEENVVQEKAKQDENQDGAGES 549
Q +L+L ++DQ+++ + EE E+ + DE ++G G++
Sbjct: 495 LPLPLMVTPQRQLLLSQETQTEFETDQREKVGD-KEDEEEGESEEDEYDEEEEGEGDN 551
Score = 84.0 bits (206), Expect = 2e-16
Identities = 41/88 (46%), Positives = 60/88 (67%), Gaps = 3/88 (3%)
Query: 416 GRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWEN 475
G RWP+ E LAL+ +R + ++ S KAPLWE IS+ M+ELGYKRS+K+CKEK+EN
Sbjct: 39 GNRWPRPETLALLRIRSEMDKAFRD--STLKAPLWEEISRKMMELGYKRSSKKCKEKFEN 96
Query: 476 INKYFRKTKDANRKRSLDSRTCPYFHLL 503
+ KY ++TK+ +S + +T +F L
Sbjct: 97 VYKYHKRTKEGRTGKS-EGKTYRFFEEL 123
Score = 73.9 bits (180), Expect = 2e-13
Identities = 39/83 (46%), Positives = 50/83 (59%), Gaps = 7/83 (8%)
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGYKRSAEKCKEKFEEESRFF 158
W E LALL+IRS M+ F D T WE +SRK+ E+GYKRS++KCKEKFE ++
Sbjct: 42 WPRPETLALLRIRSEMDKAFRDSTLKAPLWEEISRKMMELGYKRSSKKCKEKFENVYKYH 101
Query: 159 NNINHNQ--NSFGKNFRFVTELE 179
+ S GK +RF ELE
Sbjct: 102 KRTKEGRTGKSEGKTYRFFEELE 124
Score = 58.2 bits (139), Expect = 1e-08
Identities = 57/228 (25%), Positives = 98/228 (42%), Gaps = 31/228 (13%)
Query: 45 QQNHPSQHHQLPLQ-VQPNLLHPL-HPHKDDEDKEQNSTPSMNNFQIDRDQRQILPQLID 102
Q P QH+ P Q Q H + K+ ++T M N+ D +
Sbjct: 341 QPQQPQQHNHKPSQRKQYQSDHSITFESKEPRAVLLDTTIKMGNY----DNNHSVSPSSS 396
Query: 103 PWTNDEVLALLKIRSSMESWFPDF-----TWEHVSRKLAEVGYKRSAEKCKEKFEEESRF 157
W EV AL++IR ++E+ + + WE +S + +GY RSA++CKEK+E +++
Sbjct: 397 RWPKTEVEALIRIRKNLEANYQENGTKGPLWEEISAGMRRLGYNRSAKRCKEKWENINKY 456
Query: 158 FNNI---NHNQNSFGKNFRFVTELEEVYQGGGGENNKN---------LVEAEKQNEV-QD 204
F + N + K + +LE +Y E NK+ +V ++Q + Q+
Sbjct: 457 FKKVKESNKKRPLDSKTCPYFHQLEALY----NERNKSGAMPLPLPLMVTPQRQLLLSQE 512
Query: 205 KMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFK 252
E D R V K +EE E +++ + GD+ F+
Sbjct: 513 TQTEFETDQREK---VGDKEDEEEGESEEDEYDEEEEGEGDNETSEFE 557
Score = 39.7 bits (91), Expect = 0.004
Identities = 61/266 (22%), Positives = 103/266 (37%), Gaps = 44/266 (16%)
Query: 247 RFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRD----EEKFSREEAWKKQEMEKMNKE 302
R E+ K F +S +K + EE+ K++E KR +EKF + K+ E +
Sbjct: 54 RSEMDKAFRDSTLKAPL--WEEISRKMMELGYKRSSKKCKEKFENVYKYHKRTKEGRTGK 111
Query: 303 LELMAHEQAIAGDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQ 362
E + + + + + + SS + + L S+S+ ST S
Sbjct: 112 SEGKTYRFFEELEAFETLSSYQPEPESQPAKSSAVITNAPATSSLIPWISSSNPSTEKSS 171
Query: 363 NPNPETLKKTLQPIPEN-------PSSTLP-----SSSTTLVAQPRNNNP----ISSYSL 406
+P + ++QPI N PSST P S++TT V+QP +N +SS +L
Sbjct: 172 SPLKHHHQVSVQPITTNPTFLAKQPSSTTPFPFYSSNNTTTVSQPPISNDLMNNVSSLNL 231
Query: 407 ISSGERDDIGRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSA 466
SS DE + + +S K W+ + + +
Sbjct: 232 FSSSTSSSTA----SDE----------EEDHHQVKSSRKKRKYWKGL--------FTKLT 269
Query: 467 KRCKEKWENINKYFRKTKDANRKRSL 492
K EK E + K F +T + K +
Sbjct: 270 KELMEKQEKMQKRFLETLEYREKERI 295
>At5g47660 unknown protein
Length = 398
Score = 126 bits (316), Expect = 4e-29
Identities = 93/269 (34%), Positives = 128/269 (47%), Gaps = 49/269 (18%)
Query: 239 KRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEK 298
KRKR + E F E +V MM +QE+MHN+LI M K + E+ REEAW++QE E+
Sbjct: 169 KRKRETRVKLE---HFLEKLVGSMMKRQEKMHNQLINVMEKMEVERIRREEAWRQQETER 225
Query: 299 MNKELELMAHEQAIAGDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSST 358
M + E E A R +I F+ ++T +
Sbjct: 226 MTQNEEARKQEMA----RNLSLISFIR-----------------------SVTGDEIEIP 258
Query: 359 LHSQNPNPETLKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPISSYSLISSGERDDIGRR 418
+ P P Q +PE S+ R YS S GRR
Sbjct: 259 KQCEFPQP-----LQQILPEQCKDEKCESAQ------REREIKFRYSSGSGSS----GRR 303
Query: 419 WPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINK 478
WP++EV ALI+ R ++ EEK G NK +W+ IS M E GY+RSAK+CKEKWEN+NK
Sbjct: 304 WPQEEVQALISSR--SDVEEKTGI--NKGAIWDEISARMKERGYERSAKKCKEKWENMNK 359
Query: 479 YFRKTKDANRKRSLDSRTCPYFHLLTNLY 507
Y+R+ + +K+ S+T YF L N Y
Sbjct: 360 YYRRVTEGGQKQPEHSKTRSYFEKLGNFY 388
Score = 49.7 bits (117), Expect = 4e-06
Identities = 29/95 (30%), Positives = 47/95 (48%), Gaps = 7/95 (7%)
Query: 104 WTNDEVLALLKIRSSMESWF---PDFTWEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNN 160
W +EV AL+ RS +E W+ +S ++ E GY+RSA+KCKEK+E ++++
Sbjct: 304 WPQEEVQALISSRSDVEEKTGINKGAIWDEISARMKERGYERSAKKCKEKWENMNKYYRR 363
Query: 161 I---NHNQNSFGKNFRFVTELEEVYQG-GGGENNK 191
+ Q K + +L Y+ GE K
Sbjct: 364 VTEGGQKQPEHSKTRSYFEKLGNFYKTISSGEREK 398
>At1g33240 DNA-binding factor, putative
Length = 338
Score = 115 bits (288), Expect = 6e-26
Identities = 69/170 (40%), Positives = 94/170 (54%), Gaps = 20/170 (11%)
Query: 382 STLPSSSTTLVAQPRNNNPISSYS------LISSGERDDI--GRRWPKDEVLALINLRCN 433
S L + +TT + P + P ++ S E+ + RWPK E+LALINLR
Sbjct: 142 SQLEALNTTPPSHPHAHQPEQKQQQQPQQEMVMSSEQSSLPSSSRWPKAEILALINLR-- 199
Query: 434 NNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLD 493
+ E + ++ K LWE IS M +GY R+AKRCKEKWENINKY++K K++N+KR D
Sbjct: 200 SGMEPRYQDNVPKGLLWEEISTSMKRMGYNRNAKRCKEKWENINKYYKKVKESNKKRPQD 259
Query: 494 SRTCPYFHLLTNLYNQGKL---------VLQSDQKQES-NNVNVPEENVV 533
++TCPYFH L LY L L DQKQ + P+E +V
Sbjct: 260 AKTCPYFHRLDLLYRNKVLGSGGGSSTSGLPQDQKQSPVTAMKPPQEGLV 309
Score = 86.3 bits (212), Expect = 4e-17
Identities = 48/141 (34%), Positives = 77/141 (54%), Gaps = 17/141 (12%)
Query: 416 GRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWEN 475
G RWP++E LAL+ +R + ++ ++ KAPLWE +S+ +LELGYKRS+K+CKEK+EN
Sbjct: 60 GNRWPREETLALLRIRSDMDSTFRDATL--KAPLWEHVSRKLLELGYKRSSKKCKEKFEN 117
Query: 476 INKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSDQKQESNNVNVPEENVVQE 535
+ KY+++TK+ R D + +F L L + + P + ++
Sbjct: 118 VQKYYKRTKETRGGRH-DGKAYKFFSQLEAL--------------NTTPPSHPHAHQPEQ 162
Query: 536 KAKQDENQDGAGESSQVGPPS 556
K +Q Q+ S Q PS
Sbjct: 163 KQQQQPQQEMVMSSEQSSLPS 183
Score = 84.3 bits (207), Expect = 2e-16
Identities = 46/119 (38%), Positives = 68/119 (56%), Gaps = 7/119 (5%)
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGYKRSAEKCKEKFEEESRFF 158
W +E LALL+IRS M+S F D T WEHVSRKL E+GYKRS++KCKEKFE +++
Sbjct: 63 WPREETLALLRIRSDMDSTFRDATLKAPLWEHVSRKLLELGYKRSSKKCKEKFENVQKYY 122
Query: 159 NNINHNQNS--FGKNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRM 215
+ GK ++F ++LE + + + E ++Q + Q +M E S +
Sbjct: 123 KRTKETRGGRHDGKAYKFFSQLEALNTTPPSHPHAHQPEQKQQQQPQQEMVMSSEQSSL 181
Score = 78.2 bits (191), Expect = 1e-14
Identities = 52/192 (27%), Positives = 87/192 (45%), Gaps = 17/192 (8%)
Query: 62 NLLHPLHPHKDDEDKEQNSTPSMNNFQIDRDQRQILPQLIDPWTNDEVLALLKIRSSMES 121
N P HPH +++Q P + ++ LP W E+LAL+ +RS ME
Sbjct: 148 NTTPPSHPHAHQPEQKQQQQPQQE--MVMSSEQSSLPSS-SRWPKAEILALINLRSGMEP 204
Query: 122 WFPD-----FTWEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNI---NHNQNSFGKNFR 173
+ D WE +S + +GY R+A++CKEK+E ++++ + N + K
Sbjct: 205 RYQDNVPKGLLWEEISTSMKRMGYNRNAKRCKEKWENINKYYKKVKESNKKRPQDAKTCP 264
Query: 174 FVTELEEVYQ-----GGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKS-EEE 227
+ L+ +Y+ GGG + L + +KQ+ V P E + S + EEE
Sbjct: 265 YFHRLDLLYRNKVLGSGGGSSTSGLPQDQKQSPVTAMKPPQEGLVNVQQTHGSASTEEEE 324
Query: 228 VVEKGTTNDEKK 239
+E+ EKK
Sbjct: 325 PIEESPQGTEKK 336
>At5g03680 GT2 -like protein
Length = 591
Score = 112 bits (280), Expect = 5e-25
Identities = 108/437 (24%), Positives = 193/437 (43%), Gaps = 88/437 (20%)
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEV-GYKRSAEKCKEKFEEESRF 157
W E L LL+IRS ++ F + W+ VSR ++E GY+RS +KC+EKFE ++
Sbjct: 120 WPRQETLTLLEIRSRLDHKFKEANQKGPLWDEVSRIMSEEHGYQRSGKKCREKFENLYKY 179
Query: 158 FNNINHNQ--NSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQN-----------EVQD 204
+ + GK++RF +LE +Y G++N NLV N Q+
Sbjct: 180 YRKTKEGKAGRQDGKHYRFFRQLEALY----GDSN-NLVSCPNHNTQFMSSALHGFHTQN 234
Query: 205 KMDPHEEDSRMDDV-----------LVSKKSEEEV-----VEKGTTNDEKKRKRSGDDRF 248
M+ S + +V L + + E+ +G + +++KRS +
Sbjct: 235 PMNVTTTTSNIHNVDSVHGFHQSLSLSNNYNSSELELMTSSSEGNDSSSRRKKRSWKAK- 293
Query: 249 EVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAH 308
K F ++ +K+++++Q+ KL + + ++E++ +EE W+K E +++KE A
Sbjct: 294 --IKEFIDTNMKRLIERQDVWLEKLTKVIEDKEEQRMMKEEEWRKIEAARIDKEHLFWAK 351
Query: 309 EQAIAGDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPET 368
E+A R +I+ L + L S + T +N + +QN N
Sbjct: 352 ERARMEARDVAVIEALQYLTGKPLIKPLCSSPEE-----RTNGNNEIRNNSETQNENGSD 406
Query: 369 LKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPISSYSLISSGERDDIGRRWPKDEVLALI 428
T + SS W + E+L L+
Sbjct: 407 QTMTNNVCVKGSSSC-----------------------------------WGEQEILKLM 431
Query: 429 NLRCNNNNEEKE--GNSNNKAPLWERISQGMLELGY-KRSAKRCKEKWENINKYFRK-TK 484
+R + ++ +E G +++ LWE I+ +++LG+ +RSA CKEKWE I+ RK K
Sbjct: 432 EIRTSMDSTFQEILGGCSDEF-LWEEIAAKLIQLGFDQRSALLCKEKWEWISNGMRKEKK 490
Query: 485 DANRKRSLDSRTCPYFH 501
N+KR +S +C ++
Sbjct: 491 QINKKRKDNSSSCGVYY 507
Score = 75.5 bits (184), Expect = 7e-14
Identities = 39/91 (42%), Positives = 58/91 (62%), Gaps = 4/91 (4%)
Query: 418 RWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLEL-GYKRSAKRCKEKWENI 476
RWP+ E L L+ +R +++ KE +N K PLW+ +S+ M E GY+RS K+C+EK+EN+
Sbjct: 119 RWPRQETLTLLEIRSRLDHKFKE--ANQKGPLWDEVSRIMSEEHGYQRSGKKCREKFENL 176
Query: 477 NKYFRKTKDANRKRSLDSRTCPYFHLLTNLY 507
KY+RKTK+ R D + +F L LY
Sbjct: 177 YKYYRKTKEGKAGRQ-DGKHYRFFRQLEALY 206
Score = 48.1 bits (113), Expect = 1e-05
Identities = 35/116 (30%), Positives = 58/116 (49%), Gaps = 16/116 (13%)
Query: 104 WTNDEVLALLKIRSSMESWF--------PDFTWEHVSRKLAEVGY-KRSAEKCKEKFEEE 154
W E+L L++IR+SM+S F +F WE ++ KL ++G+ +RSA CKEK+E
Sbjct: 422 WGEQEILKLMEIRTSMDSTFQEILGGCSDEFLWEEIAAKLIQLGFDQRSALLCKEKWEWI 481
Query: 155 SRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHE 210
S N + + K + + VY N +N + +++ D DPH+
Sbjct: 482 S---NGMRKEKKQINKKRKDNSSSCGVYY---PRNEENPIYNNRESGYNDN-DPHQ 530
>At3g10000 hypothetical protein
Length = 496
Score = 76.6 bits (187), Expect = 3e-14
Identities = 49/152 (32%), Positives = 73/152 (47%), Gaps = 24/152 (15%)
Query: 382 STLPSSSTTLVAQPRNNNPISSYSLIS-----SGERDDIGR-RWPKDEVLALINLRCNNN 435
S LP L N+N I++ SG D G RWP+ E L L+ +R +
Sbjct: 46 SLLPRGIQGLTVAGNNSNTITTIQSGGCVGGFSGFTDGGGTGRWPRQETLMLLEVRSRLD 105
Query: 436 NEEKEGNSNNKAPLWERISQG----------------MLELGYKRSAKRCKEKWENINKY 479
++ KE +N K PLW+ +S+ LE GY RS K+C+EK+EN+ KY
Sbjct: 106 HKFKE--ANQKGPLWDEVSRSHFLTLIFSSFIFPKSPFLEHGYTRSGKKCREKFENLYKY 163
Query: 480 FRKTKDANRKRSLDSRTCPYFHLLTNLYNQGK 511
++KTK+ R D + +F L +Y + K
Sbjct: 164 YKKTKEGKSGRRQDGKNYRFFRQLEAIYGESK 195
Score = 71.2 bits (173), Expect = 1e-12
Identities = 59/263 (22%), Positives = 110/263 (41%), Gaps = 42/263 (15%)
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRK----------------LAEVGYKR 142
W E L LL++RS ++ F + W+ VSR E GY R
Sbjct: 89 WPRQETLMLLEVRSRLDHKFKEANQKGPLWDEVSRSHFLTLIFSSFIFPKSPFLEHGYTR 148
Query: 143 SAEKCKEKFEEESRFFNNINHNQNSF---GKNFRFVTELEEVYQGGGGE----NNK---- 191
S +KC+EKFE +++ ++ GKN+RF +LE +Y NN
Sbjct: 149 SGKKCREKFENLYKYYKKTKEGKSGRRQDGKNYRFFRQLEAIYGESKDSVSCYNNTQFIM 208
Query: 192 -NLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKS---------EEEVVEKGTTNDEKKRK 241
N + + + + PH ++ M + +S ++ ++ + K
Sbjct: 209 TNALHSNFRASNIHNIVPHHQNPLMTNTNTQSQSLSISNNFNSSSDLDLTSSSEGNETTK 268
Query: 242 RSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNK 301
R G E K F +++++++Q+ KL++ + ++ ++ REE W++ E E+++K
Sbjct: 269 REGMHWKEKIKEFIGVHMERLIEKQDFWLEKLMKIVEDKEHQRMLREEEWRRIEAERIDK 328
Query: 302 ELELMAHEQAIAGDRQAHIIQFL 324
E E+ R +I L
Sbjct: 329 ERSFWTKERERIEARDVAVINAL 351
>At5g01380 transcription factor GT-3a
Length = 323
Score = 67.4 bits (163), Expect = 2e-11
Identities = 56/267 (20%), Positives = 122/267 (44%), Gaps = 35/267 (13%)
Query: 101 IDPWTNDEVLALLKIRSSMESWFPD-----FTWEHVSRKLAEVGYKRSAEKCKEKFEE-E 154
I W+ +E LL IR ++ F + WE V+ K+A+ G+ RSAE+CK K++
Sbjct: 49 IPQWSIEETKELLAIREELDQTFMETKRNKLLWEVVAAKMADKGFVRSAEQCKSKWKNLV 108
Query: 155 SRFFNNINHNQNSFGKNFRFVTELEEVYQG------------GGGENNKNLVEAEKQNEV 202
+R+ ++ + F F E++ +++ + + + +E
Sbjct: 109 TRYKACETTEPDAIRQQFPFYNEIQSIFEARMQRMLWSEATEPSTSSKRKHHQFSSDDEE 168
Query: 203 QDKMDPHEE-DSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSG-----DDRFEVFKGFCE 256
++ +P+++ + + ++ ++K E EV+ T+ + +KR + G + E +
Sbjct: 169 EEVDEPNQDINEELLSLVETQKRETEVITTSTSTNPRKRAKKGKGVASGTKAETAGNTLK 228
Query: 257 SVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQ--EMEK---------MNKELEL 305
++++ M Q +M + + ++ E+ RE+ W+++ E+E+ M +E E
Sbjct: 229 DILEEFMRQTVKMEKEWRDAWEMKEIEREKREKEWRRRMAELEEERAATERRWMEREEER 288
Query: 306 MAHEQAIAGDRQAHIIQFLNKFSTSAN 332
E+A A R + I LN+ + N
Sbjct: 289 RLREEARAQKRDSLIDALLNRLNRDHN 315
Score = 40.0 bits (92), Expect = 0.003
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 5/80 (6%)
Query: 400 PISSYSLISSGERDDIGRR---WPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQG 456
P+S+ + + G G R W +E L+ +R + E N LWE ++
Sbjct: 30 PLSTTATMDPGGGGGGGERIPQWSIEETKELLAIREELDQTFMETKRNKL--LWEVVAAK 87
Query: 457 MLELGYKRSAKRCKEKWENI 476
M + G+ RSA++CK KW+N+
Sbjct: 88 MADKGFVRSAEQCKSKWKNL 107
>At2g38250 putative GT-1-like transcription factor
Length = 289
Score = 61.6 bits (148), Expect = 1e-09
Identities = 66/270 (24%), Positives = 121/270 (44%), Gaps = 46/270 (17%)
Query: 51 QHHQLPLQVQPNLLHPLHPHKDDEDKEQNSTPSMNNFQIDRDQRQILPQLIDPWTNDEVL 110
QHH L Q+Q H LH Q+ TP + + D+ PQ W+ +E
Sbjct: 5 QHHHLH-QLQYLNKHHLHT--------QSQTPEIASPVAVGDR---FPQ----WSVEETK 48
Query: 111 ALLKIRSSMESWFPD-----FTWEHVSRKLAEVGYKRSAEKCKEKFEE-ESRFFNNINHN 164
L+ IR ++ F + WE +S K+ + + RS E+CK K++ +RF
Sbjct: 49 ELIGIRGELDQTFMETKRNKLLWEVISNKMRDKSFPRSPEQCKCKWKNLVTRFKGCETME 108
Query: 165 QNSFGKNFRFVTELEEVY------------QGGGGENNKNLVEAEKQNEVQDKMDPHEED 212
+ + F F +++ ++ +GGGG + + E ++ EE+
Sbjct: 109 AETARQQFPFYDDMQNIFTTRMQRMLWAESEGGGGGTSGAARKREYSSD--------EEE 160
Query: 213 SRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNK 272
+++ LV ++ +++ N KKRK G + G E V+++ M Q M ++
Sbjct: 161 ENVNEELVDVSNDPKILNP-KKNIAKKRK-GGSNSSNSNNGVRE-VLEEFMRHQVRMESE 217
Query: 273 LIEDMVKRDEEKFSREEAWKKQEMEKMNKE 302
E R++E+ +EE W++ +ME++ KE
Sbjct: 218 WREGWEAREKERAEKEEEWRR-KMEELEKE 246
Score = 39.3 bits (90), Expect = 0.006
Identities = 26/92 (28%), Positives = 45/92 (48%), Gaps = 8/92 (8%)
Query: 418 RWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENIN 477
+W +E LI +R + E N LWE IS M + + RS ++CK KW+N+
Sbjct: 41 QWSVEETKELIGIRGELDQTFMETKRNKL--LWEVISNKMRDKSFPRSPEQCKCKWKNLV 98
Query: 478 KYFR--KTKDANRKRSLDSRTCPYFHLLTNLY 507
F+ +T +A R + P++ + N++
Sbjct: 99 TRFKGCETMEAETAR----QQFPFYDDMQNIF 126
>At3g25990 putative DNA-binding protein, GT-1
Length = 372
Score = 59.3 bits (142), Expect = 5e-09
Identities = 42/145 (28%), Positives = 66/145 (44%), Gaps = 18/145 (12%)
Query: 379 NPSSTLPSSSTTLVAQPRNNNPISSYSLI----SSGERDDI-------GRRWPKDEVLAL 427
NPS + ++ +N + + +I S GE +I W +DE L
Sbjct: 8 NPSRDI----NMMIGDVTSNGDLQPHQIILGESSGGEDHEIIKAPKKRAETWAQDETRTL 63
Query: 428 INLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDAN 487
I+LR +N SN LWE+IS+ M E G+ RS C +KW NI K F+K K
Sbjct: 64 ISLRREMDNLFNTSKSNKH--LWEQISKKMREKGFDRSPSMCTDKWRNILKEFKKAKQHE 121
Query: 488 RKRSLDSRT-CPYFHLLTNLYNQGK 511
K + T Y++ + +++ + K
Sbjct: 122 DKATSGGSTKMSYYNEIEDIFRERK 146
Score = 41.6 bits (96), Expect = 0.001
Identities = 20/91 (21%), Positives = 41/91 (44%), Gaps = 9/91 (9%)
Query: 102 DPWTNDEVLALLKIRSSMESWFPD-----FTWEHVSRKLAEVGYKRSAEKCKEKFEEESR 156
+ W DE L+ +R M++ F WE +S+K+ E G+ RS C +K+ +
Sbjct: 53 ETWAQDETRTLISLRREMDNLFNTSKSNKHLWEQISKKMREKGFDRSPSMCTDKWRNILK 112
Query: 157 FFNNINHNQNSF----GKNFRFVTELEEVYQ 183
F +++ + E+E++++
Sbjct: 113 EFKKAKQHEDKATSGGSTKMSYYNEIEDIFR 143
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 53.1 bits (126), Expect = 4e-07
Identities = 41/187 (21%), Positives = 80/187 (41%), Gaps = 5/187 (2%)
Query: 129 EHVSRKLAEVGYKRSAEKCKEKFEEESRF-----FNNINHNQNSFGKNFRFVTELEEVYQ 183
E + R+ E R E+ K + EEE++ + K EE +
Sbjct: 433 EEIERRRKEEEEARKREEAKRREEEEAKRREEEETERKKREEEEARKREEERKREEEEAK 492
Query: 184 GGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRS 243
E K EAE+ + +++ + EE ++ + +K EEV K E+KR+
Sbjct: 493 RREEERKKREEEAEQARKREEEREKEEEMAKKREEERQRKEREEVERKRREEQERKRREE 552
Query: 244 GDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKEL 303
+ E + E + K+ +++ + +E ++ ++E+ EE K++E E+ KE
Sbjct: 553 EARKREEERKREEEMAKRREQERQRKEREEVERKIREEQERKREEEMAKRREQERQKKER 612
Query: 304 ELMAHEQ 310
E M ++
Sbjct: 613 EEMERKK 619
Score = 47.4 bits (111), Expect = 2e-05
Identities = 44/176 (25%), Positives = 78/176 (44%), Gaps = 20/176 (11%)
Query: 131 VSRKLAEVGYKRSAEKCKE--KFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGE 188
+ R + V + S E K K ++ES F + + K R E+EE
Sbjct: 378 LDRDVVAVSFNLSNETIKGLLKAQKESVIFECASCAEGELSKLMR---EIEE-------R 427
Query: 189 NNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRF 248
+ E E++ + +++ EE R ++ +K+ EEE E+ +E+ RKR + +
Sbjct: 428 KRREEEEIERRRKEEEEARKREEAKRREEE-EAKRREEEETERKKREEEEARKREEERKR 486
Query: 249 EVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELE 304
E + K +++ + + E KR+EE+ EE KK+E E+ KE E
Sbjct: 487 EEEEA-------KRREEERKKREEEAEQARKREEEREKEEEMAKKREEERQRKERE 535
Score = 43.1 bits (100), Expect = 4e-04
Identities = 43/176 (24%), Positives = 75/176 (42%), Gaps = 20/176 (11%)
Query: 129 EHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGE 188
E ++ EV KR E+ +++ EEE+R + K E+ Q E
Sbjct: 528 ERQRKEREEVERKRREEQERKRREEEARKREEERKREEEMAKR------REQERQRKERE 581
Query: 189 NNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRF 248
+ + E++ + +++M E R KK EE+ K + +KR+
Sbjct: 582 EVERKIREEQERKREEEMAKRREQERQ------KKEREEMERKKREEEARKREE------ 629
Query: 249 EVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELE 304
E+ K E +K + E + E+ ++R+EE+ EEA K+ E E+ KE E
Sbjct: 630 EMAKIREEERQRKEREDVERKRRE--EEAMRREEERKREEEAAKRAEEERRKKEEE 683
>At1g13450 hypothetical protein
Length = 406
Score = 51.2 bits (121), Expect = 1e-06
Identities = 31/87 (35%), Positives = 42/87 (47%), Gaps = 8/87 (9%)
Query: 408 SSGERDDI------GRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELG 461
SSGE ++ W +DE +LI R + SN LWE+IS M E G
Sbjct: 70 SSGEDHEVKAPKKRAETWVQDETRSLIMFRRGMDGLFNTSKSNKH--LWEQISSKMREKG 127
Query: 462 YKRSAKRCKEKWENINKYFRKTKDANR 488
+ RS C +KW N+ K F+K K +R
Sbjct: 128 FDRSPTMCTDKWRNLLKEFKKAKHHDR 154
Score = 43.9 bits (102), Expect = 2e-04
Identities = 38/182 (20%), Positives = 65/182 (34%), Gaps = 26/182 (14%)
Query: 6 PDQFHQFITPRTSSSSLPLHLPFPLSTPNNTFPPFDPYNQQNHPSQHHQLPLQVQPNLLH 65
P F++ +S++S + + T N ++ NH + HH Q Q +L
Sbjct: 9 PTDFYKDDHHNSSTTSTTRDMMIDVLTTTNESVDLQSHHHHNHHN-HHLHQSQPQQQILL 67
Query: 66 PLHPHKDDEDKEQNSTPSMNNFQIDRDQRQILPQLIDPWTNDEVLALLKIRSSMESWF-- 123
+D E K + W DE +L+ R M+ F
Sbjct: 68 GESSGEDHEVKAPKKRA-------------------ETWVQDETRSLIMFRRGMDGLFNT 108
Query: 124 ---PDFTWEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQNSFGK-NFRFVTELE 179
WE +S K+ E G+ RS C +K+ + F H+ G + E+E
Sbjct: 109 SKSNKHLWEQISSKMREKGFDRSPTMCTDKWRNLLKEFKKAKHHDRGNGSAKMSYYKEIE 168
Query: 180 EV 181
++
Sbjct: 169 DI 170
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 48.1 bits (113), Expect = 1e-05
Identities = 40/199 (20%), Positives = 87/199 (43%), Gaps = 25/199 (12%)
Query: 129 EHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGE 188
E + R+ E+ + S EK +++ + ++ F+ +N + + + E E++ Q
Sbjct: 409 EELERQKVEIDH--SEEKLEKRNQAMNKKFDRVNEKEMDLEAKLKTIKEREKIIQA---- 462
Query: 189 NNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRF 248
+ + EKQ + DK + ++ + +EE++E+ + E K++
Sbjct: 463 -EEKRLSLEKQQLLSDKESLEDLQQEIEKIRAEMTKKEEMIEEECKSLEIKKEER----- 516
Query: 249 EVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAH 308
++ + Q E+ +++ + V EE S+E KQE E+ KE E++
Sbjct: 517 -----------EEYLRLQSELKSQIEKSRVH--EEFLSKEVENLKQEKERFEKEWEILDE 563
Query: 309 EQAIAGDRQAHIIQFLNKF 327
+QA+ + I + KF
Sbjct: 564 KQAVYNKERIRISEEKEKF 582
Score = 30.4 bits (67), Expect = 2.7
Identities = 35/216 (16%), Positives = 92/216 (42%), Gaps = 13/216 (6%)
Query: 141 KRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQN 200
K EK + E S+ N+ + F K + + E + VY ++ + E+
Sbjct: 527 KSQIEKSRVHEEFLSKEVENLKQEKERFEKEWEILDEKQAVYNKERIRISEEKEKFERFQ 586
Query: 201 EVQDKMDPHEEDS-------RMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKG 253
++ + EE + +DD+ + ++S E +E + ++K K + +
Sbjct: 587 LLEGERLKKEESALRVQIMQELDDIRLQRESFEANMEHERSALQEKVKLEQSKVIDDLEM 646
Query: 254 FCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAHEQAIA 313
++ ++ +++E+ L++ M + ++++ + Q+ + +N+E+E M +++
Sbjct: 647 MRRNLEIELQERKEQDEKDLLDRMAQFEDKRMAELSDINHQK-QALNREMEEMMSKRSAL 705
Query: 314 GDRQAHIIQFLNKFSTSA-----NSSSLTSMSTQLQ 344
I + +K + S L+++S L+
Sbjct: 706 QKESEEIAKHKDKLKEQQVEMHNDISELSTLSINLK 741
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 45.8 bits (107), Expect = 6e-05
Identities = 41/145 (28%), Positives = 72/145 (49%), Gaps = 12/145 (8%)
Query: 177 ELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTND 236
++EE + ++N ++E++ +V+ K+D +E ++D ++ E E VE+ T
Sbjct: 280 DIEEKTEEMKEQDNNQANKSEEEEDVKKKIDENETPEKVD----TESKEVESVEETTQEK 335
Query: 237 EKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREE----AWK 292
E++ K G +R E + E V + D Q+E + ++ VK DEEK +E K
Sbjct: 336 EEEVKEEGKERVEEEEKEKEKVKE---DDQKEKVEEEEKEKVKGDEEKEKVKEEESAEGK 392
Query: 293 KQEMEKMNKELELMAHEQAIAGDRQ 317
K+E+ K KE A+ IA Q
Sbjct: 393 KKEVVKGKKE-SPSAYNDVIASKMQ 416
Score = 30.8 bits (68), Expect = 2.1
Identities = 21/85 (24%), Positives = 40/85 (46%), Gaps = 5/85 (5%)
Query: 225 EEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQE-----EMHNKLIEDMVK 279
EE++++ +EK + D + K E VKK +D+ E + +K +E + +
Sbjct: 271 EEKLIKNEDDIEEKTEEMKEQDNNQANKSEEEEDVKKKIDENETPEKVDTESKEVESVEE 330
Query: 280 RDEEKFSREEAWKKQEMEKMNKELE 304
+EK + K+ +E+ KE E
Sbjct: 331 TTQEKEEEVKEEGKERVEEEEKEKE 355
Score = 28.9 bits (63), Expect = 7.9
Identities = 30/130 (23%), Positives = 53/130 (40%), Gaps = 7/130 (5%)
Query: 150 KFEEESRFFNNINHNQ-----NSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQD 204
K EEE I+ N+ ++ K V E + + E K VE E++ + +
Sbjct: 298 KSEEEEDVKKKIDENETPEKVDTESKEVESVEETTQEKEEEVKEEGKERVEEEEKEKEKV 357
Query: 205 KMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMD 264
K D +E ++ K EE+ K + E K+K + E + + + KM
Sbjct: 358 KEDDQKEKVEEEEKEKVKGDEEKEKVKEEESAEGKKKEVVKGKKESPSAYNDVIASKM-- 415
Query: 265 QQEEMHNKLI 274
Q+ NK++
Sbjct: 416 QENPRKNKVL 425
>At3g28770 hypothetical protein
Length = 2081
Score = 45.8 bits (107), Expect = 6e-05
Identities = 46/180 (25%), Positives = 82/180 (45%), Gaps = 10/180 (5%)
Query: 129 EHVSRKLA--EVGYKRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGG 186
E+V+ +L E K + + K +EE++ +++S KN R E EE
Sbjct: 965 EYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKN-REKKEYEEKKSKTK 1023
Query: 187 GENNKNLVEAEKQNEVQDKMDPHEEDSRMDDV----LVSKKSEEEVVEKGTTNDEKKRKR 242
E K + + Q++ +++ D E S+ + L +KK EEE EK + + K +K+
Sbjct: 1024 EEAKKE--KKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKK 1081
Query: 243 SGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKE 302
E K + KK + EE ++ E+ K+D EK + + KK+E + K+
Sbjct: 1082 EDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEE-DKKDMEKLEDQNSNKKKEDKNEKKK 1140
Score = 45.1 bits (105), Expect = 1e-04
Identities = 98/501 (19%), Positives = 196/501 (38%), Gaps = 73/501 (14%)
Query: 74 EDKEQNSTPSMNNFQIDR--DQRQILPQLIDPWTNDE--VLALLKIRSSMESWFPDFTWE 129
EDK+ + S + + D+ D +Q Q+ + D+ V A K + S E+
Sbjct: 707 EDKKLENKESQTDSKDDKSVDDKQEEAQIYGGESKDDKSVEAKGKKKESKENKKTKTNEN 766
Query: 130 HVSRKLAEV-GYKRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGE 188
V K V G K+ +EK ++ ++ES+ ++ N K +E + G +
Sbjct: 767 RVRNKEENVQGNKKESEKVEKGEKKESKDAKSVETKDN---KKLSSTENRDEAKERSGED 823
Query: 189 NNKNL--------VEAEKQNE-------VQDKMDPHE-EDSRMDDVLVSK-----KSEEE 227
N ++ VEA+++NE V +K D + +D R +V +K K EE
Sbjct: 824 NKEDKEESKDYQSVEAKEKNENGGVDTNVGNKEDSKDLKDDRSVEVKANKEESMKKKREE 883
Query: 228 VVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSR 287
V ++ ++ R + + +V KG ES VK D+++E + + +D + ++ +
Sbjct: 884 VQRNDKSSTKEVRDFANNMDIDVQKGSGES-VKYKKDEKKEGNKEENKDTINTSSKQKGK 942
Query: 288 EEAWKKQEMEKMNKELELMAHEQAIAGDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYL 347
++ KK+E + N + + D++ ++ L K +
Sbjct: 943 DKKKKKKESKNSNMKKK--------EEDKKEYVNNELKKQEDNKKE-------------- 980
Query: 348 ATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPISSYSLI 407
T+ S +S L +N + + K++ +N + + S
Sbjct: 981 ---TTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQD-- 1035
Query: 408 SSGERDDIGRRWPKDEVLALINLRCNNNNE---EKEGNSNNKAPLWERISQGMLELGYKR 464
E D R K E +L+ E EK+ + N+K+ E + K+
Sbjct: 1036 KKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKK 1095
Query: 465 SAKRCKEKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSDQKQESNN 524
+ ++K +K +K +D L+ + N K ++K++S +
Sbjct: 1096 EEDKKEKKKHEESKSRKKEEDKKDMEKLEDQ------------NSNKKKEDKNEKKKSQH 1143
Query: 525 VN-VPEENVVQEKAKQDENQD 544
V V +E+ +EK + +E +
Sbjct: 1144 VKLVKKESDKKEKKENEEKSE 1164
Score = 44.7 bits (104), Expect = 1e-04
Identities = 64/272 (23%), Positives = 108/272 (39%), Gaps = 41/272 (15%)
Query: 133 RKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTEL---EEVYQGGGGEN 189
+K E K+ +E K K +E+ + H N K E EE E+
Sbjct: 1062 KKEEETKEKKESENHKSKKKEDKK-----EHEDNKSMKKEEDKKEKKKHEESKSRKKEED 1116
Query: 190 NKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEE----EVVEKGTTND--------- 236
K++ + E QN + K D +E+ LV K+S++ E EK T +
Sbjct: 1117 KKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKN 1176
Query: 237 --EKKRKRSG-DDRFEVFKGFCESVVKKMMDQQE-------------EMHNKLIEDMVKR 280
+KK K+S D + + K ES KK+ +E + K ++ K
Sbjct: 1177 EVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKD 1236
Query: 281 DEEKFSREEAWKKQEMEKMNKELELMAHEQAIA-GDRQAHIIQFLNKFSTSANSSSLTSM 339
D++ +++ KK+ ME +KE E QA D + L + + A+S S +
Sbjct: 1237 DKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQA 1296
Query: 340 STQLQAYLATLTSNSSSSTLHSQNPNPETLKK 371
+ + ++S ++T Q N E KK
Sbjct: 1297 DSDESKNEILMQADSQATT---QRNNEEDRKK 1325
Score = 44.7 bits (104), Expect = 1e-04
Identities = 69/420 (16%), Positives = 171/420 (40%), Gaps = 48/420 (11%)
Query: 141 KRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVT------------ELEEVYQGGGGE 188
K +K EK E+++ + N+ ++ + V E E + +
Sbjct: 1113 KEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSK 1172
Query: 189 NNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRF 248
+ KN V+ +++ +D+ E++ + + KK+EE+ ++ + + KK+K + ++
Sbjct: 1173 SQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKN 1232
Query: 249 EVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAH 308
+ K ++ K+ ++E M ++ ++ E + +A + + ++ E+ + A
Sbjct: 1233 KP-KDDKKNTTKQSGGKKESMESE------SKEAENQQKSQATTQADSDESKNEILMQAD 1285
Query: 309 EQAIA-GDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLAT-LTSNSSSSTLHSQNPNP 366
QA + D QA + N+ A+S + T + + T + N + P
Sbjct: 1286 SQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKP 1345
Query: 367 ETLKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPISSYSLISSGERDDIGRRWPKDEVLA 426
+ KK ++ S S + N + + S E K+E+L
Sbjct: 1346 KDDKKNTTKQSGGKKESMESES----KEAENQQKSQATTQADSDE--------SKNEILM 1393
Query: 427 LINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENI--NKYFRKTK 484
+ + +++++ + + +K E + Q + +R+ + ++K ++ NK ++TK
Sbjct: 1394 QADSQADSHSDSQADSDESKN---EILMQADSQATTQRNNEEDRKKQTSVAENKKQKETK 1450
Query: 485 DANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSDQKQESNNVNVPEENVVQEKAKQDENQD 544
+ K D + T + GK + +E+ N +++ + + DE+++
Sbjct: 1451 EEKNKPKDDKKN-------TTEQSGGKKESMESESKEAEN---QQKSQATTQGESDESKN 1500
Score = 40.8 bits (94), Expect = 0.002
Identities = 80/428 (18%), Positives = 150/428 (34%), Gaps = 75/428 (17%)
Query: 139 GYKRSAEKCKEKFEEESRFFN---NINHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVE 195
G K+ +++ K+ E+R N N+ N+ K + + + + ++NK L
Sbjct: 750 GKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAKSVETKDNKKLSS 809
Query: 196 AEKQNEV-----QDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEV 250
E ++E +D + EE V +K+E V+ N E + D EV
Sbjct: 810 TENRDEAKERSGEDNKEDKEESKDYQSVEAKEKNENGGVDTNVGNKEDSKDLKDDRSVEV 869
Query: 251 FKGFCESVVKKMMDQQ--EEMHNKLIEDMVKR---DEEKFSREEA-WKKQEMEKMNKELE 304
ES+ KK + Q ++ K + D D +K S E +KK E ++ NKE
Sbjct: 870 KANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKE-- 927
Query: 305 LMAHEQAIAGDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNP 364
N ++ + S Q S +S + +
Sbjct: 928 --------------------------ENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEE 961
Query: 365 NP-ETLKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPISSYSLISSGERDDIGRRWPKDE 423
+ E + L+ +N T S ++ L + ++N D + K E
Sbjct: 962 DKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKE-------KKESEDSASKNREKKE 1014
Query: 424 VLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKT 483
EEK+ + +A ++ SQ ++ K++ E K
Sbjct: 1015 Y------------EEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKK 1062
Query: 484 KDANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSDQKQESNNVNVPEENVVQEKAKQDENQ 543
K+ K +S K + D+K+ +N ++ +E +EK K +E++
Sbjct: 1063 KEEETKEKKESEN-------------HKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESK 1109
Query: 544 DGAGESSQ 551
E +
Sbjct: 1110 SRKKEEDK 1117
Score = 39.7 bits (91), Expect = 0.004
Identities = 82/474 (17%), Positives = 189/474 (39%), Gaps = 57/474 (12%)
Query: 107 DEVLAL-LKIRSSMESWFPDFTWEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQ 165
DE ++L L+ ++++E+ WE ++ ++ + ++ E EES ++ HN
Sbjct: 149 DEAMSLSLEQKNAVETSITQ--WEQTITRIVKIVVEVKSKSSSEASSEES---SSTEHNN 203
Query: 166 NSFGKNFRFVT--ELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKK 223
+ G N E + G G N + +N +K++ +E + + + +K+
Sbjct: 204 VTTGSNMVETNGENSESTQEKGDGVEGSNGGDVSMENLQGNKVEDLKEGNNVVENGETKE 263
Query: 224 SEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEE 283
+ E VE +N+EK+ + G+ +S ++K ++ +E++ +++ + K D
Sbjct: 264 NNGENVE---SNNEKEVEGQGES-------IGDSAIEKNLESKEDVKSEV--EAAKNDGS 311
Query: 284 KFSRE--EAWKKQEMEKMNKELELMAHEQAIAGDRQAHIIQFLNKFSTSANSSSLTSMST 341
+ EA + ++ E E+ ++I ++ +K + + + +
Sbjct: 312 SMTENLGEAQGNNGVSTIDNEKEVEGQGESIEDSDIEKNLE--SKEDVKSEVEAAKNAGS 369
Query: 342 QLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPI 401
+ L N+ ST ET+ + E+ + + +++T + N
Sbjct: 370 SMTGKLEEAQRNNGVST-------NETMNSENKGSGESTNDKMVNATTNDEDHKKENKEE 422
Query: 402 SSYSLISSGERDDIGRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELG 461
+ + S + +++ + +E + NL NEE +GN++ +A S+
Sbjct: 423 THENNGESVKGENLENKAGNEESMKGENLENKVGNEELKGNASVEAKTNNESSKEEKREE 482
Query: 462 YKRSAK----RCKEKWENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGKLVLQSD 517
+RS + + K EN+N D+ + SL+++ + N + +
Sbjct: 483 SQRSNEVYMNKETTKGENVNIQGESIGDSTKDNSLENKEDVKPKVDANESDGNSTKERHQ 542
Query: 518 QKQESNNVNVPEENVVQ----------------------EKAKQDENQDGAGES 549
+ Q +N V+ ++N+ K K++E Q GES
Sbjct: 543 EAQVNNGVSTEDKNLDNIGADEQKKNDKSVEVTTNDGDHTKEKREETQGNNGES 596
>At1g56660 hypothetical protein
Length = 522
Score = 45.4 bits (106), Expect = 8e-05
Identities = 45/193 (23%), Positives = 85/193 (43%), Gaps = 28/193 (14%)
Query: 137 EVGYKRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVEA 196
E +K+ EK E+ EEE GK + E +E G E NK +
Sbjct: 112 EKEHKKGKEKKHEELEEEKE------------GKKKKNKKEKDE---SGPEEKNKKADKE 156
Query: 197 EKQNEV-QDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFC 255
+K +V Q+K + EED + + +S E +K ++K+++ S + + KG
Sbjct: 157 KKHEDVSQEKEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKKQKEESKSNEDKKVKGKK 216
Query: 256 ESVVKKMMDQQEE----MHNKLIEDMVKRDEEKFSR--------EEAWKKQEMEKMNKEL 303
E K +++++E H++ ++M ++D +K + EE KK + EK K+
Sbjct: 217 EKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKDESCAEEKKKKPDKEKKEKDE 276
Query: 304 ELMAHEQAIAGDR 316
++ + G +
Sbjct: 277 STEKEDKKLKGKK 289
Score = 43.1 bits (100), Expect = 4e-04
Identities = 36/146 (24%), Positives = 73/146 (49%), Gaps = 11/146 (7%)
Query: 177 ELEEVYQGGGGENNKNLVEA--EKQNEVQDKMDPHEEDSRMDDVLVS---KKSEEEVVEK 231
ELEE +G +N K E+ E++N+ DK HE+ S+ + L KK++++ ++
Sbjct: 125 ELEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKHEDVSQEKEELEEEDGKKNKKKEKDE 184
Query: 232 GTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAW 291
T ++KK+ + + E K + VK ++ E+ D+ K DEEK +
Sbjct: 185 SGTEEKKKKPKKEKKQKEESKSNEDKKVKGKKEKGEK------GDLEKEDEEKKKEHDET 238
Query: 292 KKQEMEKMNKELELMAHEQAIAGDRQ 317
++ EK +K+ + +++ A +++
Sbjct: 239 DQEMKEKDSKKNKKKEKDESCAEEKK 264
Score = 32.3 bits (72), Expect = 0.71
Identities = 32/127 (25%), Positives = 57/127 (44%), Gaps = 14/127 (11%)
Query: 189 NNKNLVEAEKQNEVQ-DKMDPHEEDSRMDDVL---------VSKKSEEEVVEKGTTNDEK 238
N +N E + +++ ++DP E+ ++ + V K +EE K + EK
Sbjct: 4 NQENAKEEKLHVKIKTQELDPKEKGENVEVEMEVKAKSIEKVKAKKDEESSGKSKKDKEK 63
Query: 239 KRKRSGDDRFEVFKGFCESVVKKMMDQQ-EEMHNKLIEDMVKRDEEKFSREEAWKKQEME 297
K+ ++ D + K + KM+ ++ EE H L VK + K E K+ E
Sbjct: 64 KKGKNVDSEVKEDKDDDKKKDGKMVSKKHEEGHGDL---EVKESDVKVEEHEKEHKKGKE 120
Query: 298 KMNKELE 304
K ++ELE
Sbjct: 121 KKHEELE 127
Score = 32.0 bits (71), Expect = 0.93
Identities = 32/145 (22%), Positives = 62/145 (42%), Gaps = 27/145 (18%)
Query: 188 ENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKK--------------------SEEE 227
E +++ E +K+ ++K + E + D L KK +E+E
Sbjct: 254 EKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEKEDEGKKTKEHDATEQE 313
Query: 228 VVEKGTTNDEKKRKRSGDD---RFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDE-- 282
+ ++ + E K+K++ D + V CE K D + E K + K+ E
Sbjct: 314 MDDEAADHKEGKKKKNKDKAKKKETVIDEVCEKETKDKDDDEGETKQKKNKKKEKKSEKG 373
Query: 283 EKFSREEAWKKQ--EMEKMNKELEL 305
EK +E+ K+ E E M+++++L
Sbjct: 374 EKDVKEDKKKENPLETEVMSRDIKL 398
Score = 30.8 bits (68), Expect = 2.1
Identities = 30/117 (25%), Positives = 53/117 (44%), Gaps = 18/117 (15%)
Query: 188 ENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDR 247
+ +N +E E + +P E DD KKS+ E G ++E K+K+ D +
Sbjct: 381 KKKENPLETEVMSRDIKLEEPEAEKKEEDDTEEKKKSKVE----GGESEEGKKKKKKDKK 436
Query: 248 FEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELE 304
K ++ KM + +EE K+D+ K + E K +E EK +K+++
Sbjct: 437 KNKKK---DTKEPKMTEDEEE----------KKDDSKDVKIEGSKAKE-EKKDKDVK 479
Score = 30.0 bits (66), Expect = 3.5
Identities = 21/98 (21%), Positives = 45/98 (45%), Gaps = 4/98 (4%)
Query: 180 EVYQGGGGENNKNLVEAEKQNEVQDKMDP---HEEDSRMDDVLVSKKSEEEVVEKGTTND 236
+V G E K + +K+N+ +D +P +E+ + DD K + E+ D
Sbjct: 418 KVEGGESEEGKKKKKKDKKKNKKKDTKEPKMTEDEEEKKDDSKDVKIEGSKAKEEKKDKD 477
Query: 237 EKKRKRSGD-DRFEVFKGFCESVVKKMMDQQEEMHNKL 273
KK+K D + + + + +M+++ E+ N++
Sbjct: 478 VKKKKGGNDIGKLKTKLAKIDEKIGALMEEKAEIENQI 515
>At3g24860 unknown protein
Length = 310
Score = 43.5 bits (101), Expect = 3e-04
Identities = 54/217 (24%), Positives = 86/217 (38%), Gaps = 41/217 (18%)
Query: 351 TSNSSSSTLHSQNPN-----PETLKKTLQPIPENPSSTLPSSSTTLVAQPRNNNPISSYS 405
T + +SS NPN P + P P NPSS P TT+VA + + ++
Sbjct: 3 TPSPTSSPPSDSNPNSAATPPHQKQPPSPPQPTNPSS--PPPHTTVVALAASTSAVA--- 57
Query: 406 LISSGERDDIGRRWPKDEVLALINLRCNNNNEEKE---GNSNNKAPLWERIS-QGMLELG 461
R W +DE L LI + +EK G K+ WE I+ G
Sbjct: 58 ------RKTQPVLWTQDETLLLIE-----SYKEKWFAIGRGPLKSTHWEEIAVAASSRSG 106
Query: 462 YKRSAKRCKEKWENINKYFRKTKDA----------NRKRSLDS------RTCPYFHLLTN 505
+R++ +C+ K E + K FR + + N+ LDS P L N
Sbjct: 107 VERTSTQCRHKIEKMRKRFRSERQSMGPISIWPFYNQMEELDSSNPAPISARPLTRLPPN 166
Query: 506 LYNQGKLVLQSDQKQESNNVNVPEENVVQEKAKQDEN 542
N+ + D+++++NN EE ++ + N
Sbjct: 167 SNNRYVDDEEEDEEEDNNNYEEEEEEDERQSKSRSIN 203
Score = 37.4 bits (85), Expect = 0.022
Identities = 64/332 (19%), Positives = 121/332 (36%), Gaps = 64/332 (19%)
Query: 30 LSTPNNTFPPFDPYNQQNHPSQHHQLPLQVQPNLLHPLHPHKDDEDKEQNSTPSMNNFQI 89
++TP+ T P N + + HQ P +P P ++ S +
Sbjct: 1 MATPSPTSSPPSDSNPNSAATPPHQKQPPSPPQPTNPSSPPPHTTVVALAASTSA----V 56
Query: 90 DRDQRQILPQLIDPWTNDEVLALLKIRSSMESWFP-------DFTWEHVS-RKLAEVGYK 141
R + +L WT DE L L I S E WF WE ++ + G +
Sbjct: 57 ARKTQPVL------WTQDETLLL--IESYKEKWFAIGRGPLKSTHWEEIAVAASSRSGVE 108
Query: 142 RSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGG------------EN 189
R++ +C+ K E+ + F + + + F ++EE+ +
Sbjct: 109 RTSTQCRHKIEKMRKRFRSERQSMGPISI-WPFYNQMEELDSSNPAPISARPLTRLPPNS 167
Query: 190 NKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTN-------------- 235
N V+ E+++E +D + EE+ + S+ + GT N
Sbjct: 168 NNRYVDDEEEDEEEDNNNYEEEEEEDERQSKSRSINYILRRPGTVNRFAGVGGGLLSWGQ 227
Query: 236 --DEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKK 293
KRKR+ D E + +V ++ E + ++++ + +F++E +
Sbjct: 228 KERSSKRKRNDGDGGERRRKGMRAVAAEIRAFAERVM------VMEKKKIEFAKETVRLR 281
Query: 294 QEMEKMNKELELMAHEQAIAGDRQAHIIQFLN 325
+EME L Q ++QF+N
Sbjct: 282 KEMEIRRINL---------IQSSQTQLLQFIN 304
>At1g20290 hypothetical protein
Length = 497
Score = 43.1 bits (100), Expect = 4e-04
Identities = 30/150 (20%), Positives = 74/150 (49%), Gaps = 10/150 (6%)
Query: 161 INHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLV 220
+N + + F ++F + V +G +V+AE++ E +++ + EE+ ++
Sbjct: 338 VNPSDSDF---YKFTPSMVHVGEG-------TIVQAEEEEEEEEEEEEEEEEEEEEEEEE 387
Query: 221 SKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKR 280
++ EEE E+ +E++ + ++ E + E ++ +++EE + E+ +
Sbjct: 388 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 447
Query: 281 DEEKFSREEAWKKQEMEKMNKELELMAHEQ 310
+EE+ EE +++E E+ +E E E+
Sbjct: 448 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 477
Score = 40.4 bits (93), Expect = 0.003
Identities = 26/124 (20%), Positives = 61/124 (48%)
Query: 187 GENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDD 246
GE E E++ E +++ + EE+ ++ ++ EEE E+ +E++ + ++
Sbjct: 356 GEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 415
Query: 247 RFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELM 306
E + E ++ +++EE + E+ + +EE+ EE +++E E+ +E E
Sbjct: 416 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 475
Query: 307 AHEQ 310
E+
Sbjct: 476 EEEE 479
Score = 40.0 bits (92), Expect = 0.003
Identities = 25/126 (19%), Positives = 62/126 (48%)
Query: 181 VYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKR 240
+ Q E + E E++ E +++ + EE+ ++ ++ EEE E+ +E++
Sbjct: 360 IVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 419
Query: 241 KRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMN 300
+ ++ E + E ++ +++EE + E+ + +EE+ EE +++E E+
Sbjct: 420 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 479
Query: 301 KELELM 306
+E + M
Sbjct: 480 EEKKYM 485
Score = 38.9 bits (89), Expect = 0.008
Identities = 27/126 (21%), Positives = 60/126 (47%)
Query: 177 ELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTND 236
E EE + E + E E++ E +++ + EE+ ++ ++ EEE E+ +
Sbjct: 367 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 426
Query: 237 EKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEM 296
E++ + ++ E + E ++ +++EE + E+ + +EE+ EE KK +
Sbjct: 427 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKYMI 486
Query: 297 EKMNKE 302
K K+
Sbjct: 487 VKKKKK 492
Score = 38.5 bits (88), Expect = 0.010
Identities = 24/124 (19%), Positives = 61/124 (48%)
Query: 175 VTELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTT 234
+ + EE + E + E E++ E +++ + EE+ ++ ++ EEE E+
Sbjct: 360 IVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 419
Query: 235 NDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQ 294
+E++ + ++ E + E ++ +++EE + E+ + +EE+ EE +++
Sbjct: 420 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 479
Query: 295 EMEK 298
E +K
Sbjct: 480 EEKK 483
Score = 37.4 bits (85), Expect = 0.022
Identities = 26/126 (20%), Positives = 60/126 (46%)
Query: 177 ELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTND 236
E EE + E + E E++ E +++ + EE+ ++ ++ EEE E+ +
Sbjct: 368 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 427
Query: 237 EKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEM 296
E++ + ++ E + E ++ +++EE + E+ + +EE+ EE K +
Sbjct: 428 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKYMIV 487
Query: 297 EKMNKE 302
+K K+
Sbjct: 488 KKKKKK 493
Score = 36.6 bits (83), Expect = 0.038
Identities = 25/120 (20%), Positives = 58/120 (47%)
Query: 177 ELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTND 236
E EE + E + E E++ E +++ + EE+ ++ ++ EEE E+ +
Sbjct: 366 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 425
Query: 237 EKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEM 296
E++ + ++ E + E ++ +++EE + E+ + +EE+ EE +K+ M
Sbjct: 426 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKYM 485
Score = 34.7 bits (78), Expect = 0.14
Identities = 24/126 (19%), Positives = 61/126 (48%)
Query: 177 ELEEVYQGGGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTND 236
E EE + E + E E++ E +++ + EE+ ++ ++ EEE E+ +
Sbjct: 365 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 424
Query: 237 EKKRKRSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEM 296
E++ + ++ E + E ++ +++EE + E+ + +EE+ EE ++++
Sbjct: 425 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKY 484
Query: 297 EKMNKE 302
+ K+
Sbjct: 485 MIVKKK 490
Score = 28.9 bits (63), Expect = 7.9
Identities = 22/98 (22%), Positives = 43/98 (43%)
Query: 145 EKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQD 204
E+ +E+ EEE + + E EE + E + E E++ E ++
Sbjct: 397 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 456
Query: 205 KMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKR 242
+ + EE+ ++ ++ EEE +K +KK+KR
Sbjct: 457 EEEEEEEEEEEEEEEEEEEEEEEEEKKYMIVKKKKKKR 494
>At5g59390 transcriptional regulator - like protein
Length = 561
Score = 42.4 bits (98), Expect = 7e-04
Identities = 48/239 (20%), Positives = 102/239 (42%), Gaps = 27/239 (11%)
Query: 141 KRSAEKCKEKFEEESRFFNNINHNQNSFGKNF-----RFVTELEEVYQGGGGENNKNLVE 195
K+S ++ ++K +E SRF ++ + KN+ + ++EE+YQ + K+L E
Sbjct: 187 KQSKQELEQKVDETSRFLESLELHNVLLNKNYQEGFQKMQMKMEELYQQVLDGHEKSLAE 246
Query: 196 AEKQNEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFC 255
E + E K+D ++ ++ E+ +E+ ++K + E K
Sbjct: 247 LEAKRE---KLDERARLIEQRAIINEEEMEKSRLER--EMNQKAMCEQNEANEEAMKLAE 301
Query: 256 ESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAHEQAIAGD 315
+ + ++E++H +++E K +E + + E+EK+ +M H GD
Sbjct: 302 KHQASSSLKEKEKLHKRIMEMEAKLNETQ------ELELEIEKLKGTTNVMKHMVGSDGD 355
Query: 316 RQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQ 374
+ I++ + K L + T L + TL ++ Q + LK+ +Q
Sbjct: 356 KD--IVEKIAK-----TQIQLDAQETALHEKMMTLARKERATNDEYQ----DVLKEMIQ 403
>At4g36520 trichohyalin like protein
Length = 1432
Score = 42.0 bits (97), Expect = 9e-04
Identities = 41/168 (24%), Positives = 78/168 (46%), Gaps = 15/168 (8%)
Query: 140 YKRSAEKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQ 199
++++A K +E ES ++ N N GK ++ E + N+ L E++
Sbjct: 585 WEQNARKLREALGNESTLEVSVELNGN--GKKMEMRSQSET-------KLNEPLKRMEEE 635
Query: 200 NEVQDKMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRK-RSGDDRFEVFKGFCESV 258
+++ EE+ R + V V K E+ ++ +EK+RK + ++ E + E+
Sbjct: 636 TRIKEAR-LREENDRRERVAVEKAENEKRLKAALEQEEKERKIKEAREKAENERRAVEAR 694
Query: 259 VK----KMMDQQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKE 302
K + M +Q+E+ +L E K +E + RE +QE E+ KE
Sbjct: 695 EKAEQERKMKEQQELELQLKEAFEKEEENRRMREAFALEQEKERRIKE 742
Score = 36.6 bits (83), Expect = 0.038
Identities = 32/146 (21%), Positives = 71/146 (47%), Gaps = 23/146 (15%)
Query: 188 ENNKNLVEAEKQNEVQDKM--------------DPHEEDSRMDDVLVSKKSEEEVVEKGT 233
EN + VEA ++ E + KM + EE+ RM + ++ +E +++
Sbjct: 685 ENERRAVEAREKAEQERKMKEQQELELQLKEAFEKEEENRRMREAFALEQEKERRIKEAR 744
Query: 234 TNDEKKRK-RSGDDRFEVFKGFCESVVKKMMDQQEEMHNKLIEDMVKRDEEKFSREEAWK 292
+E +R+ + ++ E+ E +K ++Q+E+ + I++ +R+E + +E +
Sbjct: 745 EKEENERRIKEAREKAEL-----EQRLKATLEQEEK--ERQIKERQEREENERRAKEVLE 797
Query: 293 KQEMEKMNKE-LELMAHEQAIAGDRQ 317
+ E E+ KE LE +E+ + R+
Sbjct: 798 QAENERKLKEALEQKENERRLKETRE 823
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.127 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,208,581
Number of Sequences: 26719
Number of extensions: 705109
Number of successful extensions: 4544
Number of sequences better than 10.0: 305
Number of HSP's better than 10.0 without gapping: 71
Number of HSP's successfully gapped in prelim test: 237
Number of HSP's that attempted gapping in prelim test: 3668
Number of HSP's gapped (non-prelim): 684
length of query: 557
length of database: 11,318,596
effective HSP length: 104
effective length of query: 453
effective length of database: 8,539,820
effective search space: 3868538460
effective search space used: 3868538460
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC135565.15


