

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.8 + phase: 0
(454 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
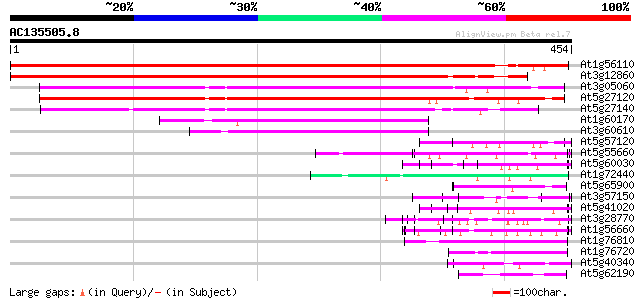
Score E
Sequences producing significant alignments: (bits) Value
At1g56110 SAR like DNA binding protein 647 0.0
At3g12860 nucleolar protein, putative 624 e-179
At3g05060 putative SAR DNA-binding protein-1 325 2e-89
At5g27120 SAR DNA-binding protein - like 324 5e-89
At5g27140 SAR DNA-binding protein - like 243 2e-64
At1g60170 unknown protein (At1g60170) 83 3e-16
At3g60610 putative protein 72 8e-13
At5g57120 unknown protein 57 3e-08
At5g55660 putative protein 55 1e-07
At5g60030 KED - like protein 53 4e-07
At1g72440 hypothetical protein 53 4e-07
At5g65900 ATP-dependent RNA helicase-like 52 5e-07
At3g57150 putative pseudouridine synthase (NAP57) 50 3e-06
At5g41020 putative protein 49 4e-06
At3g28770 hypothetical protein 49 6e-06
At1g56660 hypothetical protein 49 6e-06
At1g76810 translation initiation factor IF-2 like protein 47 2e-05
At1g76720 putative translation initiation factor IF-2 47 3e-05
At5g40340 unknown protein 46 4e-05
At5g62190 putative RNA helicase (MMI9.2) 46 5e-05
>At1g56110 SAR like DNA binding protein
Length = 522
Score = 647 bits (1669), Expect = 0.0
Identities = 333/463 (71%), Positives = 385/463 (82%), Gaps = 22/463 (4%)
Query: 1 MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQ 60
MTDELR+ LE NLPKVKEGKK KFSLG+AE K+GSHI EATKIP QSNEFV EL+RGVRQ
Sbjct: 71 MTDELRSFLELNLPKVKEGKKPKFSLGLAEPKLGSHIFEATKIPCQSNEFVLELLRGVRQ 130
Query: 61 HFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAM 120
HFD+F+ DLK GDLEK+QLGL HSYSRAKVKFNVNRVDNMVIQAIF+LDTLDKD+NSFAM
Sbjct: 131 HFDRFIKDLKPGDLEKSQLGLAHSYSRAKVKFNVNRVDNMVIQAIFMLDTLDKDINSFAM 190
Query: 121 RVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEA 180
RVREWYSWHFPELVKIVNDNYLY +V+K I+DKSKL ED I LT+++GDEDKAKE++EA
Sbjct: 191 RVREWYSWHFPELVKIVNDNYLYARVSKMIDDKSKLTEDHIPMLTEVLGDEDKAKEVIEA 250
Query: 181 AKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGAR 240
KASMG DLSP+DLINV FAQ+VMDL+DYR++L DYL TKM+DIAPNL +L+G+ VGAR
Sbjct: 251 GKASMGSDLSPLDLINVQTFAQKVMDLADYRKKLYDYLVTKMSDIAPNLAALIGEMVGAR 310
Query: 241 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGR 300
LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASA+NKGR
Sbjct: 311 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASAKNKGR 370
Query: 301 MARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIES 360
+ARYLANKCSIASRIDCF++ T+AFGEKLREQVEERL+FYDKGVAPRKN+DVMK IE+
Sbjct: 371 IARYLANKCSIASRIDCFADGATTAFGEKLREQVEERLEFYDKGVAPRKNVDVMKEVIEN 430
Query: 361 VDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK 420
+ ++ E + S KK+KKKK K + +++ E+ KSE KKKKKEKRK
Sbjct: 431 LKQEEEGKEPVDASVKKSKKKKAKGEEEEEVVA----------MEEDKSE-KKKKKEKRK 479
Query: 421 LD--QEVEVEDKV---------VEDGANADSSKKKKKKSKKKD 452
++ +E E +K E+ + S+KKKKKKSK +
Sbjct: 480 METAEENEKSEKKKTKKSKAGGEEETDDGHSTKKKKKKSKSAE 522
>At3g12860 nucleolar protein, putative
Length = 477
Score = 624 bits (1609), Expect = e-179
Identities = 320/419 (76%), Positives = 359/419 (85%), Gaps = 15/419 (3%)
Query: 1 MTDELRTVLETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQ 60
M+DELR+ LE NLPKVKEGKK KFSLGV+E KIGS I EATKIP QSNEFV EL+RGVRQ
Sbjct: 71 MSDELRSFLELNLPKVKEGKKPKFSLGVSEPKIGSCIFEATKIPCQSNEFVHELLRGVRQ 130
Query: 61 HFDKFVGDLKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAM 120
HFD+F+ DLK GDLEKAQLGL HSYSRAKVKFNVNRVDNMVIQAIF+LDTLDKD+NSFAM
Sbjct: 131 HFDRFIKDLKPGDLEKAQLGLAHSYSRAKVKFNVNRVDNMVIQAIFMLDTLDKDINSFAM 190
Query: 121 RVREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEA 180
RVREWYSWHFPELVKIVNDNYLY KV+K I DKSKL+E+ I LT+ +GDEDKA+E++EA
Sbjct: 191 RVREWYSWHFPELVKIVNDNYLYAKVSKIIVDKSKLSEEHIPMLTEALGDEDKAREVIEA 250
Query: 181 AKASMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGAR 240
KASMGQDLSPVDLINV FAQRVMDL+DYR++L DYL TKM+DIAPNL +L+G+ VGAR
Sbjct: 251 GKASMGQDLSPVDLINVQTFAQRVMDLADYRKKLYDYLVTKMSDIAPNLAALIGEMVGAR 310
Query: 241 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGR 300
LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASA+NKGR
Sbjct: 311 LISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASAKNKGR 370
Query: 301 MARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIES 360
+AR+LANKCSIASRIDCFS+ T+AFGEKLREQVEERLDFYDKGVAPRKN+DVMK E
Sbjct: 371 IARFLANKCSIASRIDCFSDNSTTAFGEKLREQVEERLDFYDKGVAPRKNVDVMK---EK 427
Query: 361 VDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKR 419
++ E E EE +KK KKKK KA +G+++ A D+ KKKKK + +
Sbjct: 428 RKTEEKEEEKEEEKSKK-KKKKSKAVEGEELT-----------ATDNGHSKKKKKTKSQ 474
>At3g05060 putative SAR DNA-binding protein-1
Length = 533
Score = 325 bits (834), Expect = 2e-89
Identities = 196/458 (42%), Positives = 275/458 (59%), Gaps = 43/458 (9%)
Query: 25 SLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHS 84
+L VA+SK+G+ I E KI N V EL+RGVR F + + L DL LGL HS
Sbjct: 85 TLAVADSKLGNVIKEKLKIDCIHNNAVMELLRGVRSQFTELISGLGDQDLAPMSLGLSHS 144
Query: 85 YSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYC 144
+R K+KF+ ++VD M+IQAI LLD LDK++N++AMRVREWY WHFPEL KI++DN LY
Sbjct: 145 LARYKLKFSSDKVDTMIIQAIGLLDDLDKELNTYAMRVREWYGWHFPELAKIISDNILYA 204
Query: 145 KVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRV 204
K K + ++ A+ ++++ DE +A ++ +AA SMG ++S +DL+++ +V
Sbjct: 205 KSVKLMGNRVNAAK---LDFSEILADEIEA-DLKDAAVISMGTEVSDLDLLHIRELCDQV 260
Query: 205 MDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGA 264
+ LS+YR +L DYL ++MN IAPNL +LVG+ VGARLISH GSL NL+K P ST+QILGA
Sbjct: 261 LSLSEYRAQLYDYLKSRMNTIAPNLTALVGELVGARLISHGGSLLNLSKQPGSTVQILGA 320
Query: 265 EKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTS 324
EKALFRALKT+ TPKYGLIFH+S +G+A+ ++KG+++R LA K +A R+D + +
Sbjct: 321 EKALFRALKTKHATPKYGLIFHASLVGQAAPKHKGKISRSLAAKTVLAIRVDALGDSQDN 380
Query: 325 AFGEKLREQVEERL-DFYDKGVAPRKNIDVMKSAIESVDNKDTEM--------------- 368
G + R ++E RL + K + K IE V NKD +M
Sbjct: 381 TMGLENRAKLEARLRNLEGKDLGRLSGSSKGKPKIE-VYNKDKKMGSGGLITPAKTYNTA 439
Query: 369 ----------ETEEVSAKKTKKKKQKA-------ADGDDMAVDKAAEITNGDAEDHKSEK 411
++EE S KK KKKK+K + + K AE + E+
Sbjct: 440 ADSLLGETSAKSEEPSKKKDKKKKKKVEEEKPEEEEPSEKKKKKKAEAETEAVVEVAKEE 499
Query: 412 KKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSK 449
KKK K+KRK + E++ E A K+KKKKSK
Sbjct: 500 KKKNKKKRKHE-----EEETTETPAKKKDKKEKKKKSK 532
>At5g27120 SAR DNA-binding protein - like
Length = 533
Score = 324 bits (831), Expect = 5e-89
Identities = 194/461 (42%), Positives = 277/461 (60%), Gaps = 48/461 (10%)
Query: 25 SLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHS 84
+L VA+SK+G+ I E KI N V EL+RG+R + + L DL LGL HS
Sbjct: 84 TLAVADSKLGNIIKEKLKIVCVHNNAVMELLRGIRSQLTELISGLGDQDLGPMSLGLSHS 143
Query: 85 YSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYC 144
+R K+KF+ ++VD M+IQAI LLD LDK++N++AMRVREW+ WHFPEL KIV DN LY
Sbjct: 144 LARYKLKFSSDKVDTMIIQAIGLLDDLDKELNTYAMRVREWFGWHFPELAKIVQDNILYA 203
Query: 145 KVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRV 204
K K + ++ A+ ++++ DE +A E+ EAA SMG ++S +DL+++ +V
Sbjct: 204 KAVKLMGNRINAAK---LDFSEILADEIEA-ELKEAAVISMGTEVSDLDLLHIRELCDQV 259
Query: 205 MDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGA 264
+ L++YR +L DYL ++MN IAPNL +LVG+ VGARLISH GSL NLAK P ST+QILGA
Sbjct: 260 LSLAEYRAQLYDYLKSRMNTIAPNLTALVGELVGARLISHGGSLLNLAKQPGSTVQILGA 319
Query: 265 EKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTS 324
EKALFRALKT+ TPKYGLIFH+S +G+A+ +NKG+++R LA K +A R D + +
Sbjct: 320 EKALFRALKTKHATPKYGLIFHASVVGQAAPKNKGKISRSLAAKSVLAIRCDALGDSQDN 379
Query: 325 AFGEKLREQVEERL---------------------DFYDKG--------VAPRKNIDVMK 355
G + R ++E RL + YDK + P K +
Sbjct: 380 TMGVENRLKLEARLRTLEGKDLGRLSGSAKGKPKIEVYDKDKKKGSGGLITPAKTYNTAA 439
Query: 356 SAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVD----KAAEITNGDAEDHKSE- 410
++ D+E +E KK KKKK+KA D ++ + K + +AE +E
Sbjct: 440 DSLLQTPTVDSENGVKE---KKDKKKKKKADDEEEAKTEEPSKKKSNKKKTEAEPETAEE 496
Query: 411 --KKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSK 449
KK+KKK+++ ++E E+ K E S KKKKKK++
Sbjct: 497 PAKKEKKKKRKHEEEETEMPAKKKE-----KSEKKKKKKTE 532
>At5g27140 SAR DNA-binding protein - like
Length = 445
Score = 243 bits (619), Expect = 2e-64
Identities = 153/406 (37%), Positives = 230/406 (55%), Gaps = 24/406 (5%)
Query: 26 LGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHSY 85
L VA+ K+G I E I N+ V EL+RGVR + + L DL L L H
Sbjct: 57 LAVADPKLGDIITEKLDIECVHNDAVMELLRGVRSQLTELLSGLDDNDLAPVSLELSHIL 116
Query: 86 SRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYCK 145
+R K+K ++ M+I +I LLD LDK++N++ V E Y HFPEL IV DN LY K
Sbjct: 117 ARYKLKITSDKT--MIILSISLLDDLDKELNTYTTSVCELYGLHFPELANIVQDNILYAK 174
Query: 146 VAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRVM 205
V K + ++ A ++++ DE +A E+ EA+ S ++S +DL+++ +V+
Sbjct: 175 VVKLMGNRINAAT---LDFSEILADEVEA-ELKEASMVSTRTEVSDLDLMHIQELCDQVL 230
Query: 206 DLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGAE 265
+++ + L D L KMN IAPNL +LVG+ VGARLISH GSL NL+K P ST+QILGAE
Sbjct: 231 SIAEDKTLLCDDLKNKMNKIAPNLTALVGELVGARLISHCGSLWNLSKLPWSTIQILGAE 290
Query: 266 KALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTSA 325
K L++ALKT+ TPKYGLI+H+ + +A+ NKG++AR LA K ++A R D F +
Sbjct: 291 KTLYKALKTKQATPKYGLIYHAPLVRQAAPENKGKIARSLAAKSALAIRCDAFGNGQDNT 350
Query: 326 FGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK---KKK 382
G + R ++E RL + G ++ + E V++KDT+ E ++ KT+ KK+
Sbjct: 351 MGVESRLKLEARLRNLEGG-----DLGACEEE-EEVNDKDTKKEADDEEEPKTEECSKKR 404
Query: 383 QKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVE 428
+K A+ E A+ K E K K + + + V ++
Sbjct: 405 KKEAE---------LETVEDPAKKSKQEVKDVVKTNKYVHKNVNIK 441
>At1g60170 unknown protein (At1g60170)
Length = 485
Score = 83.2 bits (204), Expect = 3e-16
Identities = 58/220 (26%), Positives = 106/220 (47%), Gaps = 8/220 (3%)
Query: 122 VREWYSWHFPELVKIVNDNYLYCKVAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAA 181
+++ Y F EL +V+ Y V K I +++ LA L DL A +V +
Sbjct: 117 IKDKYKLKFQELESLVHHPIDYACVVKKIGNETDLA------LVDLADLLPSAIIMVVSV 170
Query: 182 KA--SMGQDLSPVDLINVHMFAQRVMDLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGA 239
A + G L L V R +DL R+++ +++ +KM IAPNL ++VG +V A
Sbjct: 171 TALTTKGSALPEDVLQKVLEACDRALDLDSARKKVLEFVESKMGSIAPNLSAIVGSAVAA 230
Query: 240 RLISHAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKG 299
+L+ AG L+ LAK P+ +Q+LG ++ + + + G + + +
Sbjct: 231 KLMGTAGGLSALAKMPACNVQVLGHKRKNLAGFSSATSQSRVGYLEQTEIYQSTPPGLQA 290
Query: 300 RMARYLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLD 339
R R +A K ++A+R+D G+ RE++ ++++
Sbjct: 291 RAGRLVAAKSTLAARVDATRGDPLGISGKAFREEIRKKIE 330
>At3g60610 putative protein
Length = 442
Score = 71.6 bits (174), Expect = 8e-13
Identities = 52/195 (26%), Positives = 89/195 (44%), Gaps = 6/195 (3%)
Query: 146 VAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRVM 205
V FI++K KL ++ESL D K DL+ VD + + +M
Sbjct: 103 VHNFIKEKYKLRFQELESLVHHPIDY-----ACVVKKIGNETDLTLVDFAGLDLTPAIIM 157
Query: 206 DLS-DYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGA 264
+S L M IAPNL ++VG +V A+L+ AG L+ LAK P+ +Q+LG
Sbjct: 158 AISVTASTTKGSALPEDMGSIAPNLSAIVGSAVAAKLMGTAGGLSALAKMPACNVQVLGH 217
Query: 265 EKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTS 324
++ + + + G + + + R R +A K ++A+R+D E
Sbjct: 218 KRKNLVGFSSATSQSRVGYLEQTEIFQSTPPGLEARAGRLVAAKSTLAARVDATREDPLG 277
Query: 325 AFGEKLREQVEERLD 339
G+ RE++ +++D
Sbjct: 278 ISGKAFREEIRKKID 292
>At5g57120 unknown protein
Length = 330
Score = 56.6 bits (135), Expect = 3e-08
Identities = 38/100 (38%), Positives = 55/100 (55%), Gaps = 8/100 (8%)
Query: 359 ESVDNKDTEMETEEV--SAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKK 416
E+ N +TE E + KK KKKK+ ++ ++ + T+ ED EKKKKK+
Sbjct: 70 EAAANGNTEANVVEAVENVKKDKKKKKNKETKVEVTEEEKVKETDAVIEDGVKEKKKKKE 129
Query: 417 EKRKLDQEVEVE--DKVVEDGANADSSKKKKKKSKKKDAE 454
K K+ +E +V+ D V+EDG +KKKKKSK K E
Sbjct: 130 TKVKVTEEEKVKETDAVIEDGV----KEKKKKKSKSKSVE 165
Score = 45.1 bits (105), Expect = 8e-05
Identities = 39/132 (29%), Positives = 60/132 (44%), Gaps = 14/132 (10%)
Query: 332 EQVEERLDFYDKGVAPRKNIDVMKSAIESVDN-KDTEMETEE-VSAKKTKKKKQK--AAD 387
E+V+E + GV +K K + + K+T+ E+ V KK KK K K AD
Sbjct: 108 EKVKETDAVIEDGVKEKKKKKETKVKVTEEEKVKETDAVIEDGVKEKKKKKSKSKSVEAD 167
Query: 388 GDDMAVDK-----AAEITNGDAEDHKSEKKKKKKEKRKLD-----QEVEVEDKVVEDGAN 437
D V K E T + ED E K++KKE+ ++ QE V++ ++ N
Sbjct: 168 DDKEKVSKKRKRSEPEETKEETEDDDEESKRRKKEENVVENDEGVQETPVKETETKENGN 227
Query: 438 ADSSKKKKKKSK 449
A+ S+ K K
Sbjct: 228 AEKSETKSTNQK 239
Score = 39.3 bits (90), Expect = 0.005
Identities = 47/186 (25%), Positives = 82/186 (43%), Gaps = 41/186 (22%)
Query: 301 MARYLANKCSIASRIDCFSEKGTSAFGEK---------LREQVEERLDFYDKGVAPRKNI 351
+A+YL +C + CF + + A EK L E E L+ D A N
Sbjct: 22 VAQYL-ERCGFSK---CFKKLLSEAEIEKKELNTSLPDLEEIFSEFLNKRDHEAAANGNT 77
Query: 352 DV-MKSAIESVD-------NKDTEME-TEEVSAKKT--------KKKKQKAADGDDMAVD 394
+ + A+E+V NK+T++E TEE K+T K+KK+K + +
Sbjct: 78 EANVVEAVENVKKDKKKKKNKETKVEVTEEEKVKETDAVIEDGVKEKKKKKETKVKVTEE 137
Query: 395 KAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVE-----------DKVVEDGANADSSKK 443
+ + T+ ED EKKKKK + + ++ + + E ++ E+ + D K
Sbjct: 138 EKVKETDAVIEDGVKEKKKKKSKSKSVEADDDKEKVSKKRKRSEPEETKEETEDDDEESK 197
Query: 444 KKKKSK 449
++KK +
Sbjct: 198 RRKKEE 203
Score = 35.4 bits (80), Expect = 0.065
Identities = 27/121 (22%), Positives = 52/121 (42%), Gaps = 6/121 (4%)
Query: 334 VEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAV 393
V E ++ K +KN + E K+T+ E+ +K KKK+ K + V
Sbjct: 81 VVEAVENVKKDKKKKKNKETKVEVTEEEKVKETDAVIEDGVKEKKKKKETK------VKV 134
Query: 394 DKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDA 453
+ ++ DA K+KKKK+ + E + + + V ++ K++++ D
Sbjct: 135 TEEEKVKETDAVIEDGVKEKKKKKSKSKSVEADDDKEKVSKKRKRSEPEETKEETEDDDE 194
Query: 454 E 454
E
Sbjct: 195 E 195
>At5g55660 putative protein
Length = 759
Score = 54.7 bits (130), Expect = 1e-07
Identities = 54/206 (26%), Positives = 87/206 (42%), Gaps = 8/206 (3%)
Query: 248 LTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLAN 307
L + P +T +L EK + +K + K SS R++ K N
Sbjct: 442 LVEFLEKPHATTDVLVNEKE--KGVKRKRTPKKSSPAAGSSSSKRSAKSQKKTEEATRTN 499
Query: 308 KCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTE 367
K S+A D E+ E+ ++VEE + + G+ P K+ D ES +N ++E
Sbjct: 500 KKSVAHSDDESEEEKEDDEEEEKEQEVEEEEEENENGI-PDKSEDEAPQLSESEENVESE 558
Query: 368 METEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEV 427
E+EE +TKKKK+ + D + A KS KK +KR + +
Sbjct: 559 EESEE----ETKKKKRGSRTSSDKKESAGKSRSKKTAVPTKSSPPKKATQKRSAGKRKKS 614
Query: 428 EDKVVEDGANADSSKKKKKKSKKKDA 453
+D + A S +KK +K K+ A
Sbjct: 615 DDD-SDTSPKASSKRKKTEKPAKEQA 639
Score = 47.4 bits (111), Expect = 2e-05
Identities = 45/157 (28%), Positives = 72/157 (45%), Gaps = 31/157 (19%)
Query: 328 EKLREQVEERLDFYDKGVA--PRKNIDVMKSAIESVDNKDTE-METEEVSAKKTKKKKQK 384
+K+ E +++ D GV+ + VMK ++ES DNKD E E E+ K +K +
Sbjct: 95 DKMDEDTDDKNLKADDGVSGVATEEDAVMKESVESADNKDAENPEGEQEKESKEEKLEGG 154
Query: 385 AADGDDMA--------------VDKAAEITNGDAEDHKSEKKKKKKEKRKLDQE-----V 425
A+G++ VD+A ++ N D ED K E K+K E ++E
Sbjct: 155 KANGNEEGDTEEKLVGGDKGDDVDEAEKVENVD-EDDKEEALKEKNEAELAEEEETNKGE 213
Query: 426 EVEDKVVEDGANADS--------SKKKKKKSKKKDAE 454
EV++ ED AD+ KK + K + +D E
Sbjct: 214 EVKEANKEDDVEADTKVAEPEVEDKKTESKDENEDKE 250
Score = 44.3 bits (103), Expect = 1e-04
Identities = 38/143 (26%), Positives = 67/143 (46%), Gaps = 19/143 (13%)
Query: 327 GEKLREQVEERL-----------DFYDKGVAPRKNIDVMKSA-IESVDNKDTEM------ 368
GE+ +E EE+L D +K V K DV ++ +E+VD D E
Sbjct: 140 GEQEKESKEEKLEGGKANGNEEGDTEEKLVGGDKGDDVDEAEKVENVDEDDKEEALKEKN 199
Query: 369 ETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVE 428
E E ++T K ++ + V+ ++ + ED K+E K + ++K + ++E E E
Sbjct: 200 EAELAEEEETNKGEEVKEANKEDDVEADTKVAEPEVEDKKTESKDENEDKEE-EKEDEKE 258
Query: 429 DKVVEDGANADSSKKKKKKSKKK 451
D+ E + + K+ KKS K+
Sbjct: 259 DEKEESNDDKEDKKEDIKKSNKR 281
Score = 35.8 bits (81), Expect = 0.050
Identities = 34/131 (25%), Positives = 58/131 (43%), Gaps = 24/131 (18%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAAD 387
E L+E+ E L ++ K +V ++ E DT++ EV KKT+ K +
Sbjct: 193 EALKEKNEAELAEEEE---TNKGEEVKEANKEDDVEADTKVAEPEVEDKKTESKDE---- 245
Query: 388 GDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKK 447
N D E+ K ++K+ +KE+ D+E + ED + ++K + K
Sbjct: 246 -------------NEDKEEEKEDEKEDEKEESNDDKEDKKEDIKKSNKRGKGKTEKTRGK 292
Query: 448 SK----KKDAE 454
+K KKD E
Sbjct: 293 TKSDEEKKDIE 303
Score = 35.0 bits (79), Expect = 0.085
Identities = 30/138 (21%), Positives = 67/138 (47%), Gaps = 18/138 (13%)
Query: 329 KLREQVEERLDFYDKGVAPRKNIDVMKSAIES-VDNKDTEMETEEVSAK-------KTKK 380
K + V + ++F +K P DV+ + E V K T ++ + K++K
Sbjct: 434 KKEDIVTKLVEFLEK---PHATTDVLVNEKEKGVKRKRTPKKSSPAAGSSSSKRSAKSQK 490
Query: 381 KKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDK-------VVE 433
K ++A + +V + + + + ED + E+K+++ E+ + + E + DK + E
Sbjct: 491 KTEEATRTNKKSVAHSDDESEEEKEDDEEEEKEQEVEEEEEENENGIPDKSEDEAPQLSE 550
Query: 434 DGANADSSKKKKKKSKKK 451
N +S ++ ++++KKK
Sbjct: 551 SEENVESEEESEEETKKK 568
Score = 30.8 bits (68), Expect = 1.6
Identities = 25/120 (20%), Positives = 54/120 (44%), Gaps = 22/120 (18%)
Query: 354 MKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADG------DDMAVDKAAEITNGDAEDH 407
++ +++ K+ + E S K+ ++ K++ G DD E GD E
Sbjct: 20 LQKTSDAISGKEVQ---ENASGKEVQESKKEEDTGLEKMEIDDEGKQHEGESETGDKEVE 76
Query: 408 KSEKKKK-------KKEKRKLDQEVEVEDKVVEDGANADSS------KKKKKKSKKKDAE 454
+E++KK + E K+D++ + ++ +DG + ++ K+ + + KDAE
Sbjct: 77 VTEEEKKDVGEDKEQPEADKMDEDTDDKNLKADDGVSGVATEEDAVMKESVESADNKDAE 136
>At5g60030 KED - like protein
Length = 292
Score = 52.8 bits (125), Expect = 4e-07
Identities = 37/136 (27%), Positives = 64/136 (46%), Gaps = 2/136 (1%)
Query: 319 SEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
+ K EK++E++E+ D+ RK K+ E V ++ ++E E+ SA+
Sbjct: 158 NNKDEDVVDEKVKEKLEDEQKSADR--KERKKKKSKKNNDEDVVDEKEKLEDEQKSAEIK 215
Query: 379 KKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA 438
+KKK K D D + E E K +KKK+K ++ + +E + + K D
Sbjct: 216 EKKKNKDEDVVDEKEKEKLEDEQRSGERKKEKKKKRKSDEEIVSEERKSKKKRKSDEEMG 275
Query: 439 DSSKKKKKKSKKKDAE 454
+K KKK K K+ +
Sbjct: 276 SEERKSKKKRKLKEID 291
Score = 51.2 bits (121), Expect = 1e-06
Identities = 31/111 (27%), Positives = 59/111 (52%), Gaps = 3/111 (2%)
Query: 342 DKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITN 401
+K + K+ DV+ + + + E +EE +K +KKK+K +D+ +K E
Sbjct: 117 EKKMKKSKDADVVDEKVN--EKLEAEQRSEERRERKKEKKKKKNNKDEDVVDEKVKEKLE 174
Query: 402 GDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ + +++KKKK K+ D++V E + +ED + K+KKK+K +D
Sbjct: 175 DEQKSADRKERKKKKSKKNNDEDVVDEKEKLED-EQKSAEIKEKKKNKDED 224
Score = 49.7 bits (117), Expect = 3e-06
Identities = 30/78 (38%), Positives = 47/78 (59%), Gaps = 2/78 (2%)
Query: 379 KKKKQKAADGDDMAVDKA--AEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGA 436
K KK K AD D V++ AE + + + K EKKKKK K + + +V++K+ ++
Sbjct: 119 KMKKSKDADVVDEKVNEKLEAEQRSEERRERKKEKKKKKNNKDEDVVDEKVKEKLEDEQK 178
Query: 437 NADSSKKKKKKSKKKDAE 454
+AD ++KKKKSKK + E
Sbjct: 179 SADRKERKKKKSKKNNDE 196
Score = 45.4 bits (106), Expect = 6e-05
Identities = 27/93 (29%), Positives = 52/93 (55%), Gaps = 6/93 (6%)
Query: 368 METEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGD--AEDHKSEKKKKKKEKRKLDQEV 425
+ E V ++ +KK K + D+ +K E + +E+ + KK+KKK+K D++V
Sbjct: 105 VSVESVYGRERDEKKMKKSKDADVVDEKVNEKLEAEQRSEERRERKKEKKKKKNNKDEDV 164
Query: 426 ---EVEDKVVEDGANADSSKKKKKK-SKKKDAE 454
+V++K+ ++ +AD ++KKKK K D +
Sbjct: 165 VDEKVKEKLEDEQKSADRKERKKKKSKKNNDED 197
Score = 43.5 bits (101), Expect = 2e-04
Identities = 35/125 (28%), Positives = 62/125 (49%), Gaps = 10/125 (8%)
Query: 332 EQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDM 391
E+V E+L+ + R+ K + +NKD ++ E+V K+ + +QK+AD +
Sbjct: 131 EKVNEKLEAEQRSEERRER---KKEKKKKKNNKDEDVVDEKV--KEKLEDEQKSADRKER 185
Query: 392 AVDKAAEITNGDAEDHKS-----EKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKK 446
K+ + + D D K +K + KEK+K E V++K E + S ++KK
Sbjct: 186 KKKKSKKNNDEDVVDEKEKLEDEQKSAEIKEKKKNKDEDVVDEKEKEKLEDEQRSGERKK 245
Query: 447 KSKKK 451
+ KKK
Sbjct: 246 EKKKK 250
Score = 35.0 bits (79), Expect = 0.085
Identities = 22/98 (22%), Positives = 46/98 (46%)
Query: 355 KSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKK 414
K S D + ET++ S K+++ DG + + ++ + ++KK
Sbjct: 61 KETKPSSDRETKSTETKQSSDAKSERNVIDEFDGRKIRYRNSEAVSVESVYGRERDEKKM 120
Query: 415 KKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
KK K + +V +K+ + + + ++KK+K KKK+
Sbjct: 121 KKSKDADVVDEKVNEKLEAEQRSEERRERKKEKKKKKN 158
>At1g72440 hypothetical protein
Length = 1056
Score = 52.8 bits (125), Expect = 4e-07
Identities = 56/240 (23%), Positives = 92/240 (38%), Gaps = 37/240 (15%)
Query: 244 HAGSLTNLAKCPSSTLQILGAEKALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMAR 303
H GS +K + +++GAE G+ L+FH ++ + ++ + + +
Sbjct: 823 HGGSQIEPSKKLDMSNRVIGAEILSL----AEGDVAPEDLVFHKFYVNKMTSTKQSKKKK 878
Query: 304 -------------YLANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKN 350
Y N D S+ A E E++E LD D +
Sbjct: 879 KKKLPEEEAAEELYDVNDGDGGENYD--SDVEFEAGDESDNEEIENMLDDVDDNAVEEEG 936
Query: 351 IDVMKSAIESVDNKDTEMETEEVSAKK--TKKKKQKAADGDDMAVDKAAEITNGD----A 404
+ ++ V +D E +VS + T D DD VD GD
Sbjct: 937 GEYDYDDLDGVAGEDDEELVADVSDAEMDTDMDMDLIDDEDDNNVDDDGTGDGGDDDSDG 996
Query: 405 EDHKSEKKKKKKEKRK------------LDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+D +S+KKKK+K KRK +DQ+ + + K + + KKKKKKSK +
Sbjct: 997 DDGRSKKKKKEKRKRKSPFASLEEYKHLIDQDEKEDSKTKRKATSEPTKKKKKKKSKASE 1056
>At5g65900 ATP-dependent RNA helicase-like
Length = 633
Score = 52.4 bits (124), Expect = 5e-07
Identities = 34/92 (36%), Positives = 44/92 (46%), Gaps = 10/92 (10%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
E D K+ +E KK KKK +K D DD + AE E KKKKK+
Sbjct: 44 EDGDAKENNALIDEEPKKKKKKKNKKRGDTDDGEDEAVAE----------EEPKKKKKKN 93
Query: 419 RKLDQEVEVEDKVVEDGANADSSKKKKKKSKK 450
+KL Q + D+ E A + KKKKKK +K
Sbjct: 94 KKLQQRGDTNDEEDEVIAEEEEPKKKKKKQRK 125
Score = 47.8 bits (112), Expect = 1e-05
Identities = 32/98 (32%), Positives = 54/98 (54%), Gaps = 12/98 (12%)
Query: 360 SVDNKDTEMETEEVSAKKTKKK-KQKAADGDDMAVDKAAEITNGDAE------DHKSEKK 412
++D + E EE+ KK KK+ + +A A+++ + +GDA+ D + +KK
Sbjct: 3 NLDMEQHSSENEEIKKKKHKKRARDEAKKLKQPAMEEEPDHEDGDAKENNALIDEEPKKK 62
Query: 413 KKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKK 450
KKKK K++ D + ++ V E + KKKKKK+KK
Sbjct: 63 KKKKNKKRGDTDDGEDEAVAE-----EEPKKKKKKNKK 95
Score = 38.9 bits (89), Expect = 0.006
Identities = 35/114 (30%), Positives = 53/114 (45%), Gaps = 9/114 (7%)
Query: 328 EKLREQVEERLDFYDKGVAPRKN--IDVMKSAIESVDNK---DTEMETEEVSAK---KTK 379
+KL++ E ++ G A N ID + NK DT+ +E A+ K K
Sbjct: 30 KKLKQPAMEEEPDHEDGDAKENNALIDEEPKKKKKKKNKKRGDTDDGEDEAVAEEEPKKK 89
Query: 380 KKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVE 433
KKK K D+ E+ + E+ K +KKK++K+ +E EVEDK E
Sbjct: 90 KKKNKKLQQRGDTNDEEDEVI-AEEEEPKKKKKKQRKDTEAKSEEEEVEDKEEE 142
Score = 34.7 bits (78), Expect = 0.11
Identities = 27/70 (38%), Positives = 34/70 (48%), Gaps = 17/70 (24%)
Query: 399 ITNGDAEDHKSE----KKKKKK-----EKRKLDQEVEVEDKVVEDG--------ANADSS 441
+ N D E H SE KKKK K E +KL Q E+ EDG + +
Sbjct: 1 MANLDMEQHSSENEEIKKKKHKKRARDEAKKLKQPAMEEEPDHEDGDAKENNALIDEEPK 60
Query: 442 KKKKKKSKKK 451
KKKKKK+KK+
Sbjct: 61 KKKKKKNKKR 70
Score = 30.4 bits (67), Expect = 2.1
Identities = 27/109 (24%), Positives = 51/109 (46%), Gaps = 12/109 (11%)
Query: 339 DFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAE 398
D D+ VA + K + DT E +EV A++ + KK+K D
Sbjct: 75 DGEDEAVAEEEPKKKKKKNKKLQQRGDTNDEEDEVIAEEEEPKKKKKKQRKD-------- 126
Query: 399 ITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKK 447
T +E+ E+ + K+E++KL++ + +K E + +D++ K K+
Sbjct: 127 -TEAKSEE---EEVEDKEEEKKLEETSIMTNKTFESLSLSDNTYKSIKE 171
>At3g57150 putative pseudouridine synthase (NAP57)
Length = 565
Score = 50.1 bits (118), Expect = 3e-06
Identities = 36/90 (40%), Positives = 49/90 (54%), Gaps = 20/90 (22%)
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K+ E E E K +KKKK+K DK E E+ SEKK+KKK+K K
Sbjct: 462 KEVEGEEAEEKVKSSKKKKKK---------DKEEE----KEEEAGSEKKEKKKKKDK--- 505
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKKKDA 453
+++V+E+ A+ S KKKKKKSK +A
Sbjct: 506 ----KEEVIEEVASPKSEKKKKKKSKDTEA 531
Score = 49.7 bits (117), Expect = 3e-06
Identities = 39/115 (33%), Positives = 56/115 (47%), Gaps = 18/115 (15%)
Query: 351 IDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMA----------VDKAAEIT 400
+ KS + V+ ++ E E+V + K KKKK K + ++ A DK E+
Sbjct: 454 VTTKKSKTKEVEGEEAE---EKVKSSKKKKKKDKEEEKEEEAGSEKKEKKKKKDKKEEVI 510
Query: 401 NGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKK-KKSKKKDAE 454
+ KSEKKKKKK K D E V+ + ++ KKKK KK K KD+E
Sbjct: 511 E-EVASPKSEKKKKKKSK---DTEAAVDAEDESAAEKSEKKKKKKDKKKKNKDSE 561
Score = 49.3 bits (116), Expect = 4e-06
Identities = 34/105 (32%), Positives = 55/105 (52%), Gaps = 6/105 (5%)
Query: 327 GEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMET-EEVSAKKTKKKKQKA 385
GE+ E+V+ K K + E KD + E EEV++ K++KKK+K
Sbjct: 466 GEEAEEKVKSSKKKKKKDKEEEKEEEAGSEKKEKKKKKDKKEEVIEEVASPKSEKKKKKK 525
Query: 386 ADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDK 430
+ + AVD E + KSEKKKKKK+K+K +++ E +++
Sbjct: 526 SKDTEAAVDAEDE-----SAAEKSEKKKKKKDKKKKNKDSEDDEE 565
>At5g41020 putative protein
Length = 588
Score = 49.3 bits (116), Expect = 4e-06
Identities = 38/115 (33%), Positives = 55/115 (47%), Gaps = 27/115 (23%)
Query: 342 DKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITN 401
DK RK D A E++D +EV K KKK +D +D+ +D +
Sbjct: 161 DKKKGKRKRDDTDLGAEENID--------KEVKRKNNKKKPSVDSDVEDINLDSTND--- 209
Query: 402 GDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA--DSSKKKKKKSKKKDAE 454
KKK+KK+K+ D E E E+G N+ D+ K++KKK KKK +E
Sbjct: 210 --------GKKKRKKKKQSEDSETE------ENGLNSTKDAKKRRKKKKKKKQSE 250
Score = 46.6 bits (109), Expect = 3e-05
Identities = 45/131 (34%), Positives = 56/131 (42%), Gaps = 20/131 (15%)
Query: 332 EQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETE----EVSAKKTKKKKQKAAD 387
E + E DK +KN K D + T E+E EV K KKK +K
Sbjct: 24 EFISEHSSMKDKEKKRKKNKRENKDGFTGEDMEITGRESEKLGDEVFIVKKKKKSKKPIR 83
Query: 388 GDDMAVD-------KAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADS 440
D AVD K ++ T D+E +KK KEK K E KV D D
Sbjct: 84 IDSEAVDAVKKKSKKRSKETKADSEAEDDGVEKKSKEKSK-------ETKV--DSEAHDG 134
Query: 441 SKKKKKKSKKK 451
K+KKKKSKK+
Sbjct: 135 VKRKKKKSKKE 145
Score = 44.3 bits (103), Expect = 1e-04
Identities = 32/123 (26%), Positives = 58/123 (47%), Gaps = 12/123 (9%)
Query: 332 EQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDM 391
E++ + + K +K I + A+++V K + +K+TK + DG +
Sbjct: 63 EKLGDEVFIVKKKKKSKKPIRIDSEAVDAVKKKSKKR------SKETKADSEAEDDGVEK 116
Query: 392 AVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
+ ++ T D+E H K+KKKK K+ E V+E+ ++ S KKK K K+
Sbjct: 117 KSKEKSKETKVDSEAHDGVKRKKKKSKK------ESGGDVIENTESSKVSDKKKGKRKRD 170
Query: 452 DAE 454
D +
Sbjct: 171 DTD 173
Score = 43.9 bits (102), Expect = 2e-04
Identities = 33/110 (30%), Positives = 53/110 (48%), Gaps = 12/110 (10%)
Query: 355 KSAIESVDNKDTEMETEEVSAKKTKKKKQKAAD-GDDMAVDKAAEITNG--------DAE 405
KS ES + E+ +VS KK K+K+ D G + +DK + N D E
Sbjct: 141 KSKKESGGDVIENTESSKVSDKKKGKRKRDDTDLGAEENIDKEVKRKNNKKKPSVDSDVE 200
Query: 406 D---HKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
D + KKK++K+K ++ E E+ + +A +KKKKK K+ +
Sbjct: 201 DINLDSTNDGKKKRKKKKQSEDSETEENGLNSTKDAKKRRKKKKKKKQSE 250
Score = 43.9 bits (102), Expect = 2e-04
Identities = 27/105 (25%), Positives = 53/105 (49%), Gaps = 13/105 (12%)
Query: 363 NKDTEMETEEVSAKKTKKKKQKAADGDDMAVD-------------KAAEITNGDAEDHKS 409
+K+T++++E K KKKK K G D+ + + + T+ AE++
Sbjct: 122 SKETKVDSEAHDGVKRKKKKSKKESGGDVIENTESSKVSDKKKGKRKRDDTDLGAEENID 181
Query: 410 EKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
++ K+K K+K + +VED ++ + +KKKK+S+ + E
Sbjct: 182 KEVKRKNNKKKPSVDSDVEDINLDSTNDGKKKRKKKKQSEDSETE 226
Score = 37.0 bits (84), Expect = 0.022
Identities = 26/110 (23%), Positives = 47/110 (42%), Gaps = 16/110 (14%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKK---- 414
E ++ + M+ +E KK K++ + G+DM + GD +KKK
Sbjct: 23 EEFISEHSSMKDKEKKRKKNKRENKDGFTGEDMEITGRESEKLGDEVFIVKKKKKSKKPI 82
Query: 415 ----------KKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
KK+ +K +E + + + +DG + K+K K K D+E
Sbjct: 83 RIDSEAVDAVKKKSKKRSKETKADSEAEDDG--VEKKSKEKSKETKVDSE 130
Score = 34.3 bits (77), Expect = 0.15
Identities = 22/76 (28%), Positives = 40/76 (51%), Gaps = 1/76 (1%)
Query: 376 KKTKKKKQKA-ADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVED 434
KK KKKK A D ++ + +E ++ ++ K +K K++ + +++E+ + E
Sbjct: 5 KKNKKKKSDAKVDSEETGEEFISEHSSMKDKEKKRKKNKRENKDGFTGEDMEITGRESEK 64
Query: 435 GANADSSKKKKKKSKK 450
+ KKKKKSKK
Sbjct: 65 LGDEVFIVKKKKKSKK 80
Score = 33.5 bits (75), Expect = 0.25
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query: 408 KSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKK-KSKKKD 452
+ +K KKKK K+D E E+ + E + D KK+KK K + KD
Sbjct: 3 EEKKNKKKKSDAKVDSEETGEEFISEHSSMKDKEKKRKKNKRENKD 48
Score = 30.0 bits (66), Expect = 2.7
Identities = 16/43 (37%), Positives = 25/43 (57%)
Query: 412 KKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
++KK +K+K D +V+ E+ E + S K K+KK KK E
Sbjct: 3 EEKKNKKKKSDAKVDSEETGEEFISEHSSMKDKEKKRKKNKRE 45
>At3g28770 hypothetical protein
Length = 2081
Score = 48.9 bits (115), Expect = 6e-06
Identities = 35/105 (33%), Positives = 57/105 (53%), Gaps = 10/105 (9%)
Query: 352 DVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNG--DAEDHKS 409
+ K +S D K E ++EE +K+KK+K+++ D + K E T ++E+HKS
Sbjct: 1025 EAKKEKKKSQDKKREEKDSEE---RKSKKEKEESRD---LKAKKKEEETKEKKESENHKS 1078
Query: 410 EKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+KK+ KKE + EDK ++ + SK +KK+ KKD E
Sbjct: 1079 KKKEDKKEHEDNKSMKKEEDK--KEKKKHEESKSRKKEEDKKDME 1121
Score = 47.4 bits (111), Expect = 2e-05
Identities = 46/147 (31%), Positives = 67/147 (45%), Gaps = 22/147 (14%)
Query: 328 EKLREQVEERLDFYDKGV---APRKNIDVMKSAIESVD---------NKDTEMETEEVSA 375
+K RE+V+ K V A +IDV K + ESV NK+ +T S+
Sbjct: 878 KKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTSS 937
Query: 376 K------KTKKKKQKAADGDDMAVDKAAEITN--GDAEDHKSEKKKKKKEKRKLDQEVEV 427
K K KKK+ K ++ DK + N ED+K E K + K K + +
Sbjct: 938 KQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNK 997
Query: 428 EDKVVEDGANADSSKK--KKKKSKKKD 452
E K ED A+ + KK ++KKSK K+
Sbjct: 998 EKKESEDSASKNREKKEYEEKKSKTKE 1024
Score = 45.4 bits (106), Expect = 6e-05
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 9/105 (8%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKK---- 414
E DNK + E ++ KK ++ K + + D ++K + + ++ K+EKKK
Sbjct: 1086 EHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVK 1145
Query: 415 --KKE---KRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
KKE K K + E + E K +E + + KK+K KD +
Sbjct: 1146 LVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQ 1190
Score = 44.7 bits (104), Expect = 1e-04
Identities = 36/142 (25%), Positives = 72/142 (50%), Gaps = 7/142 (4%)
Query: 319 SEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
SE S EK +E E++ ++ +K K + + + ++ E EE K
Sbjct: 1002 SEDSASKNREK-KEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKA 1060
Query: 379 KKKKQKAADGDDMAVDKAAEITNG-DAEDHKS-----EKKKKKKEKRKLDQEVEVEDKVV 432
KKK+++ + + K+ + + + ED+KS +KK+KKK + ++ E + K +
Sbjct: 1061 KKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDM 1120
Query: 433 EDGANADSSKKKKKKSKKKDAE 454
E + +S+KKK+ K++KK ++
Sbjct: 1121 EKLEDQNSNKKKEDKNEKKKSQ 1142
Score = 42.0 bits (97), Expect = 7e-04
Identities = 33/132 (25%), Positives = 58/132 (43%), Gaps = 6/132 (4%)
Query: 323 TSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKK 382
T + KL+E+ ++ + + + KN + E + K E + KK++ KK
Sbjct: 982 TKSENSKLKEENKDNKEKKESEDSASKN----REKKEYEEKKSKTKEEAKKEKKKSQDKK 1037
Query: 383 QKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSK 442
++ D ++ K E + K E+ K+KKE + + + K ED N K
Sbjct: 1038 REEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHED--NKSMKK 1095
Query: 443 KKKKKSKKKDAE 454
++ KK KKK E
Sbjct: 1096 EEDKKEKKKHEE 1107
Score = 42.0 bits (97), Expect = 7e-04
Identities = 34/160 (21%), Positives = 72/160 (44%), Gaps = 13/160 (8%)
Query: 305 LANKCSIASRIDCFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNK 364
+ +K S ++ G+S GE+ +E ++ ++ DK + +++ K +SVD+K
Sbjct: 673 MESKEDTKSEVEVKKNDGSSEKGEEGKENNKDSME--DKKLENKESQTDSKDD-KSVDDK 729
Query: 365 DTEME----------TEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKK 414
E + + E KK + K+ K ++ V E G+ ++ + +K +
Sbjct: 730 QEEAQIYGGESKDDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGE 789
Query: 415 KKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
KKE + ++K + N D +K++ + K+D E
Sbjct: 790 KKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKE 829
Score = 41.6 bits (96), Expect = 0.001
Identities = 35/132 (26%), Positives = 56/132 (41%), Gaps = 8/132 (6%)
Query: 327 GEKLREQVEERLDFYD-KGVAPRKNIDVMKSAIESVDNKDTEMETE-EVSAKKT---KKK 381
GE +E EE D+ + +N V + D+KD + + EV A K KKK
Sbjct: 821 GEDNKEDKEESKDYQSVEAKEKNENGGVDTNVGNKEDSKDLKDDRSVEVKANKEESMKKK 880
Query: 382 KQKAADGDDMAVDKAAEITNG---DAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA 438
+++ D + + + N D + E K KK+++K + E +D +
Sbjct: 881 REEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQK 940
Query: 439 DSSKKKKKKSKK 450
KKKKKK K
Sbjct: 941 GKDKKKKKKESK 952
Score = 41.6 bits (96), Expect = 0.001
Identities = 31/127 (24%), Positives = 65/127 (50%), Gaps = 4/127 (3%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKK--KQKA 385
E +E V L + + K E+ DNK+ + E+E+ ++K +KK ++K
Sbjct: 961 EDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKE-KKESEDSASKNREKKEYEEKK 1019
Query: 386 ADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK-LDQEVEVEDKVVEDGANADSSKKK 444
+ + A + + + E+ SE++K KKEK + D + + +++ ++ +++ K K
Sbjct: 1020 SKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSK 1079
Query: 445 KKKSKKK 451
KK+ KK+
Sbjct: 1080 KKEDKKE 1086
Score = 39.3 bits (90), Expect = 0.005
Identities = 29/125 (23%), Positives = 60/125 (47%), Gaps = 13/125 (10%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTE--METEEVSAKKTKKKKQKA 385
EKL +Q + +++ ++K + + K+ E ET+E+ + K++K +
Sbjct: 1121 EKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNE--- 1177
Query: 386 ADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKR-KLDQEVEVEDKVVEDGANADSSKKK 444
VDK + ++ D + K ++ K+ +EK+ K ++E + VE+ +KK+
Sbjct: 1178 -------VDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKE 1230
Query: 445 KKKSK 449
K K K
Sbjct: 1231 KNKPK 1235
Score = 38.1 bits (87), Expect = 0.010
Identities = 25/108 (23%), Positives = 53/108 (48%), Gaps = 4/108 (3%)
Query: 349 KNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHK 408
KN ++ K + + + E++ +E + K+T K + ++ D + + D+
Sbjct: 952 KNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENK--DNKEKKESEDSASKN 1009
Query: 409 SEKKKKKKEKRKLDQEVEVEDKVVEDGANA--DSSKKKKKKSKKKDAE 454
EKK+ +++K K +E + E K +D DS ++K KK K++ +
Sbjct: 1010 REKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRD 1057
Score = 38.1 bits (87), Expect = 0.010
Identities = 24/100 (24%), Positives = 52/100 (52%), Gaps = 8/100 (8%)
Query: 362 DNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKL 421
DNK ++E K+ K ++ + +D A K E + + K++++ KK++K+
Sbjct: 976 DNKKETTKSENSKLKEENKDNKEKKESEDSA-SKNREKKEYEEKKSKTKEEAKKEKKKSQ 1034
Query: 422 DQEVEVED-------KVVEDGANADSSKKKKKKSKKKDAE 454
D++ E +D K E+ + + KK+++ +KK++E
Sbjct: 1035 DKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESE 1074
Score = 37.7 bits (86), Expect = 0.013
Identities = 31/123 (25%), Positives = 60/123 (48%), Gaps = 27/123 (21%)
Query: 349 KNIDVMKSAIESVDNKDTEMETEEVSAK--------KTKKKKQKAADGDDMAVDKAAEIT 400
K+ + KS E +++D + + +E K K+KKK+ K D+ ++ K
Sbjct: 1041 KDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKE---- 1096
Query: 401 NGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA---DSSKKKK--------KKSK 449
ED K +KK ++ + RK +++ + +K+ + +N D ++KKK K+S
Sbjct: 1097 ----EDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESD 1152
Query: 450 KKD 452
KK+
Sbjct: 1153 KKE 1155
Score = 36.6 bits (83), Expect = 0.029
Identities = 26/136 (19%), Positives = 65/136 (47%), Gaps = 11/136 (8%)
Query: 319 SEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKT 378
+EK S + ++++ +++ ++ + K I+ KS VD K+ + ++ +K
Sbjct: 1136 NEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQ---QKK 1192
Query: 379 KKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA 438
K+K+ K ++ + ++ E++K +K+ KK++ + D + ++
Sbjct: 1193 KEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDK--------KNTTKQ 1244
Query: 439 DSSKKKKKKSKKKDAE 454
KK+ +S+ K+AE
Sbjct: 1245 SGGKKESMESESKEAE 1260
Score = 35.8 bits (81), Expect = 0.050
Identities = 20/98 (20%), Positives = 51/98 (51%), Gaps = 5/98 (5%)
Query: 362 DNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNG-----DAEDHKSEKKKKKK 416
++K+ + +++ V K + K++ + ++ + K E + D ++ KS K ++KK
Sbjct: 1133 EDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKK 1192
Query: 417 EKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+++++ + E + K E+ +S ++ KK K+ E
Sbjct: 1193 KEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKE 1230
Score = 34.7 bits (78), Expect = 0.11
Identities = 29/144 (20%), Positives = 65/144 (45%), Gaps = 24/144 (16%)
Query: 331 REQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKK-------TKKKKQ 383
+E+ ++ +++ + +K D K +E ++++++ + E+ + KK KK+
Sbjct: 1095 KEEDKKEKKKHEESKSRKKEED--KKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESD 1152
Query: 384 KAADGDDMAVDKAAEITNGDAEDHKSEKKKKK---------------KEKRKLDQEVEVE 428
K ++ + EI + ++ ++ +KK+KK E++KL + E
Sbjct: 1153 KKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDR 1212
Query: 429 DKVVEDGANADSSKKKKKKSKKKD 452
K N + KK+K+K KD
Sbjct: 1213 KKQTSVEENKKQKETKKEKNKPKD 1236
Score = 32.3 bits (72), Expect = 0.55
Identities = 33/161 (20%), Positives = 71/161 (43%), Gaps = 38/161 (23%)
Query: 328 EKLREQVEERLDFY-DKGVAPRKNIDVMKSAI------ESVDNKDT---EMETEEVSAKK 377
E+ RE+ + + Y +K +N+++ +I S++NK+ +++ E
Sbjct: 477 EEKREESQRSNEVYMNKETTKGENVNIQGESIGDSTKDNSLENKEDVKPKVDANESDGNS 536
Query: 378 TKKKKQKAADGDDMAV----------------DKAAEITNGDAEDHKSEKK--------- 412
TK++ Q+A + ++ DK+ E+T D DH EK+
Sbjct: 537 TKERHQEAQVNNGVSTEDKNLDNIGADEQKKNDKSVEVTTNDG-DHTKEKREETQGNNGE 595
Query: 413 --KKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
K + + K D++ +D+ V N ++S ++K++ +K
Sbjct: 596 SVKNENLENKEDKKELKDDESVGAKTNNETSLEEKREQTQK 636
Score = 32.3 bits (72), Expect = 0.55
Identities = 41/176 (23%), Positives = 69/176 (38%), Gaps = 46/176 (26%)
Query: 321 KGTSAFGEKLREQ--VEERLDFYDKGVAPRKNIDVMKSAIESVD-NKDTEMET------- 370
K + G K + +EE+ + KG N ++ + + D NK+ E+
Sbjct: 612 KDDESVGAKTNNETSLEEKREQTQKGHDNSINSKIVDNKGGNADSNKEKEVHVGDSTNDN 671
Query: 371 ---------EEVSAKKTKKKKQKAADG-----------------------DDMAVD---K 395
EV KK +K +G DD +VD +
Sbjct: 672 NMESKEDTKSEVEVKKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDSKDDKSVDDKQE 731
Query: 396 AAEITNGDAEDHKS-EKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKK 450
A+I G+++D KS E K KKKE ++ + E++V N +KK+ +K +K
Sbjct: 732 EAQIYGGESKDDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEK 787
Score = 29.3 bits (64), Expect = 4.7
Identities = 24/128 (18%), Positives = 47/128 (35%), Gaps = 4/128 (3%)
Query: 329 KLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADG 388
K + VEE + K D K+ + K ME+E A+ +K +
Sbjct: 1213 KKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQAD 1272
Query: 389 DDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKL----DQEVEVEDKVVEDGANADSSKKK 444
D + ++ + A+ H + + K ++ D + + ED S +
Sbjct: 1273 SDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAEN 1332
Query: 445 KKKSKKKD 452
KK+ + K+
Sbjct: 1333 KKQKETKE 1340
Score = 29.3 bits (64), Expect = 4.7
Identities = 26/136 (19%), Positives = 52/136 (38%), Gaps = 1/136 (0%)
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
E S E +++ + D K K + K D K E K+TK
Sbjct: 1169 ESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETK 1228
Query: 380 KKKQKAADG-DDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA 438
K+K K D + + + ++E ++E ++K + + D + + +++ + A
Sbjct: 1229 KEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQA 1288
Query: 439 DSSKKKKKKSKKKDAE 454
DS + S + E
Sbjct: 1289 DSHSDSQADSDESKNE 1304
Score = 29.3 bits (64), Expect = 4.7
Identities = 27/122 (22%), Positives = 48/122 (39%), Gaps = 23/122 (18%)
Query: 354 MKSAIESVDNKDTEMETEEVSAKKTKKK------------KQKAADGDD------MAVDK 395
M+S + +N+ T + + ++K + AD D+ M D
Sbjct: 1363 MESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADS 1422
Query: 396 AAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA---DSSKKKKKKSKKKD 452
A + ED K K+ E +K + E ++K +D N KK+ +S+ K+
Sbjct: 1423 QATTQRNNEEDRK--KQTSVAENKKQKETKEEKNKPKDDKKNTTEQSGGKKESMESESKE 1480
Query: 453 AE 454
AE
Sbjct: 1481 AE 1482
Score = 29.3 bits (64), Expect = 4.7
Identities = 27/122 (22%), Positives = 48/122 (39%), Gaps = 23/122 (18%)
Query: 354 MKSAIESVDNKDTEMETEEVSAKKTKKK------------KQKAADGDD------MAVDK 395
M+S + +N+ T + + ++K + AD D+ M D
Sbjct: 1252 MESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADS 1311
Query: 396 AAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANA---DSSKKKKKKSKKKD 452
A + ED K K+ E +K + E ++K +D N KK+ +S+ K+
Sbjct: 1312 QATTQRNNEEDRK--KQTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKE 1369
Query: 453 AE 454
AE
Sbjct: 1370 AE 1371
>At1g56660 hypothetical protein
Length = 522
Score = 48.9 bits (115), Expect = 6e-06
Identities = 41/140 (29%), Positives = 67/140 (47%), Gaps = 16/140 (11%)
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
EK A EK E V + + ++ D K+ + D TE + ++ KK K
Sbjct: 148 EKNKKADKEKKHEDVSQEKEELEEE-------DGKKNKKKEKDESGTEEKKKK--PKKEK 198
Query: 380 KKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVED-----KVVED 434
K+K+++ +D V E G+ D + E ++KKKE + DQE++ +D K +D
Sbjct: 199 KQKEESKSNEDKKVKGKKE--KGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKD 256
Query: 435 GANADSSKKKKKKSKKKDAE 454
+ A+ KKK K KK+ E
Sbjct: 257 ESCAEEKKKKPDKEKKEKDE 276
Score = 48.5 bits (114), Expect = 7e-06
Identities = 31/95 (32%), Positives = 52/95 (54%), Gaps = 4/95 (4%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGD-DMAVDKAAEITNGDAEDHKSEKKKKKKE 417
E+ D D E ET++ KK KKK++K+ G+ D+ DK E + K ++ +
Sbjct: 347 ETKDKDDDEGETKQ---KKNKKKEKKSEKGEKDVKEDKKKENPLETEVMSRDIKLEEPEA 403
Query: 418 KRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
++K + + E + K +G ++ KKKKKK KKK+
Sbjct: 404 EKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKN 438
Score = 48.5 bits (114), Expect = 7e-06
Identities = 47/161 (29%), Positives = 69/161 (42%), Gaps = 29/161 (18%)
Query: 321 KGTSAFGEK--LREQVEERLDFYDKGVAPRKNIDVMKSAIESVDN--------------- 363
KG GEK L ++ EE+ +D+ K D K+ + D
Sbjct: 213 KGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKDESCAEEKKKKPDKEKK 272
Query: 364 -KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEIT----NGDAEDHKSEKKKKKKEK 418
KD E E+ K K K +K D+ K + T + +A DHK KKKK K+K
Sbjct: 273 EKDESTEKEDKKLKGKKGKGEKPEKEDEGKKTKEHDATEQEMDDEAADHKEGKKKKNKDK 332
Query: 419 RK-----LDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
K +D+ E E K +D + +K+KK K K+K +E
Sbjct: 333 AKKKETVIDEVCEKETKDKDD--DEGETKQKKNKKKEKKSE 371
Score = 44.3 bits (103), Expect = 1e-04
Identities = 31/111 (27%), Positives = 53/111 (46%), Gaps = 14/111 (12%)
Query: 349 KNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHK 408
K DV E K E + EE+ +K KKK+ + D+ ++ ++ K
Sbjct: 102 KESDVKVEEHEKEHKKGKEKKHEELEEEKEGKKKKNKKEKDESGPEE---------KNKK 152
Query: 409 SEKKKKKKEKRKLDQEVEVED-----KVVEDGANADSSKKKKKKSKKKDAE 454
++K+KK ++ + +E+E ED K +D + + KKK KK KK+ E
Sbjct: 153 ADKEKKHEDVSQEKEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKKQKEE 203
Score = 43.5 bits (101), Expect = 2e-04
Identities = 39/137 (28%), Positives = 65/137 (46%), Gaps = 10/137 (7%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNI----DVMKSAIESVDNKDTEMETEEVSAKKTKKKKQ 383
EK E++EE + K K+ + K A + ++D E EE+ + KK K+
Sbjct: 120 EKKHEELEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKHEDVSQEKEELEEEDGKKNKK 179
Query: 384 KAADG----DDMAVDKAAEITNGDAEDHKSEKKKKKKEK-RKLDQEVEVEDKVVE-DGAN 437
K D + K + +++ ++ +K K KKEK K D E E E+K E D +
Sbjct: 180 KEKDESGTEEKKKKPKKEKKQKEESKSNEDKKVKGKKEKGEKGDLEKEDEEKKKEHDETD 239
Query: 438 ADSSKKKKKKSKKKDAE 454
+ +K KK+KKK+ +
Sbjct: 240 QEMKEKDSKKNKKKEKD 256
Score = 43.5 bits (101), Expect = 2e-04
Identities = 29/99 (29%), Positives = 45/99 (45%), Gaps = 3/99 (3%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADG---DDMAVDKAAEITNGDAEDHKSEKKKKK 415
E +N + EME + S +K K KK + + G D K + + ED +KKK
Sbjct: 26 EKGENVEVEMEVKAKSIEKVKAKKDEESSGKSKKDKEKKKGKNVDSEVKEDKDDDKKKDG 85
Query: 416 KEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
K K +E + +V E + +K+ KK K+K E
Sbjct: 86 KMVSKKHEEGHGDLEVKESDVKVEEHEKEHKKGKEKKHE 124
Score = 43.5 bits (101), Expect = 2e-04
Identities = 35/128 (27%), Positives = 61/128 (47%), Gaps = 9/128 (7%)
Query: 328 EKLREQVEERLDFYDKGVAP-RKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAA 386
+K ++ E++ + +K V +K + +++ + S D K E E E+ T++KK+
Sbjct: 360 QKKNKKKEKKSEKGEKDVKEDKKKENPLETEVMSRDIKLEEPEAEKKEEDDTEEKKKSKV 419
Query: 387 DGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVED---KVVEDGANADSSKK 443
+G + K + +D K KKK KE + + E E +D V +G+ A KK
Sbjct: 420 EGGESEEGKKKK-----KKDKKKNKKKDTKEPKMTEDEEEKKDDSKDVKIEGSKAKEEKK 474
Query: 444 KKKKSKKK 451
K KKK
Sbjct: 475 DKDVKKKK 482
Score = 43.1 bits (100), Expect = 3e-04
Identities = 50/160 (31%), Positives = 70/160 (43%), Gaps = 25/160 (15%)
Query: 319 SEKGTSAFGEKLREQVE-ERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKK 377
S +A EKL +++ + LD +KG ++V +IE V K E E+ S K
Sbjct: 2 SSNQENAKEEKLHVKIKTQELDPKEKGENVEVEMEVKAKSIEKVKAKKDE-ESSGKSKKD 60
Query: 378 TKKKKQKAADG------------DDMAVDKAAEITNGDAEDHKS-------EKKKKK-KE 417
+KKK K D D V K E +GD E +S EK+ KK KE
Sbjct: 61 KEKKKGKNVDSEVKEDKDDDKKKDGKMVSKKHEEGHGDLEVKESDVKVEEHEKEHKKGKE 120
Query: 418 KRKLDQEVEVEDKVVEDGANADSS---KKKKKKSKKKDAE 454
K+ + E E E K ++ D S +K KK K+K E
Sbjct: 121 KKHEELEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKHE 160
Score = 39.3 bits (90), Expect = 0.005
Identities = 24/95 (25%), Positives = 51/95 (53%), Gaps = 4/95 (4%)
Query: 364 KDTEMETEEVSAK-KTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKK---KKKEKR 419
K+ +ETE +S K ++ + + + DD K +++ G++E+ K +KKK K K+K
Sbjct: 383 KENPLETEVMSRDIKLEEPEAEKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKD 442
Query: 420 KLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+ ++ +++ +D + + K K +KKD +
Sbjct: 443 TKEPKMTEDEEEKKDDSKDVKIEGSKAKEEKKDKD 477
Score = 32.3 bits (72), Expect = 0.55
Identities = 29/105 (27%), Positives = 49/105 (46%), Gaps = 6/105 (5%)
Query: 349 KNIDVMKSAIESVDNKDTE----METEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDA 404
++I + + E + DTE + E +++ KKKK+K + K ++T D
Sbjct: 394 RDIKLEEPEAEKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKDTKEPKMTE-DE 452
Query: 405 EDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSK 449
E+ K + K K E K +E + +D + G N D K K K +K
Sbjct: 453 EEKKDDSKDVKIEGSKAKEEKKDKDVKKKKGGN-DIGKLKTKLAK 496
>At1g76810 translation initiation factor IF-2 like protein
Length = 1280
Score = 47.4 bits (111), Expect = 2e-05
Identities = 33/132 (25%), Positives = 63/132 (47%), Gaps = 10/132 (7%)
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
E+ T+A +K +++ +E+ K +A SV+ K+ + E + K
Sbjct: 347 EEETAAAKKKKKKKEKEK----------EKKAAAAAAATSSVEVKEEKQEESVTEPLQPK 396
Query: 380 KKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANAD 439
KK K + E+ A ++E++KKK+E+ KL +E E + E A A+
Sbjct: 397 KKDAKGKAAEKKIPKHVREMQEALARRQEAEERKKKEEEEKLRKEEEERRRQEELEAQAE 456
Query: 440 SSKKKKKKSKKK 451
+K+K+K+ +K+
Sbjct: 457 EAKRKRKEKEKE 468
Score = 37.4 bits (85), Expect = 0.017
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 12/100 (12%)
Query: 367 EMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSE---------KKKKKKE 417
E E EE +A KKKK+K + + A AA ++ + ++ K E KKK K
Sbjct: 343 EKEGEEETAAAKKKKKKKEKEKEKKAAAAAAATSSVEVKEEKQEESVTEPLQPKKKDAKG 402
Query: 418 K---RKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
K +K+ + V + + A+ KKK+++ K + E
Sbjct: 403 KAAEKKIPKHVREMQEALARRQEAEERKKKEEEEKLRKEE 442
Score = 36.2 bits (82), Expect = 0.038
Identities = 27/82 (32%), Positives = 42/82 (50%), Gaps = 10/82 (12%)
Query: 382 KQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRK---------LDQEVEVEDKVV 432
++KAA + +A + A G+ E ++KKKKKKEK K VEV+++
Sbjct: 326 EEKAAQPEPVAPVENAGEKEGEEETAAAKKKKKKKEKEKEKKAAAAAAATSSVEVKEEKQ 385
Query: 433 EDGANADSSKKKKKKSKKKDAE 454
E+ + + KKK +K K AE
Sbjct: 386 EESV-TEPLQPKKKDAKGKAAE 406
Score = 35.8 bits (81), Expect = 0.050
Identities = 23/94 (24%), Positives = 45/94 (47%)
Query: 356 SAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKK 415
S++E + K E TE + KK K + A V + E E + +KK+++
Sbjct: 376 SSVEVKEEKQEESVTEPLQPKKKDAKGKAAEKKIPKHVREMQEALARRQEAEERKKKEEE 435
Query: 416 KEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSK 449
++ RK ++E ++++ A +K+K+K K
Sbjct: 436 EKLRKEEEERRRQEELEAQAEEAKRKRKEKEKEK 469
Score = 33.9 bits (76), Expect = 0.19
Identities = 24/104 (23%), Positives = 47/104 (45%), Gaps = 1/104 (0%)
Query: 352 DVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKA-AEITNGDAEDHKSE 410
D + ++ D K+ E+ S KK K K ++ +DK A + A + +
Sbjct: 262 DSVADETKTSDTKNVEVVETGKSKKKKKNNKSGRTVQEEEDLDKLLAALGETPAAERPAS 321
Query: 411 KKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+++ + + VE+ ++G ++ KKKKK K+K+ E
Sbjct: 322 STPVEEKAAQPEPVAPVENAGEKEGEEETAAAKKKKKKKEKEKE 365
Score = 33.1 bits (74), Expect = 0.32
Identities = 30/128 (23%), Positives = 54/128 (41%), Gaps = 27/128 (21%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKK---------- 377
EKLR++ EER + + +++ E K E E E++ KK
Sbjct: 436 EKLRKEEEER-----------RRQEELEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTAKQ 484
Query: 378 -TKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEK-----KKKKKEKRKLDQEVEVEDKV 431
T+ +K++A +A + + D + S++ KKK ++ +D V+ ED+V
Sbjct: 485 KTEAQKREAFKNQLLAAGGGLPVADNDGDATSSKRPIYANKKKSSRQKGIDTSVQGEDEV 544
Query: 432 VEDGANAD 439
AD
Sbjct: 545 EPKENQAD 552
Score = 32.0 bits (71), Expect = 0.72
Identities = 24/81 (29%), Positives = 40/81 (48%), Gaps = 8/81 (9%)
Query: 357 AIESVDNKDT--EMETEEVSAKKTKKKKQK------AADGDDMAVDKAAEITNGDAEDHK 408
A S D+++T + E+ EV+ KK +K A+ GDD D+ + E +
Sbjct: 221 ASNSRDDENTIEDEESPEVTFSGKKKSSKKKGGSVLASVGDDSVADETKTSDTKNVEVVE 280
Query: 409 SEKKKKKKEKRKLDQEVEVED 429
+ K KKKK+ K + V+ E+
Sbjct: 281 TGKSKKKKKNNKSGRTVQEEE 301
Score = 30.8 bits (68), Expect = 1.6
Identities = 31/125 (24%), Positives = 59/125 (46%), Gaps = 13/125 (10%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEME---TEEVSAKKTKKKKQK 384
EK E V E L P+K K+A + + EM+ A++ KKK+++
Sbjct: 383 EKQEESVTEPLQ-------PKKKDAKGKAAEKKIPKHVREMQEALARRQEAEERKKKEEE 435
Query: 385 AADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKK 444
+ + E AE+ K +K+K+KEK KL ++ ++E K++ ++ K++
Sbjct: 436 EKLRKEEEERRRQEELEAQAEEAK--RKRKEKEKEKLLRK-KLEGKLLTAKQKTEAQKRE 492
Query: 445 KKKSK 449
K++
Sbjct: 493 AFKNQ 497
>At1g76720 putative translation initiation factor IF-2
Length = 1224
Score = 46.6 bits (109), Expect = 3e-05
Identities = 31/96 (32%), Positives = 53/96 (54%), Gaps = 3/96 (3%)
Query: 356 SAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKK 415
S++E+ + K E TE + KK K K KAA+ E+ A ++E++KKK
Sbjct: 352 SSVEAKEEKQEESVTEPLQPKK-KDAKGKAAE--KKIPKHVREMQEALARRQEAEERKKK 408
Query: 416 KEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
+E+ KL +E E + E A A+ +K+K+K+ +K+
Sbjct: 409 EEEEKLRKEEEERRRQEELEAQAEEAKRKRKEKEKE 444
Score = 32.7 bits (73), Expect = 0.42
Identities = 34/124 (27%), Positives = 57/124 (45%), Gaps = 19/124 (15%)
Query: 348 RKNIDVMKSAIESVDNKDT------EMETEEVSAKKTKK-KKQKAADGDDMAVDKAAEIT 400
+KN + + +E D+ D E E A T + +K +A G V+ A E
Sbjct: 261 KKNKNKVARTLEEEDDLDKLLAELGETPAAERPASSTPEVEKVQAQPGPVAPVENAGE-K 319
Query: 401 NGDAEDHKSEKKKKKKEKRKLDQE----------VEVEDKVVEDGANADSSKKKKKKSKK 450
G+ E ++ KKKK+K++ D+E VE +++ E+ + + KKK +K
Sbjct: 320 EGEKETVETAAAKKKKKKKEKDKEKKAAAAATSSVEAKEEKQEESV-TEPLQPKKKDAKG 378
Query: 451 KDAE 454
K AE
Sbjct: 379 KAAE 382
Score = 32.3 bits (72), Expect = 0.55
Identities = 31/120 (25%), Positives = 52/120 (42%), Gaps = 27/120 (22%)
Query: 328 EKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKK---------- 377
EKLR++ EER + + +++ E K E E E++ KK
Sbjct: 412 EKLRKEEEER-----------RRQEELEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTAKQ 460
Query: 378 -TKKKKQKAADGDDMAVDKAAEITN--GDAEDHKSE---KKKKKKEKRKLDQEVEVEDKV 431
T+ +K++A +A + + + GDA K KKK ++ D V+VED+V
Sbjct: 461 KTEAQKREAFKNQLLAAGRGLPVADDVGDATSSKRPIYANKKKPSRQKGNDTSVQVEDEV 520
Score = 30.4 bits (67), Expect = 2.1
Identities = 29/107 (27%), Positives = 43/107 (40%), Gaps = 14/107 (13%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
E VD + ++ S KKT+ K + +A+ E T GD E + KKK
Sbjct: 164 EEVDGDEEQVSPITFSGKKTRSSKSSKKTTNKVALPDEEEGTMGDEESLEITFSGKKKGS 223
Query: 419 RKLDQEVEVEDKVVEDGANADSSK------------KKKKKSKKKDA 453
L +D V ++ + A + KKKKK+K K A
Sbjct: 224 IVLAS--LGDDSVADETSQAKTPDTKSVEVIETGKIKKKKKNKNKVA 268
Score = 30.4 bits (67), Expect = 2.1
Identities = 23/131 (17%), Positives = 63/131 (47%), Gaps = 2/131 (1%)
Query: 320 EKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTK 379
+K +A + + E++ + + + P+K K+A + + EM+ E ++ ++
Sbjct: 344 KKAAAAATSSVEAKEEKQEESVTEPLQPKKKDAKGKAAEKKIPKHVREMQ-EALARRQEA 402
Query: 380 KKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLD-QEVEVEDKVVEDGANA 438
++++K + + + ++ + E E K+K+KEK K ++E K++
Sbjct: 403 EERKKKEEEEKLRKEEEERRRQEELEAQAEEAKRKRKEKEKEKLLRKKLEGKLLTAKQKT 462
Query: 439 DSSKKKKKKSK 449
++ K++ K++
Sbjct: 463 EAQKREAFKNQ 473
Score = 28.9 bits (63), Expect = 6.1
Identities = 22/72 (30%), Positives = 34/72 (46%), Gaps = 5/72 (6%)
Query: 380 KKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANAD 439
+ K+KA D+ DK + T + +KK KK +K D + + + V E+G
Sbjct: 37 ESKKKAVQSDEE--DKYSINTEEEKVVITGKKKSNKKVTQKHDDDDDFTEAVPENGF--- 91
Query: 440 SSKKKKKKSKKK 451
KKKK K K +
Sbjct: 92 VGKKKKSKGKNR 103
Score = 28.5 bits (62), Expect = 8.0
Identities = 21/72 (29%), Positives = 34/72 (47%), Gaps = 12/72 (16%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAA------EITNG------DAED 406
+S DN D +++ +E A+ KK+ KA D D A ++T G + ED
Sbjct: 573 KSWDNVDLKIDDKEEEAQPVVKKELKAHDSDHETEKPTAKPAGMSKLTTGAVKAISEVED 632
Query: 407 HKSEKKKKKKEK 418
++ K+ KK K
Sbjct: 633 AATQTKRAKKGK 644
>At5g40340 unknown protein
Length = 1008
Score = 46.2 bits (108), Expect = 4e-05
Identities = 31/103 (30%), Positives = 56/103 (54%), Gaps = 13/103 (12%)
Query: 355 KSAIESVDNKDTEMETEEVSAKKTKKKK---QKAADGDDMAVDKAAEITNGDAEDHKSEK 411
+S E + ++T+ E E + K+ K+KK +K +DG++ + +E T K E+
Sbjct: 738 ESKKEGGEGEETQKEANESTKKERKRKKSESKKQSDGEEETQKEPSEST-------KKER 790
Query: 412 KKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
K+K E +K + VE E+ E + +S+KK++K+ K K E
Sbjct: 791 KRKNPESKKKAEAVEEEETRKE---SVESTKKERKRKKPKHDE 830
Score = 42.4 bits (98), Expect = 5e-04
Identities = 31/97 (31%), Positives = 48/97 (48%), Gaps = 3/97 (3%)
Query: 360 SVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEK--KKKKKE 417
S D+ D E E K K K+K D A KA EI E+++++K K KK+
Sbjct: 674 SYDSSDKEKEELSEMGKPVTKGKEKK-DKKGKAKQKAEEIEVTGKEENETDKHGKMKKER 732
Query: 418 KRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
KRK + + + E A+ S KK++K KK +++
Sbjct: 733 KRKKSESKKEGGEGEETQKEANESTKKERKRKKSESK 769
Score = 40.0 bits (92), Expect = 0.003
Identities = 37/174 (21%), Positives = 74/174 (42%), Gaps = 10/174 (5%)
Query: 273 KTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTSAFGEKLRE 332
K T K+G + ++ ++ +G + + +++ + +K S E
Sbjct: 717 KEENETDKHGKMKKERKRKKSESKKEGGEGEETQKEANESTKKERKRKKSESKKQSDGEE 776
Query: 333 QVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMA 392
+ ++ K RKN + K A E+V+ ++T E+ E + K+ K+KK K +
Sbjct: 777 ETQKEPSESTKKERKRKNPESKKKA-EAVEEEETRKESVESTKKERKRKKPKHDE----- 830
Query: 393 VDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKK 446
E+ N + K +KKK++ + +K + E E + SS KK+
Sbjct: 831 ----EEVPNETEKPEKKKKKKREGKSKKKETETEFSGAELYVTFGPGSSLPKKE 880
Score = 39.3 bits (90), Expect = 0.005
Identities = 25/96 (26%), Positives = 47/96 (48%), Gaps = 4/96 (4%)
Query: 359 ESVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
ES D E ET++ ++ TKK++++ + E ++K++K+K+
Sbjct: 767 ESKKQSDGEEETQKEPSESTKKERKRKNPESKKKAEAVEEEETRKESVESTKKERKRKKP 826
Query: 419 RKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
+ ++EV E + E KK++ KSKKK+ E
Sbjct: 827 KHDEEEVPNETEKPE----KKKKKKREGKSKKKETE 858
Score = 38.5 bits (88), Expect = 0.008
Identities = 33/138 (23%), Positives = 53/138 (37%), Gaps = 13/138 (9%)
Query: 322 GTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVD-----NKDTEMETEEVSAK 376
G+ +K +E++ E KG + K E ++ +T+ + +
Sbjct: 673 GSYDSSDKEKEELSEMGKPVTKGKEKKDKKGKAKQKAEEIEVTGKEENETDKHGKMKKER 732
Query: 377 KTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGA 436
K KK + K G+ K A E K E+K+KK E +K E K E
Sbjct: 733 KRKKSESKKEGGEGEETQKEAN------ESTKKERKRKKSESKKQSDGEEETQK--EPSE 784
Query: 437 NADSSKKKKKKSKKKDAE 454
+ +K+K KK AE
Sbjct: 785 STKKERKRKNPESKKKAE 802
>At5g62190 putative RNA helicase (MMI9.2)
Length = 671
Score = 45.8 bits (107), Expect = 5e-05
Identities = 28/87 (32%), Positives = 46/87 (52%), Gaps = 21/87 (24%)
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K ++T E+ +KK KK+++ ++++ D E+ + +K KKK +KRK +
Sbjct: 17 KKMALDTPELDSKKGKKEQK-------------LKLSDSDEEESEKKKSKKKDKKRKASE 63
Query: 424 EVEVEDKVVEDGANADSSKKKKKKSKK 450
E ED +DSS +KKK SKK
Sbjct: 64 E--------EDEVKSDSSSEKKKSSKK 82
Score = 32.7 bits (73), Expect = 0.42
Identities = 22/66 (33%), Positives = 35/66 (52%), Gaps = 9/66 (13%)
Query: 364 KDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ 423
K ++ + EE KK+KKK +K KA+E + D SEKKK K+ + +
Sbjct: 38 KLSDSDEEESEKKKSKKKDKKR---------KASEEEDEVKSDSSSEKKKSSKKVKLGVE 88
Query: 424 EVEVED 429
+VEV++
Sbjct: 89 DVEVDN 94
Score = 28.9 bits (63), Expect = 6.1
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 4/50 (8%)
Query: 406 DHKSEKKKKKK---EKRKLDQEV-EVEDKVVEDGANADSSKKKKKKSKKK 451
D K EKK KKK + +LD + + E K+ ++ + S+KKK K K K
Sbjct: 8 DKKEEKKMKKKMALDTPELDSKKGKKEQKLKLSDSDEEESEKKKSKKKDK 57
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,692,329
Number of Sequences: 26719
Number of extensions: 440145
Number of successful extensions: 6419
Number of sequences better than 10.0: 504
Number of HSP's better than 10.0 without gapping: 171
Number of HSP's successfully gapped in prelim test: 341
Number of HSP's that attempted gapping in prelim test: 3159
Number of HSP's gapped (non-prelim): 1839
length of query: 454
length of database: 11,318,596
effective HSP length: 103
effective length of query: 351
effective length of database: 8,566,539
effective search space: 3006855189
effective search space used: 3006855189
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC135505.8


