

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.7 - phase: 0 /pseudo
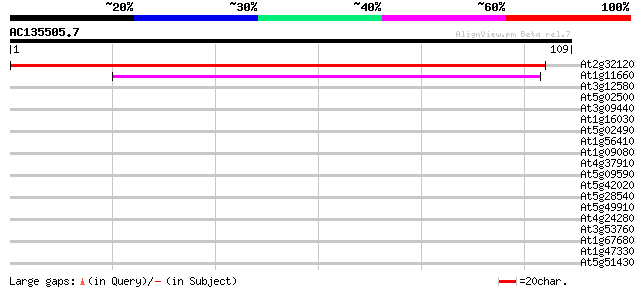
(109 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g32120 70kD heat shock protein 107 1e-24
At1g11660 heat-shock protein like 38 7e-04
At3g12580 putative protein 37 0.002
At5g02500 dnaK-type molecular chaperone hsc70.1 36 0.003
At3g09440 heat-shock protein (At-hsc70-3) 36 0.003
At1g16030 unknown protein 35 0.008
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 34 0.010
At1g56410 hypothetical protein 34 0.010
At1g09080 putative luminal binding protein 34 0.010
At4g37910 heat shock protein 70 like protein 34 0.014
At5g09590 heat shock protein 70 (Hsc70-5) 33 0.018
At5g42020 luminal binding protein (dbj|BAA13948.1) 32 0.040
At5g28540 luminal binding protein 32 0.040
At5g49910 heat shock protein 70 (Hsc70-7) 31 0.12
At4g24280 hsp 70-like protein 31 0.12
At3g53760 putative protein 26 2.9
At1g67680 putative protein 26 2.9
At1g47330 unknown protein 25 6.4
At5g51430 putative protein 25 8.3
>At2g32120 70kD heat shock protein
Length = 563
Score = 107 bits (267), Expect = 1e-24
Identities = 56/104 (53%), Positives = 73/104 (69%)
Query: 1 MHTLDIKVRPFIAAIANKACRSTTAVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFT 60
+ TLDI VRPFIAA+ N A RSTT E+L FLV+L+ E ++K P+R VV TVPVSF+
Sbjct: 119 VQTLDIGVRPFIAALVNNAWRSTTPEEVLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFS 178
Query: 61 RLRRTQIERASAWADLDDVELMPQPIAVALFYAQQQLQTSASSL 104
R + T+ ERA A A L + LMP+P A+AL YAQQQ T+ ++
Sbjct: 179 RFQLTRFERACAMAGLHVLRLMPEPTAIALLYAQQQQMTTHDNM 222
>At1g11660 heat-shock protein like
Length = 773
Score = 38.1 bits (87), Expect = 7e-04
Identities = 27/83 (32%), Positives = 39/83 (46%)
Query: 21 RSTTAVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVE 80
+S + V++LG L LK E +K P+ V +P FT +R A+A A L +
Sbjct: 110 QSFSPVQILGMLLSHLKQIAEKSLKTPVSDCVIGIPSYFTNSQRLAYLDAAAIAGLRPLR 169
Query: 81 LMPQPIAVALFYAQQQLQTSASS 103
LM A AL Y + A+S
Sbjct: 170 LMHDSTATALGYGIYKTDLVANS 192
>At3g12580 putative protein
Length = 650
Score = 37.0 bits (84), Expect = 0.002
Identities = 25/100 (25%), Positives = 46/100 (46%), Gaps = 3/100 (3%)
Query: 9 RPFIAAIANKACRSTTAVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIE 68
+P I + +A E+ L+K++ E + P++ V TVP F +R +
Sbjct: 104 KPMIVVNHKGEEKQFSAEEISSMVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATK 163
Query: 69 RASAWADLDDVELMPQPIAVALFYAQQQLQTSASSLEDMN 108
A + L+ + ++ +P A A+ Y L ASS+ + N
Sbjct: 164 DAGVISGLNVMRIINEPTAAAIAYG---LDKKASSVGEKN 200
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 36.2 bits (82), Expect = 0.003
Identities = 21/76 (27%), Positives = 37/76 (48%)
Query: 25 AVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQ 84
A E+ L+K++ E + + I+ V TVP F +R + A A L+ + ++ +
Sbjct: 120 AEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINE 179
Query: 85 PIAVALFYAQQQLQTS 100
P A A+ Y + TS
Sbjct: 180 PTAAAIAYGLDKKATS 195
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 36.2 bits (82), Expect = 0.003
Identities = 23/92 (25%), Positives = 41/92 (44%)
Query: 9 RPFIAAIANKACRSTTAVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIE 68
+P I + +A E+ L+K++ E + I+ V TVP F +R +
Sbjct: 104 KPMIVVNYKGEDKEFSAEEISSMILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATK 163
Query: 69 RASAWADLDDVELMPQPIAVALFYAQQQLQTS 100
A A L+ + ++ +P A A+ Y + TS
Sbjct: 164 DAGVIAGLNVMRIINEPTAAAIAYGLDKKATS 195
>At1g16030 unknown protein
Length = 646
Score = 34.7 bits (78), Expect = 0.008
Identities = 23/93 (24%), Positives = 41/93 (43%)
Query: 9 RPFIAAIANKACRSTTAVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIE 68
+P I + + E+ LVK+K E + ++ V TVP F +R +
Sbjct: 103 KPMIVVSYKNEEKQFSPEEISSMVLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATK 162
Query: 69 RASAWADLDDVELMPQPIAVALFYAQQQLQTSA 101
A A + L+ + ++ +P A A+ Y + T A
Sbjct: 163 DAGAISGLNVLRIINEPTAAAIAYGLDKKGTKA 195
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 34.3 bits (77), Expect = 0.010
Identities = 22/92 (23%), Positives = 40/92 (42%)
Query: 9 RPFIAAIANKACRSTTAVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIE 68
+P I + A E+ L+K++ E + ++ V TVP F +R +
Sbjct: 104 KPMIVVEYKGEEKQFAAEEISSMVLIKMREIAEAFLGTTVKNAVVTVPAYFNDSQRQATK 163
Query: 69 RASAWADLDDVELMPQPIAVALFYAQQQLQTS 100
A A L+ + ++ +P A A+ Y + TS
Sbjct: 164 DAGVIAGLNVLRIINEPTAAAIAYGLDKKATS 195
>At1g56410 hypothetical protein
Length = 617
Score = 34.3 bits (77), Expect = 0.010
Identities = 21/76 (27%), Positives = 36/76 (46%)
Query: 25 AVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQ 84
A E+ L+K++ E + I+ V TVP F +R + A A L+ + ++ +
Sbjct: 120 AEEISSMVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINE 179
Query: 85 PIAVALFYAQQQLQTS 100
P A A+ Y + TS
Sbjct: 180 PTAAAIAYGLDKKATS 195
>At1g09080 putative luminal binding protein
Length = 678
Score = 34.3 bits (77), Expect = 0.010
Identities = 22/84 (26%), Positives = 38/84 (45%)
Query: 9 RPFIAAIANKACRSTTAVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIE 68
+P+I + + E+ L K+K + E + I+ V TVP F +R +
Sbjct: 146 KPYIQVKVKGEEKLFSPEEISAMILTKMKETAEAFLGKKIKDAVITVPAYFNDAQRQATK 205
Query: 69 RASAWADLDDVELMPQPIAVALFY 92
A A A L+ V ++ +P A+ Y
Sbjct: 206 DAGAIAGLNVVRIINEPTGAAIAY 229
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 33.9 bits (76), Expect = 0.014
Identities = 21/60 (35%), Positives = 29/60 (48%)
Query: 33 LVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQPIAVALFY 92
L K+K + E + I K V TVP F +R + A A LD ++ +P A AL Y
Sbjct: 170 LTKMKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIINEPTAAALSY 229
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 33.5 bits (75), Expect = 0.018
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 1/65 (1%)
Query: 29 LGKF-LVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQPIA 87
+G F L K+K + E + + K V TVP F +R + A A LD ++ +P A
Sbjct: 170 IGAFILTKMKETAEAYLGKSVTKAVVTVPAYFNDAQRQATKDAGRIAGLDVERIINEPTA 229
Query: 88 VALFY 92
AL Y
Sbjct: 230 AALSY 234
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 32.3 bits (72), Expect = 0.040
Identities = 19/66 (28%), Positives = 31/66 (46%)
Query: 27 ELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQPI 86
E+ L K+K + E + I+ V TVP F +R + A A L+ ++ +P
Sbjct: 150 EISAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 209
Query: 87 AVALFY 92
A A+ Y
Sbjct: 210 AAAIAY 215
>At5g28540 luminal binding protein
Length = 669
Score = 32.3 bits (72), Expect = 0.040
Identities = 19/66 (28%), Positives = 31/66 (46%)
Query: 27 ELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQPI 86
E+ L K+K + E + I+ V TVP F +R + A A L+ ++ +P
Sbjct: 150 EISAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 209
Query: 87 AVALFY 92
A A+ Y
Sbjct: 210 AAAIAY 215
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 30.8 bits (68), Expect = 0.12
Identities = 20/68 (29%), Positives = 32/68 (46%)
Query: 25 AVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQ 84
A E+ + L KL + + K V TVP F +RT + A A L+ + ++ +
Sbjct: 188 AEEISAQVLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINE 247
Query: 85 PIAVALFY 92
P A +L Y
Sbjct: 248 PTAASLAY 255
>At4g24280 hsp 70-like protein
Length = 718
Score = 30.8 bits (68), Expect = 0.12
Identities = 20/68 (29%), Positives = 32/68 (46%)
Query: 25 AVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQ 84
A E+ + L KL + + K V TVP F +RT + A A L+ + ++ +
Sbjct: 188 AEEISAQVLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINE 247
Query: 85 PIAVALFY 92
P A +L Y
Sbjct: 248 PTAASLAY 255
>At3g53760 putative protein
Length = 738
Score = 26.2 bits (56), Expect = 2.9
Identities = 19/69 (27%), Positives = 36/69 (51%), Gaps = 5/69 (7%)
Query: 8 VRPFIAAIANKACRSTTAVELLGKFLVKLKFSVEIRVKIPIRKVVFTVPVSFTRLRRTQI 67
VR ++ S T+V+ G + L++SV+ +++ + VF + RL+RTQ+
Sbjct: 485 VRSKVSLTGKANLTSDTSVD--GWDAIALEYSVDWPMQLFFTQEVFQYLI---RLKRTQM 539
Query: 68 ERASAWADL 76
E +WA +
Sbjct: 540 ELEKSWASV 548
>At1g67680 putative protein
Length = 664
Score = 26.2 bits (56), Expect = 2.9
Identities = 16/36 (44%), Positives = 20/36 (55%), Gaps = 3/36 (8%)
Query: 77 DDVELMPQPIAVALFYAQQ---QLQTSASSLEDMNK 109
+++E+ PIAV L Y QQ Q Q S SS D K
Sbjct: 235 EEIEIELAPIAVQLAYVQQILGQTQESTSSYVDFIK 270
>At1g47330 unknown protein
Length = 527
Score = 25.0 bits (53), Expect = 6.4
Identities = 18/67 (26%), Positives = 32/67 (46%), Gaps = 10/67 (14%)
Query: 28 LLGKFLVKLKFSVEIRVKIPIRK--------VVFTVPV--SFTRLRRTQIERASAWADLD 77
++G LVK +V+ R ++P+RK V T+P+ ++ A + DLD
Sbjct: 251 IIGLILVKNLLAVDARKEVPLRKMSMRKIPRVSETMPLYDILNEFQKGHSHIAVVYKDLD 310
Query: 78 DVELMPQ 84
+ E P+
Sbjct: 311 EQEQSPE 317
>At5g51430 putative protein
Length = 836
Score = 24.6 bits (52), Expect = 8.3
Identities = 9/38 (23%), Positives = 20/38 (51%)
Query: 48 IRKVVFTVPVSFTRLRRTQIERASAWADLDDVELMPQP 85
+ ++V+ V +S R R ++ R W+ +++ P P
Sbjct: 655 VNELVYDVLISKVRQRLGEVSRLPIWSSVEEQTAFPLP 692
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.134 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,897,827
Number of Sequences: 26719
Number of extensions: 59846
Number of successful extensions: 170
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 154
Number of HSP's gapped (non-prelim): 19
length of query: 109
length of database: 11,318,596
effective HSP length: 85
effective length of query: 24
effective length of database: 9,047,481
effective search space: 217139544
effective search space used: 217139544
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC135505.7


