

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.15 - phase: 0 /pseudo
(403 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
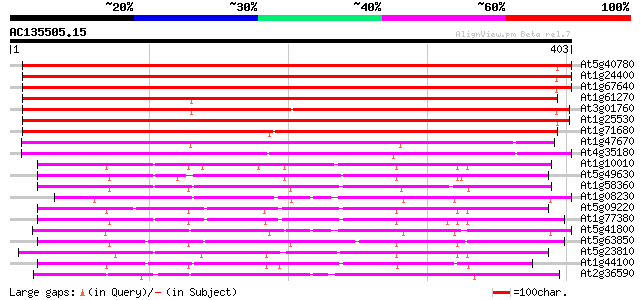
Score E
Sequences producing significant alignments: (bits) Value
At5g40780 amino acid permease 596 e-171
At1g24400 amino acid transporter-like protein 2 (AATL2) 584 e-167
At1g67640 putative amino acid permease 583 e-167
At1g61270 amino acid permease 494 e-140
At3g01760 putative amino acid permease 491 e-139
At1g25530 hypothetical protein 467 e-132
At1g71680 amino acid permease, putative 442 e-124
At1g47670 unknown protein 286 2e-77
At4g35180 amino acid permease - like protein 203 2e-52
At1g10010 putative amino acid permease 176 2e-44
At5g49630 amino acid permease 6 (emb|CAA65051.1) 173 1e-43
At1g58360 unknown protein 169 2e-42
At1g08230 unknown protein 167 1e-41
At5g09220 amino acid transport protein AAP2 163 2e-40
At1g77380 amino acid permease AAP3 161 7e-40
At5g41800 amino acid permease-like protein; proline transporter-... 159 3e-39
At5g63850 amino acid transporter AAP4 (pir||S51169) 150 1e-36
At5g23810 amino acid transporter 150 1e-36
At1g44100 amino acid permease AAP5 148 5e-36
At2g36590 putative proline transporter 113 2e-25
>At5g40780 amino acid permease
Length = 446
Score = 596 bits (1536), Expect = e-171
Identities = 276/405 (68%), Positives = 332/405 (81%), Gaps = 11/405 (2%)
Query: 10 SRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHE 69
SRNAK YSAFHNVTAMVGA VLG PYAMSQLGWG GI +LVLSW+ TLYT WQM+EMHE
Sbjct: 33 SRNAKWWYSAFHNVTAMVGAGVLGLPYAMSQLGWGPGIAVLVLSWVITLYTLWQMVEMHE 92
Query: 70 SVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDD 129
V GKRFD+YHEL QHAFGE+LGL+IVVPQQL+VE+G+ IVYMV G KSLKK HE++CDD
Sbjct: 93 MVPGKRFDRYHELGQHAFGEKLGLYIVVPQQLIVEIGVCIVYMVTGGKSLKKFHELVCDD 152
Query: 130 CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALPDV 189
C+PIK TYFI++FA V +VLSHLP+FNS++G+SL AA MSLSYSTIAW +S +G DV
Sbjct: 153 CKPIKLTYFIMIFASVHFVLSHLPNFNSISGVSLAAAVMSLSYSTIAWASSASKGVQEDV 212
Query: 190 QYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAY 249
QY + T AG +F F+ LGD+AF YAGHNV+LEIQ+TIPSTPEKPSK MWRG+I+AY
Sbjct: 213 QYGYKAKTTAGTVFNFFSGLGDVAFAYAGHNVVLEIQATIPSTPEKPSKGPMWRGVIVAY 272
Query: 250 LVVALCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVF 309
+VVALCYFPV + GY FGN V+DNIL+SL+KP WLI ANIFVV+HV+GSYQ+YA+PVF
Sbjct: 273 IVVALCYFPVALVGYYIFGNGVEDNILMSLKKPAWLIATANIFVVIHVIGSYQIYAMPVF 332
Query: 310 HMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFL 369
M+E+ L +K+NF+P+ LRF +RN YV+ TM + +TFPFFGGLL+FFGGF FAPTTYFL
Sbjct: 333 DMMETLLVKKLNFRPTTTLRFFVRNFYVAATMFVGMTFPFFGGLLAFFGGFAFAPTTYFL 392
Query: 370 PCIMWIFIYKPKLFSLSWCANWV-----------SPIGALRQVIL 403
PC++W+ IYKPK +SLSW ANWV SPIG LR +++
Sbjct: 393 PCVIWLAIYKPKKYSLSWWANWVCIVFGLFLMVLSPIGGLRTIVI 437
>At1g24400 amino acid transporter-like protein 2 (AATL2)
Length = 441
Score = 584 bits (1505), Expect = e-167
Identities = 273/405 (67%), Positives = 328/405 (80%), Gaps = 11/405 (2%)
Query: 10 SRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHE 69
SRNAK YSAFHNVTAMVGA VL PYAMS LGWG G+TI+V+SWI TLYT WQM+EMHE
Sbjct: 28 SRNAKWWYSAFHNVTAMVGAGVLSLPYAMSNLGWGPGVTIMVMSWIITLYTLWQMVEMHE 87
Query: 70 SVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDD 129
V GKR D+YHEL QHAFGE+LGLWIVVPQQL+VEVG+DIVYMV G SLKK+H+++C D
Sbjct: 88 IVPGKRLDRYHELGQHAFGEKLGLWIVVPQQLIVEVGVDIVYMVTGGASLKKVHQLVCPD 147
Query: 130 CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALPDV 189
C+ I+TT++I++FA V +V+SHLP+FNS++ ISL AA MSL+YSTIAW AS+H+G PDV
Sbjct: 148 CKEIRTTFWIMIFASVHFVISHLPNFNSISIISLAAAVMSLTYSTIAWAASVHKGVHPDV 207
Query: 190 QYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAY 249
YS R ST G +F NALGD+AF YAGHNV+LEIQ+TIPSTPE PSKV MWRG+I+AY
Sbjct: 208 DYSPRASTDVGKVFNFLNALGDVAFAYAGHNVVLEIQATIPSTPEMPSKVPMWRGVIVAY 267
Query: 250 LVVALCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVF 309
+VVA+CYFPV GY FGNSVDDNIL++LEKP WLI AN+FVV+HV+GSYQ++A+PVF
Sbjct: 268 IVVAICYFPVAFLGYYIFGNSVDDNILITLEKPIWLIAMANMFVVIHVIGSYQIFAMPVF 327
Query: 310 HMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFL 369
MLE+ L +KMNF PS LRF R+LYV+ TM++AI PFFGGLL FFGGF FAPTTY+L
Sbjct: 328 DMLETVLVKKMNFNPSFKLRFITRSLYVAFTMIVAICVPFFGGLLGFFGGFAFAPTTYYL 387
Query: 370 PCIMWIFIYKPKLFSLSWCANW-----------VSPIGALRQVIL 403
PCIMW+ + KPK F LSW ANW ++PIG LR +I+
Sbjct: 388 PCIMWLVLKKPKRFGLSWTANWFCIIVGVLLTILAPIGGLRTIII 432
>At1g67640 putative amino acid permease
Length = 441
Score = 583 bits (1502), Expect = e-167
Identities = 270/405 (66%), Positives = 328/405 (80%), Gaps = 11/405 (2%)
Query: 10 SRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHE 69
SRNAK YSAFHNVTAMVGA VL PYAMS LGWG G+TI+++SW+ T YT WQM++MHE
Sbjct: 28 SRNAKWWYSAFHNVTAMVGAGVLSLPYAMSNLGWGPGVTIMIMSWLITFYTLWQMVQMHE 87
Query: 70 SVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDD 129
V GKRFD+YHEL QHAFGE+LGLWIVVPQQL+VEVG+DIVYMV G KSLKK+H++LC D
Sbjct: 88 MVPGKRFDRYHELGQHAFGEKLGLWIVVPQQLIVEVGVDIVYMVTGGKSLKKIHDLLCTD 147
Query: 130 CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALPDV 189
C+ I+TTY+I++FA + +VL+HLP+FNS++ +SL AA MSLSYSTIAW S+ +G P+V
Sbjct: 148 CKNIRTTYWIMIFASIHFVLAHLPNFNSISIVSLAAAVMSLSYSTIAWATSVKKGVHPNV 207
Query: 190 QYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAY 249
YSSR ST +GN+F NALGD+AF YAGHNV+LEIQ+TIPSTPEKPSK++MW+G+++AY
Sbjct: 208 DYSSRASTTSGNVFNFLNALGDVAFAYAGHNVVLEIQATIPSTPEKPSKIAMWKGVVVAY 267
Query: 250 LVVALCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVF 309
+VVA+CYFPV Y FGNSVDDNIL++LEKP WLI AN FVVVHV+GSYQ+YA+PVF
Sbjct: 268 IVVAICYFPVAFVCYYIFGNSVDDNILMTLEKPIWLIAIANAFVVVHVIGSYQIYAMPVF 327
Query: 310 HMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFL 369
MLE+FL +KM F PS LRF R LYV+ TM +AI PFFGGLL FFGGF FAPTTY+L
Sbjct: 328 DMLETFLVKKMMFAPSFKLRFITRTLYVAFTMFVAICIPFFGGLLGFFGGFAFAPTTYYL 387
Query: 370 PCIMWIFIYKPKLFSLSWCANW-----------VSPIGALRQVIL 403
PCIMW+ I KPK + LSWC NW ++PIG LR +I+
Sbjct: 388 PCIMWLCIKKPKKYGLSWCINWFCIVVGVILTILAPIGGLRTIII 432
>At1g61270 amino acid permease
Length = 418
Score = 494 bits (1272), Expect = e-140
Identities = 231/386 (59%), Positives = 296/386 (75%), Gaps = 2/386 (0%)
Query: 10 SRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHE 69
SRNA YSAFHNVTA+VGA VLG PYAMS+LGWG G+ +L+LSW+ TLYT WQMIEMHE
Sbjct: 23 SRNANWYYSAFHNVTAIVGAGVLGLPYAMSELGWGPGVVVLILSWVITLYTFWQMIEMHE 82
Query: 70 SVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDD 129
GKRFD+YHEL Q AFG++LGL+IVVP QL+VE IVYMV G +SLKK+H++ D
Sbjct: 83 MFEGKRFDRYHELGQAAFGKKLGLYIVVPLQLLVETSACIVYMVTGGESLKKIHQLSVGD 142
Query: 130 --CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALP 187
C +K +FI++FA Q+VLS L +FNS++G+SLVAA MS+SYSTIAW+AS+ +G
Sbjct: 143 YECRKLKVRHFILIFASSQFVLSLLKNFNSISGVSLVAAVMSMSYSTIAWVASLTKGVAN 202
Query: 188 DVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMII 247
+V+Y + ALG++AF YAGHNV+LEIQ+TIPSTPE PSK MW+G I+
Sbjct: 203 NVEYGYKRRNNTSVPLAFLGALGEMAFAYAGHNVVLEIQATIPSTPENPSKRPMWKGAIV 262
Query: 248 AYLVVALCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYAVP 307
AY++VA CYFPV + G+ FGN+V++NIL +L P+ LII ANIFV++H++GSYQVYA+P
Sbjct: 263 AYIIVAFCYFPVALVGFWTFGNNVEENILKTLRGPKGLIIVANIFVIIHLMGSYQVYAMP 322
Query: 308 VFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTY 367
VF M+ES + +K +F P+R LRF IR +V+ TM +A+ P F LLSFFGGF+FAPTTY
Sbjct: 323 VFDMIESVMIKKWHFSPTRVLRFTIRWTFVAATMGIAVALPHFSALLSFFGGFIFAPTTY 382
Query: 368 FLPCIMWIFIYKPKLFSLSWCANWVS 393
F+PCI+W+ + KPK FSLSWC NW S
Sbjct: 383 FIPCIIWLILKKPKRFSLSWCINWAS 408
>At3g01760 putative amino acid permease
Length = 455
Score = 491 bits (1263), Expect = e-139
Identities = 235/408 (57%), Positives = 309/408 (75%), Gaps = 16/408 (3%)
Query: 10 SRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHE 69
SRNA YSAFHNVTA+VGA VLG PYAMS+LGWG G+ +L+LSW+ TLYT WQMIEMHE
Sbjct: 31 SRNANWYYSAFHNVTAIVGAGVLGLPYAMSELGWGPGVVVLILSWVITLYTLWQMIEMHE 90
Query: 70 SVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDD 129
G+RFD+YHEL Q AFG++LGL+I+VP QL+VE+ + IVYMV G KSLK +H++ D
Sbjct: 91 MFEGQRFDRYHELGQAAFGKKLGLYIIVPLQLLVEISVCIVYMVTGGKSLKNVHDLALGD 150
Query: 130 ---CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGAL 186
C ++ +FI++FA Q+VLS L +FNS++G+SLVAA MS+SYSTIAW+AS+ +GA
Sbjct: 151 GDKCTKLRIQHFILIFASSQFVLSLLKNFNSISGVSLVAAVMSVSYSTIAWVASLRKGAT 210
Query: 187 P-DVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGM 245
V+Y R T + + +ALG++AF YAGHNV+LEIQ+TIPSTPE PSK MW+G
Sbjct: 211 TGSVEYGYRKRTTSVPL-AFLSALGEMAFAYAGHNVVLEIQATIPSTPENPSKRPMWKGA 269
Query: 246 IIAYLVVALCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYA 305
++AY++VA CYFPV + G++ FGNSV+++IL SL KP L+I AN+FVV+H++GSYQVYA
Sbjct: 270 VVAYIIVAFCYFPVALVGFKTFGNSVEESILESLTKPTALVIVANMFVVIHLLGSYQVYA 329
Query: 306 VPVFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPT 365
+PVF M+ES + +F P+R LRF IR +V+ TM +A+ P++ LLSFFGGFVFAPT
Sbjct: 330 MPVFDMIESVMIRIWHFSPTRVLRFTIRWTFVAATMGIAVGLPYYSALLSFFGGFVFAPT 389
Query: 366 TYFLPCIMWIFIYKPKLFSLSWCANW-----------VSPIGALRQVI 402
TYF+PCIMW+ + KPK FSLSWC NW ++PIG L ++I
Sbjct: 390 TYFIPCIMWLILKKPKRFSLSWCMNWFCIIFGLVLMIIAPIGGLAKLI 437
>At1g25530 hypothetical protein
Length = 440
Score = 467 bits (1201), Expect = e-132
Identities = 223/404 (55%), Positives = 289/404 (71%), Gaps = 11/404 (2%)
Query: 10 SRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHE 69
SR AK YS FH VTAM+GA VL PYAM+ LGWG G +L ++W TL T WQM+++HE
Sbjct: 27 SRPAKWWYSTFHTVTAMIGAGVLSLPYAMAYLGWGPGTFVLAMTWGLTLNTMWQMVQLHE 86
Query: 70 SVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDD 129
V G RFD+Y +L ++AFG +LG WIV+PQQL+V+VG +IVYMV G K LK+ EI C
Sbjct: 87 CVPGTRFDRYIDLGRYAFGPKLGPWIVLPQQLIVQVGCNIVYMVTGGKCLKQFVEITCST 146
Query: 130 CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALPDV 189
C P++ +Y+I+ F V ++LS LP+FNSVAG+SL AA MSL YSTIAW SI G +PDV
Sbjct: 147 CTPVRQSYWILGFGGVHFILSQLPNFNSVAGVSLAAAVMSLCYSTIAWGGSIAHGRVPDV 206
Query: 190 QYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAY 249
Y + + F +FNALG I+F +AGH V LEIQ+T+PSTPE+PSKV MW+G+I AY
Sbjct: 207 SYDYKATNPGDFTFRVFNALGQISFAFAGHAVALEIQATMPSTPERPSKVPMWQGVIGAY 266
Query: 250 LVVALCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVF 309
+V A+CYFPV + Y AFG VDDN+L++L++P WLI AAN+ VVVHV+GSYQV+A+PVF
Sbjct: 267 VVNAVCYFPVALICYWAFGQDVDDNVLMNLQRPAWLIAAANLMVVVHVIGSYQVFAMPVF 326
Query: 310 HMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFL 369
+LE + K FK LRF R +YV+ T+ + ++FPFFG LL FFGGF FAPT++FL
Sbjct: 327 DLLERMMVNKFGFKHGVVLRFFTRTIYVAFTLFIGVSFPFFGDLLGFFGGFGFAPTSFFL 386
Query: 370 PCIMWIFIYKPKLFSLSWCANWV-----------SPIGALRQVI 402
P IMW+ I KP+ FS++W NW+ S IG LR +I
Sbjct: 387 PSIMWLIIKKPRRFSVTWFVNWISIIVGVFIMLASTIGGLRNII 430
>At1g71680 amino acid permease, putative
Length = 434
Score = 442 bits (1136), Expect = e-124
Identities = 213/386 (55%), Positives = 279/386 (72%), Gaps = 3/386 (0%)
Query: 10 SRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHE 69
SR AK YSAFHNVTAMVGA VLG P+AMSQLGWG G+ +++SW T Y+ WQM+++HE
Sbjct: 36 SREAKWYYSAFHNVTAMVGAGVLGLPFAMSQLGWGPGLVAIIMSWAITFYSLWQMVQLHE 95
Query: 70 SVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDD 129
+V GKR D+Y EL Q AFG +LG WIV+PQQL+V++ DIVY V G KSLKK E+L +
Sbjct: 96 AVPGKRLDRYPELGQEAFGPKLGYWIVMPQQLLVQIASDIVYNVTGGKSLKKFVELLFPN 155
Query: 130 CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGA--LP 187
E I+ TY+I+ FA +Q VLS P FNS+ +SL+AA MS YS IA +ASI +G P
Sbjct: 156 LEHIRQTYYILGFAALQLVLSQSPDFNSIKIVSLLAALMSFLYSMIASVASIAKGTEHRP 215
Query: 188 DVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMII 247
Y R T A +F FN +G IAF +AGH+V+LEIQ+TIPSTPE PSK MW+G+++
Sbjct: 216 ST-YGVRGDTVASMVFDAFNGIGTIAFAFAGHSVVLEIQATIPSTPEVPSKKPMWKGVVV 274
Query: 248 AYLVVALCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYAVP 307
AY++V +CY V I GY AFG V+D++L+SLE+P WLI AAN V +HV+GSYQV+A+
Sbjct: 275 AYIIVIICYLFVAISGYWAFGAHVEDDVLISLERPAWLIAAANFMVFIHVIGSYQVFAMI 334
Query: 308 VFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTY 367
VF +ES+L + + F PS LR R+ YV++ ++A+ PFFGGLL FFGG VF+ T+Y
Sbjct: 335 VFDTIESYLVKTLKFTPSTTLRLVARSTYVALICLVAVCIPFFGGLLGFFGGLVFSSTSY 394
Query: 368 FLPCIMWIFIYKPKLFSLSWCANWVS 393
FLPCI+W+ + +PK FS W +WVS
Sbjct: 395 FLPCIIWLIMKRPKRFSAHWWCSWVS 420
>At1g47670 unknown protein
Length = 519
Score = 286 bits (731), Expect = 2e-77
Identities = 142/394 (36%), Positives = 230/394 (58%), Gaps = 12/394 (3%)
Query: 9 KSRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMH 68
+SRN Y+AFHN+ A VG L P A + LGW GI L +++ LYT W ++++H
Sbjct: 92 ESRNGNAHYAAFHNLNAGVGFQALVLPVAFAFLGWSWGILSLTIAYCWQLYTLWILVQLH 151
Query: 69 ESVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCD 128
E+V GKR+++Y EL+Q AFGERLG+W+ + + + G ++IG +++K +I+C
Sbjct: 152 EAVPGKRYNRYVELAQAAFGERLGVWLALFPTVYLSAGTATALILIGGETMKLFFQIVCG 211
Query: 129 ---DCEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGA 185
P+ T + ++F + VLS LP+ NS+AG+SL+ A +++YST+ W+ S+ +
Sbjct: 212 PLCTSNPLTTVEWYLVFTSLCIVLSQLPNLNSIAGLSLIGAVTAITYSTMVWVLSVSQPR 271
Query: 186 LPDVQYSS-RYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRG 244
+ Y + +G++F + NALG IAF + GHN++LEIQST+PST + P+ V MWRG
Sbjct: 272 PATISYEPLSMPSTSGSLFAVLNALGIIAFAFRGHNLVLEIQSTMPSTFKHPAHVPMWRG 331
Query: 245 MIIAYLVVALCYFPVTIFGYRAFGNSVDDNILLSL-------EKPRWLIIAANIFVVVHV 297
I+Y ++ALC FP++I G+ A+GN + +L+ + PR L+ A + VV
Sbjct: 332 AKISYFLIALCIFPISIGGFWAYGNLMPSGGMLAALYAFHIHDIPRGLLATAFLLVVFSC 391
Query: 298 VGSYQVYAVPVFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFF 357
+ S+Q+Y++P F E+ + N S ++R R + ++ + + PF L
Sbjct: 392 LSSFQIYSMPAFDSFEAGYTSRTNKPCSIWVRSGFRVFFGFVSFFIGVALPFLSSLAGLL 451
Query: 358 GGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCANW 391
GG P T+ PC MW+ I KP +S +W +W
Sbjct: 452 GGLTL-PVTFAYPCFMWVLIKKPAKYSFNWYFHW 484
>At4g35180 amino acid permease - like protein
Length = 456
Score = 203 bits (516), Expect = 2e-52
Identities = 121/405 (29%), Positives = 206/405 (49%), Gaps = 12/405 (2%)
Query: 9 KSRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMH 68
+SR + FH + + +G V+ P A + LGW G IL + ++ LYT W ++++H
Sbjct: 34 ESRKGNVYTATFHLLCSGIGLQVILLPAAFAALGWVWGTIILTVGFVWKLYTTWLLVQLH 93
Query: 69 ESVSGKRFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCD 128
E+V G R +Y L+ +FG +LG + + + + G + ++ G KS+++L +I+ D
Sbjct: 94 EAVPGIRISRYVRLAIASFGVKLGKLLGIFPVMYLSGGACTILVITGGKSIQQLLQIMSD 153
Query: 129 D-CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALP 187
D P+ + ++F+ + ++S P+ NS+ G+SL+ A M ++Y T+ WI + +
Sbjct: 154 DNTAPLTSVQCFLVFSCIAMIMSQFPNLNSLFGVSLIGAFMGIAYCTVIWILPVASDS-Q 212
Query: 188 DVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMII 247
Q S Y+T + IFNA+G IA Y G+N++LEIQ T+PS + PS +MWR ++I
Sbjct: 213 RTQVSVSYATMDKSFVHIFNAIGLIALVYRGNNLVLEIQGTLPSDSKNPSCKTMWRAVMI 272
Query: 248 AYLVVALCYFPVTIFGYRAFGNSVDDN--------ILLSLEKPRWLIIAANIFVVVHVVG 299
++ +VA+C FP+T Y A+G+ + L + E + ++ + +
Sbjct: 273 SHALVAICMFPLTFAVYWAYGDKIPATGGPVGNYLKLYTQEHSKRAACFIHLTFIFSCLC 332
Query: 300 SYQVYAVPVFHMLESFLAEKMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFFGG 359
SY + +P +E K S +R +R + +A+ FPF L G
Sbjct: 333 SYPINLMPACDNIEMVYITKKKKPASIIVRMMLRVFLSLVCFTIAVGFPFLPYLAVLIGA 392
Query: 360 FVFAPTTYFLPCIMWIFIYKPKLFSLSWCAN-WVSPIGALRQVIL 403
T+ PC MWI I KP+ S W N V +GA V+L
Sbjct: 393 IALL-VTFTYPCFMWISIKKPQRKSPMWLFNVLVGCLGASLSVLL 436
>At1g10010 putative amino acid permease
Length = 475
Score = 176 bits (447), Expect = 2e-44
Identities = 124/404 (30%), Positives = 196/404 (47%), Gaps = 38/404 (9%)
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMH---ESVSGKRFD 77
H +TA++G+ VL +A++QLGW G T+LV I T YT+ + + + +S++G R
Sbjct: 38 HIITAVIGSGVLSLAWAIAQLGWVAGTTVLVAFAIITYYTSTLLADCYRSPDSITGTRNY 97
Query: 78 KYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILC--DDCEPIKT 135
Y + + G + V Q + + VG+ I Y + + SL + + C D K
Sbjct: 98 NYMGVVRSYLGGKKVQLCGVAQYVNL-VGVTIGYTITASISLVAIGKSNCYHDKGHKAKC 156
Query: 136 TY----FIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAW---IASIHRGALPD 188
+ ++ F VQ +LS LP+F+ ++ +S++AA MS SY++I IA++ G +
Sbjct: 157 SVSNYPYMAAFGIVQIILSQLPNFHKLSFLSIIAAVMSFSYASIGIGLAIATVASGKIGK 216
Query: 189 VQYSSRYS----TKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRG 244
+ + T + ++ +F A+GDIAF YA +++EIQ T+ S+P P M R
Sbjct: 217 TELTGTVIGVDVTASEKVWKLFQAIGDIAFSYAFTTILIEIQDTLRSSP--PENKVMKRA 274
Query: 245 MIIAYLVVALCYFPVTIFGYRAFGNSVDDNIL--LSLEKPRWLIIAANIFVVVHVVGSYQ 302
++ + Y GY AFGN + L +P WLI AN + +H++G+YQ
Sbjct: 275 SLVGVSTTTVFYILCGCIGYAAFGNQAPGDFLTDFGFYEPYWLIDFANACIALHLIGAYQ 334
Query: 303 VYAVPVFHMLESFLAEKM---NFKPSRF--------------LRFAIRNLYVSITMVLAI 345
VYA P F +E +K NF + R R YV +T +A+
Sbjct: 335 VYAQPFFQFVEENCNKKWPQSNFINKEYSSKVPLLGKCRVNLFRLVWRTCYVVLTTFVAM 394
Query: 346 TFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCA 389
FPFF +L G F F P T + P M I K K +S W A
Sbjct: 395 IFPFFNAILGLLGAFAFWPLTVYFPVAMHIAQAKVKKYSRRWLA 438
>At5g49630 amino acid permease 6 (emb|CAA65051.1)
Length = 481
Score = 173 bits (439), Expect = 1e-43
Identities = 124/407 (30%), Positives = 195/407 (47%), Gaps = 46/407 (11%)
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHES---VSGKRFD 77
H +TA++G+ VL +A++QLGW G +L+ T +T+ + + + S V+GKR
Sbjct: 43 HIITAVIGSGVLSLAWAIAQLGWVAGPAVLMAFSFITYFTSTMLADCYRSPDPVTGKRNY 102
Query: 78 KYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSL----------KKLHEILC 127
Y E+ + G R + Q + +GI I Y + + S+ K H + C
Sbjct: 103 TYMEVVRSYLGGRKVQLCGLAQYGNL-IGITIGYTITASISMVAVKRSNCFHKNGHNVKC 161
Query: 128 DDCEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALP 187
T F+++FA +Q +LS +P+F++++ +S++AA MS Y++I SI + A
Sbjct: 162 ----ATSNTPFMIIFAIIQIILSQIPNFHNLSWLSILAAVMSFCYASIGVGLSIAKAAGG 217
Query: 188 DVQYSSRYS--------TKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKV 239
+ + + A I+ F A+GDIAF YA V++EIQ T+ + P +K
Sbjct: 218 GEHVRTTLTGVTVGIDVSGAEKIWRTFQAIGDIAFAYAYSTVLIEIQDTLKAGPPSENK- 276
Query: 240 SMWRGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNIL--LSLEKPRWLIIAANIFVVVHV 297
+M R ++ Y GY AFGN N L +P WLI AN+ + VH+
Sbjct: 277 AMKRASLVGVSTTTFFYMLCGCVGYAAFGNDAPGNFLTGFGFYEPFWLIDFANVCIAVHL 336
Query: 298 VGSYQVYAVPVFHMLESFLAEKM--------NFK---------PSRFLRFAIRNLYVSIT 340
+G+YQV+ P+F +ES A++ +K FLR R YV +T
Sbjct: 337 IGAYQVFCQPIFQFVESQSAKRWPDNKFITGEYKIHVPCCGDFSINFLRLVWRTSYVVVT 396
Query: 341 MVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSW 387
V+A+ FPFF L G F P T + P M I K FS +W
Sbjct: 397 AVVAMIFPFFNDFLGLIGAASFWPLTVYFPIEMHIAQKKIPKFSFTW 443
>At1g58360 unknown protein
Length = 485
Score = 169 bits (429), Expect = 2e-42
Identities = 123/407 (30%), Positives = 197/407 (48%), Gaps = 44/407 (10%)
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHES---VSGKRFD 77
H +TA++G+ VL +A++QLGW G +IL++ T +T+ + + + + V+GKR
Sbjct: 47 HIITAVIGSGVLSLAWAIAQLGWIAGTSILLIFSFITYFTSTMLADCYRAPDPVTGKRNY 106
Query: 78 KYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILC-------DDC 130
Y ++ + G R V Q + +G+ + Y + + SL + + C DC
Sbjct: 107 TYMDVVRSYLGGRKVQLCGVAQYGNL-IGVTVGYTITASISLVAVGKSNCFHDKGHTADC 165
Query: 131 EPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALPDVQ 190
I ++ +F +Q +LS +P+F+ ++ +S++AA MS +Y+TI +I A V
Sbjct: 166 T-ISNYPYMAVFGIIQVILSQIPNFHKLSFLSIMAAVMSFTYATIGIGLAIATVAGGKVG 224
Query: 191 YSSRYSTKAG-------NIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWR 243
+S T G I+ F A+GDIAF YA V++EIQ T+ S+P + +M R
Sbjct: 225 KTSMTGTAVGVDVTAAQKIWRSFQAVGDIAFAYAYATVLIEIQDTLRSSPAENK--AMKR 282
Query: 244 GMIIAYLVVALCYFPVTIFGYRAFGNSVDDNILLSLE--KPRWLIIAANIFVVVHVVGSY 301
++ Y GY AFGN+ + L +P WLI AN + VH++G+Y
Sbjct: 283 ASLVGVSTTTFFYILCGCIGYAAFGNNAPGDFLTDFGFFEPFWLIDFANACIAVHLIGAY 342
Query: 302 QVYAVPVFHMLESFLAEKMNFKPSRFL-------------------RFAIRNLYVSITMV 342
QV+A P+F +E N+ ++F+ R R YV IT V
Sbjct: 343 QVFAQPIFQFVEKKC--NRNYPDNKFITSEYSVNVPFLGKFNISLFRLVWRTAYVVITTV 400
Query: 343 LAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCA 389
+A+ FPFF +L G F P T + P M I K K +S W A
Sbjct: 401 VAMIFPFFNAILGLIGAASFWPLTVYFPVEMHIAQTKIKKYSARWIA 447
>At1g08230 unknown protein
Length = 422
Score = 167 bits (422), Expect = 1e-41
Identities = 119/399 (29%), Positives = 198/399 (48%), Gaps = 36/399 (9%)
Query: 33 GFPYAMSQLGWGLGITILVLSWICTLY--TAWQMIEMHESVSGKRFDKYHELSQHAFGER 90
G + + GW GI+ LV T Y T + H + G R+ ++ +++ H +
Sbjct: 21 GSLFVLKSKGWAAGISCLVGGAAVTFYSYTLLSLTLEHHASLGNRYLRFRDMAHHILSPK 80
Query: 91 LGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDDCEPIKTTYFIVLFAFVQYVLS 150
G + V P Q+ V G+ I ++G + LK ++ ++ + E +K F+++F + VL+
Sbjct: 81 WGRYYVGPIQMAVCYGVVIANALLGGQCLKAMYLVVQPNGE-MKLFEFVIIFGCLLLVLA 139
Query: 151 HLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALPDVQYSSRYSTKAGN----IFGIF 206
PSF+S+ I+ ++ + L YS A ASI+ G P+ + T G+ +FGIF
Sbjct: 140 QFPSFHSLRYINSLSLLLCLLYSASAAAASIYIGKEPNAP--EKDYTIVGDPETRVFGIF 197
Query: 207 NALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAYLVVALCYFPVTIFGYRA 266
NA+ IA Y G+ +I EIQ+TI + P K M +G+ + YLVV + +F V I GY A
Sbjct: 198 NAMAIIATTY-GNGIIPEIQATISA----PVKGKMMKGLCMCYLVVIMTFFTVAITGYWA 252
Query: 267 FGNSVDDNILLSLEK--------PRWLIIAANIFVVVHVVGSYQVYAVPVFHMLESFLAE 318
FG + I + P W I N+F V+ + VY P+ +LES +++
Sbjct: 253 FGKKANGLIFTNFLNAETNHYFVPTWFIFLVNLFTVLQLSAVAVVYLQPINDILESVISD 312
Query: 319 --KMNFKPSRFL-RFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWI 375
K F + R +R+L+V + ++A PFFG + S G F F P + LP + +
Sbjct: 313 PTKKEFSIRNVIPRLVVRSLFVVMATIVAAMLPFFGDVNSLLGAFGFIPLDFVLPVVFFN 372
Query: 376 FIYKP-KLFSLSW----------CANWVSPIGALRQVIL 403
F +KP K + W C ++ + A+RQ+I+
Sbjct: 373 FTFKPSKKSFIFWINTVIAVVFSCLGVIAMVAAVRQIII 411
>At5g09220 amino acid transport protein AAP2
Length = 493
Score = 163 bits (412), Expect = 2e-40
Identities = 122/405 (30%), Positives = 193/405 (47%), Gaps = 44/405 (10%)
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMH---ESVSGKRFD 77
H +TA++G+ VL +A++QLGW G +++L + TLY++ + + + ++VSGKR
Sbjct: 56 HIITAVIGSGVLSLAWAIAQLGWIAGPAVMLLFSLVTLYSSTLLSDCYRTGDAVSGKRNY 115
Query: 78 KYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILC-------DDC 130
Y + + G I Q + GI I Y + + S+ + C D C
Sbjct: 116 TYMDAVRSILGG-FKFKICGLIQYLNLFGIAIGYTIAASISMMAIKRSNCFHKSGGKDPC 174
Query: 131 EPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASI--------H 182
Y IV F + +LS +P F+ + IS+VAA MS +YS I I
Sbjct: 175 HMSSNPYMIV-FGVAEILLSQVPDFDQIWWISIVAAVMSFTYSAIGLALGIVQVAANGVF 233
Query: 183 RGALPDVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMW 242
+G+L + + T+ I+ F ALGDIAF Y+ V++EIQ T+ S P + +M
Sbjct: 234 KGSLTGISIGT--VTQTQKIWRTFQALGDIAFAYSYSVVLIEIQDTVRSPPAESK--TMK 289
Query: 243 RGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNIL--LSLEKPRWLIIAANIFVVVHVVGS 300
+ I+ V + Y GY AFG++ N+L P WL+ AN +VVH+VG+
Sbjct: 290 KATKISIAVTTIFYMLCGSMGYAAFGDAAPGNLLTGFGFYNPFWLLDIANAAIVVHLVGA 349
Query: 301 YQVYAVPVFHMLESFLAEKM---NFKPSRF---------------LRFAIRNLYVSITMV 342
YQV+A P+F +E +AE+ +F F R R+ +V T V
Sbjct: 350 YQVFAQPIFAFIEKSVAERYPDNDFLSKEFEIRIPGFKSPYKVNVFRMVYRSGFVVTTTV 409
Query: 343 LAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSW 387
+++ PFF ++ G F P T + P M+I K + +S W
Sbjct: 410 ISMLMPFFNDVVGILGALGFWPLTVYFPVEMYIKQRKVEKWSTRW 454
>At1g77380 amino acid permease AAP3
Length = 476
Score = 161 bits (407), Expect = 7e-40
Identities = 120/415 (28%), Positives = 193/415 (45%), Gaps = 43/415 (10%)
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHES---VSGKRFD 77
H +TA++G+ VL +A +QLGW G +++L T +T+ + + S +SGKR
Sbjct: 40 HIITAVIGSGVLSLAWATAQLGWLAGPVVMLLFSAVTYFTSSLLAACYRSGDPISGKRNY 99
Query: 78 KYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILC-------DDC 130
Y + + G + Q L + G+ I Y + A S+ + C D C
Sbjct: 100 TYMDAVRSNLGGVKVTLCGIVQYLNI-FGVAIGYTIASAISMMAIKRSNCFHKSGGKDPC 158
Query: 131 EPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIH-------- 182
Y I F VQ + S +P F+ + +S++AA MS +YS+ I
Sbjct: 159 HMNSNPYMIA-FGLVQILFSQIPDFDQLWWLSILAAVMSFTYSSAGLALGIAQVVVNGKV 217
Query: 183 RGALPDVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMW 242
+G+L + + T+ I+ F ALGDIAF Y+ +++EIQ T+ S P + +M
Sbjct: 218 KGSLTGISIGA--VTETQKIWRTFQALGDIAFAYSYSIILIEIQDTVKSPPSEEK--TMK 273
Query: 243 RGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNIL--LSLEKPRWLIIAANIFVVVHVVGS 300
+ +++ V + Y GY AFG+ N+L P WL+ AN +V+H++G+
Sbjct: 274 KATLVSVSVTTMFYMLCGCMGYAAFGDLSPGNLLTGFGFYNPYWLLDIANAAIVIHLIGA 333
Query: 301 YQVYAVPVFHMLE----------SFLAEKM-----NFKPSRF--LRFAIRNLYVSITMVL 343
YQVY P+F +E F+A+ + FKP R R R ++V IT V+
Sbjct: 334 YQVYCQPLFAFIEKQASIQFPDSEFIAKDIKIPIPGFKPLRLNVFRLIWRTVFVIITTVI 393
Query: 344 AITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCANWVSPIGAL 398
++ PFF ++ G F P T + P M+I K +S W V +G L
Sbjct: 394 SMLLPFFNDVVGLLGALGFWPLTVYFPVEMYIAQKKIPRWSTRWVCLQVFSLGCL 448
>At5g41800 amino acid permease-like protein; proline
transporter-like protein
Length = 452
Score = 159 bits (401), Expect = 3e-39
Identities = 118/417 (28%), Positives = 199/417 (47%), Gaps = 41/417 (9%)
Query: 17 YSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEM--HESVSGK 74
++ FH TA+VG +L PYA LGW LG L + T Y + M ++ H SG+
Sbjct: 33 HAGFHLTTAIVGPTILTLPYAFRGLGWWLGFVCLTTMGLVTFYAYYLMSKVLDHCEKSGR 92
Query: 75 RFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDDCEPIK 134
R ++ EL+ G L ++V+ Q + GI I +++ + L ++ L +K
Sbjct: 93 RHIRFRELAADVLGSGLMFYVVIFIQTAINTGIGIGAILLAGQCLDIMYSSLFPQ-GTLK 151
Query: 135 TTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGA---LPDVQY 191
FI + V VLS LPSF+S+ I+ + +SL Y+ + A I+ G P +Y
Sbjct: 152 LYEFIAMVTVVMMVLSQLPSFHSLRHINCASLLLSLGYTFLVVGACINLGLSKNAPKREY 211
Query: 192 SSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAYLV 251
S +S +G +F F ++ IA + G+ ++ EIQ+T+ P+ M +G+++ Y V
Sbjct: 212 SLEHSD-SGKVFSAFTSISIIAAIF-GNGILPEIQATLAP----PATGKMLKGLLLCYSV 265
Query: 252 VALCYFPVTIFGYRAFGNSVDDNILLSLEK-------PRWLIIAANIFVVVHVVGSYQVY 304
+ ++ I GY FGN+ NIL +L P +I A IFV++ + VY
Sbjct: 266 IFFTFYSAAISGYWVFGNNSSSNILKNLMPDEGPTLAPIVVIGLAVIFVLLQLFAIGLVY 325
Query: 305 AVPVFHMLESFLAE-------KMNFKPSRFLRFAIRNLYVSITMVLAITFPFFGGLLSFF 357
+ + ++E A+ K N P R +R LY++ +A PFFG + +
Sbjct: 326 SQVAYEIMEKKSADTTKGIFSKRNLVP----RLILRTLYMAFCGFMAAMLPFFGDINAVV 381
Query: 358 GGFVFAPTTYFLPCIMWIFIYKPKLFSLSW-----------CANWVSPIGALRQVIL 403
G F F P + LP +++ YKP S ++ CA + ++R+++L
Sbjct: 382 GAFGFIPLDFVLPMLLYNMTYKPTRRSFTYWINMTIMVVFTCAGLMGAFSSIRKLVL 438
>At5g63850 amino acid transporter AAP4 (pir||S51169)
Length = 466
Score = 150 bits (379), Expect = 1e-36
Identities = 118/416 (28%), Positives = 191/416 (45%), Gaps = 44/416 (10%)
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMHES---VSGKRFD 77
H +TA++G+ VL +A+ QLGW G T+++L T Y++ + + + + VSGKR
Sbjct: 29 HIITAVIGSGVLSLAWAIGQLGWIAGPTVMLLFSFVTYYSSTLLSDCYRTGDPVSGKRNY 88
Query: 78 KYHELSQHAFGE-RLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILC-------DD 129
Y + + G R + ++ Q + GI + Y + + S+ + C +
Sbjct: 89 TYMDAVRSILGGFRFKICGLI--QYLNLFGITVGYTIAASISMMAIKRSNCFHESGGKNP 146
Query: 130 CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHRGALPDV 189
C Y I +F + +LS + F+ + +S+VAA MS +YS I I + A V
Sbjct: 147 CHMSSNPYMI-MFGVTEILLSQIKDFDQIWWLSIVAAIMSFTYSAIGLALGIIQVAANGV 205
Query: 190 QYSSRYSTKAG------NIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWR 243
S G I+ F ALGDIAF Y+ V++EIQ T+ S P + + +
Sbjct: 206 VKGSLTGISIGAVTQTQKIWRTFQALGDIAFAYSYSVVLIEIQDTVRSPPAESKTMKIAT 265
Query: 244 GMIIAYLVVALCYFPVTIFGYRAFGNSVDDNIL--LSLEKPRWLIIAANIFVVVHVVGSY 301
+ IA V Y GY AFG+ N+L P WL+ AN +V+H+VG+Y
Sbjct: 266 RISIA--VTTTFYMLCGCMGYAAFGDKAPGNLLTGFGFYNPFWLLDVANAAIVIHLVGAY 323
Query: 302 QVYAVPVFHMLESFLAEKM-------------------NFKPSRFLRFAIRNLYVSITMV 342
QV+A P+F +E A + +K + F R R+ +V +T V
Sbjct: 324 QVFAQPIFAFIEKQAAARFPDSDLVTKEYEIRIPGFRSPYKVNVF-RAVYRSGFVVLTTV 382
Query: 343 LAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSLSWCANWVSPIGAL 398
+++ PFF ++ G F P T + P M+I K + +S+ W + G L
Sbjct: 383 ISMLMPFFNDVVGILGALGFWPLTVYFPVEMYIRQRKVERWSMKWVCLQMLSCGCL 438
>At5g23810 amino acid transporter
Length = 467
Score = 150 bits (379), Expect = 1e-36
Identities = 115/422 (27%), Positives = 195/422 (45%), Gaps = 48/422 (11%)
Query: 7 NIKSRNAKCCYSAFHNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIE 66
++ +R + H +T ++GA VL +A ++LGW G L+ TL +A+ + +
Sbjct: 22 SVTARTGTLWTAVAHIITGVIGAGVLSLAWATAELGWIAGPAALIAFAGVTLLSAFLLSD 81
Query: 67 MHESVSGK----RFDKYHELSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKL 122
+ R + Y + + G++ + V + + G I Y ++ A + +
Sbjct: 82 CYRFPDPNNGPLRLNSYSQAVKLYLGKKNEIVCGVVVYISL-FGCGIAYTIVIATCSRAI 140
Query: 123 HEILCDDCEPIKTT--------YFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYST 174
+ C T YF+VLF Q +S +P+F+++ +SLVAA MS +YS
Sbjct: 141 MKSNCYHRNGHNATCSYGDNNNYFMVLFGLTQIFMSQIPNFHNMVWLSLVAAIMSFTYSF 200
Query: 175 IAWIASIHR--------GALPDVQYSSRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQ 226
I ++ + G++ + +R ++ +F ALG+IAF Y ++LEIQ
Sbjct: 201 IGIGLALGKIIENRKIEGSIRGIPAENR----GEKVWIVFQALGNIAFSYPFSIILLEIQ 256
Query: 227 STIPSTPEKPSKVSMWRGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNIL--LSLEKPRW 284
T+ S P + K +M + +A + +F FGY AFG+S N+L +P W
Sbjct: 257 DTLRSPPAE--KQTMKKASTVAVFIQTFFFFCCGCFGYAAFGDSTPGNLLTGFGFYEPFW 314
Query: 285 LIIAANIFVVVHVVGSYQVYAVPVFHMLESFLAEKM--NFKPSRF--------------- 327
L+ AN +V+H+VG YQVY+ P+F E L +K N +RF
Sbjct: 315 LVDFANACIVLHLVGGYQVYSQPIFAAAERSLTKKYPENKFIARFYGFKLPLLRGETVRL 374
Query: 328 --LRFAIRNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWIFIYKPKLFSL 385
+R +R +YV IT +A+ FP+F +L G F P + P M I K + ++
Sbjct: 375 NPMRMCLRTMYVLITTGVAVMFPYFNEVLGVVGALAFWPLAVYFPVEMCILQKKIRSWTR 434
Query: 386 SW 387
W
Sbjct: 435 PW 436
>At1g44100 amino acid permease AAP5
Length = 480
Score = 148 bits (374), Expect = 5e-36
Identities = 111/399 (27%), Positives = 188/399 (46%), Gaps = 51/399 (12%)
Query: 21 HNVTAMVGAAVLGFPYAMSQLGWGLGITILVLSWICTLYTAWQMIEMH---ESVSGKRFD 77
H +TA++G+ VL +A++Q+GW G ++L T YT+ + + +SV+GKR
Sbjct: 38 HIITAVIGSGVLSLAWAVAQIGWIGGPVAMLLFSFVTFYTSTLLCSCYRSGDSVTGKRNY 97
Query: 78 KYHE-LSQHAFGERLGLWIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILC-------DD 129
Y + + + G ++ + VV Q + G I Y + A SL + C D
Sbjct: 98 TYMDAIHSNLGGIKVKVCGVV--QYVNLFGTAIGYTIASAISLVAIQRTSCQQMNGPNDP 155
Query: 130 CEPIKTTYFIVLFAFVQYVLSHLPSFNSVAGISLVAAAMSLSYSTIAWIASIHR------ 183
C + +++ F VQ + S +P F+ + +S+VAA MS +YS I + +
Sbjct: 156 CH-VNGNVYMIAFGIVQIIFSQIPDFDQLWWLSIVAAVMSFAYSAIGLGLGVSKVVENKE 214
Query: 184 --GALPDVQYS----SRYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPS 237
G+L V S T + I+ F +LG+IAF Y+ +++EIQ T+ S P + +
Sbjct: 215 IKGSLTGVTVGTVTLSGTVTSSQKIWRTFQSLGNIAFAYSYSMILIEIQDTVKSPPAEVN 274
Query: 238 KVSMWRGMIIAYLVVALCYFPVTIFGYRAFGNSVDDNILL--SLEKPRWLIIAANIFVVV 295
+M + ++ V + Y GY AFG++ N+L P WL+ AN+ +V+
Sbjct: 275 --TMRKATFVSVAVTTVFYMLCGCVGYAAFGDNAPGNLLAHGGFRNPYWLLDIANLAIVI 332
Query: 296 HVVGSYQVYAVPVFHMLESFLAEKMNFKPSRFL-------------------RFAIRNLY 336
H+VG+YQVY P+F +E + + F S F+ R R +
Sbjct: 333 HLVGAYQVYCQPLFAFVEKEASRR--FPESEFVTKEIKIQLFPGKPFNLNLFRLVWRTFF 390
Query: 337 VSITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPCIMWI 375
V T ++++ PFF ++ G F P T + P M+I
Sbjct: 391 VMTTTLISMLMPFFNDVVGLLGAIGFWPLTVYFPVEMYI 429
>At2g36590 putative proline transporter
Length = 436
Score = 113 bits (282), Expect = 2e-25
Identities = 96/384 (25%), Positives = 178/384 (46%), Gaps = 16/384 (4%)
Query: 18 SAFHNVTAMVGAAVLGFP-YAMSQLGWGLGITILVLSWICTLYTAWQMIEMHESVSGKRF 76
+AF T++ A VLG+ M LGW G+ L+L+ +LY + ++HE GKR
Sbjct: 33 AAFVLTTSINSAYVLGYSGTVMVPLGWIGGVVGLILATAISLYANTLVAKLHE-FGGKRH 91
Query: 77 DKYHELSQHAFGERLGL--WIVVPQQLMVEVGIDIVYMVIGAKSLKKLHEILCDDCEPIK 134
+Y +L+ +G + W++ L + I+ ++++ +LK ++ + DD +K
Sbjct: 92 IRYRDLAGFIYGRKAYCLTWVLQYVNLFM---INCGFIILAGSALKAVYVLFRDD-HAMK 147
Query: 135 TTYFIVLFAFVQYVLSHLPSFNSVAGISL-VAAAMSLSYSTIAWIASIHRGALPDVQYSS 193
+FI + + V + S GI L V+ +SL Y +A + S+ G +
Sbjct: 148 LPHFIAIAGLICAVFAIGIPHLSALGIWLAVSTILSLIYIVVAIVLSVKDGVKAPSRDYE 207
Query: 194 RYSTKAGNIFGIFNALGDIAFGYAGHNVILEIQSTIPSTPEKPSKVSMWRGMIIAYLVVA 253
+ +F I A + F + ++ EIQ+T+ ++P +M + + + V
Sbjct: 208 IQGSPLSKLFTITGAAATLVFVF-NTGMLPEIQATV----KQPVVKNMMKALYFQFTVGV 262
Query: 254 LCYFPVTIFGYRAFGNSVDDNILLSLEKPRWLIIAANIFVVVHVVGSYQVYAVPVFHMLE 313
L F V GY A+G+S +L ++ P W+ ANI ++ V S ++A P + ++
Sbjct: 263 LPMFAVVFIGYWAYGSSTSPYLLNNVNGPLWVKALANISAILQSVISLHIFASPTYEYMD 322
Query: 314 SFLAEKMNFKPSRFLRFAI--RNLYVSITMVLAITFPFFGGLLSFFGGFVFAPTTYFLPC 371
+ K N + L F I R Y++++ +L+ PF G +S G P T+ L
Sbjct: 323 TKFGIKGNPLALKNLLFRIMARGGYIAVSTLLSALLPFLGDFMSLTGAVSTFPLTFILAN 382
Query: 372 IMWIFIYKPKLFSLSWCANWVSPI 395
M+ KL +L +W++ +
Sbjct: 383 HMYYKAKNNKLNTLQKLCHWLNVV 406
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.329 0.141 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,816,326
Number of Sequences: 26719
Number of extensions: 363250
Number of successful extensions: 1107
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 953
Number of HSP's gapped (non-prelim): 64
length of query: 403
length of database: 11,318,596
effective HSP length: 102
effective length of query: 301
effective length of database: 8,593,258
effective search space: 2586570658
effective search space used: 2586570658
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135505.15


