

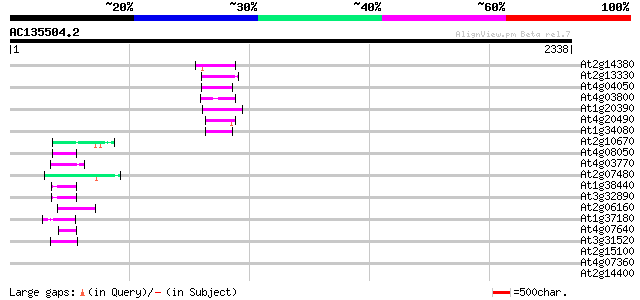
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135504.2 - phase: 0 /pseudo
(2338 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g14380 putative retroelement pol polyprotein 73 2e-12
At2g13330 F14O4.9 72 3e-12
At4g04050 putative transposon protein 70 1e-11
At4g03800 70 2e-11
At1g20390 hypothetical protein 66 2e-10
At4g20490 putative protein 61 7e-09
At1g34080 putative protein 60 2e-08
At2g10670 pseudogene 50 1e-05
At4g08050 49 3e-05
At4g03770 hypothetical protein 49 3e-05
At2g07480 F9A16.15 49 3e-05
At1g38440 hypothetical protein 49 5e-05
At3g32890 Athila ORF 1, putative 48 8e-05
At2g06160 putative Athila retroelement ORF1 protein 47 1e-04
At1g37180 hypothetical protein 46 2e-04
At4g07640 putative athila transposon protein 46 3e-04
At3g31520 hypothetical protein 45 4e-04
At2g15100 putative retroelement pol polyprotein 44 0.001
At4g07360 44 0.001
At2g14400 putative retroelement pol polyprotein 44 0.001
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 73.2 bits (178), Expect = 2e-12
Identities = 51/195 (26%), Positives = 88/195 (44%), Gaps = 27/195 (13%)
Query: 774 EQAYVDHEVTVDRFGDIVGNITSCNN---------------LW------------FSEDE 806
+Q + D V R I+G +T+C + LW F+ ++
Sbjct: 277 DQQHNDAPVPRRRVNMIMGGLTACRDSFRSIKEYIKSGAATLWSSPATKEMTPLTFTSED 336
Query: 807 MPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVK 866
+ HN L I ++ ++ +L+DTG+S+NV+ K L ++ + ++ S +
Sbjct: 337 LFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLT 396
Query: 867 AFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKL 926
FDG+ G I LP+ +G F V+D Y+ +LG PW HD A+ S+ HQ +
Sbjct: 397 GFDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCI 456
Query: 927 KFVKNGKLVTIHGEE 941
K + + TI G +
Sbjct: 457 KIPTSIGIETIRGNQ 471
>At2g13330 F14O4.9
Length = 889
Score = 72.4 bits (176), Expect = 3e-12
Identities = 48/156 (30%), Positives = 77/156 (48%), Gaps = 6/156 (3%)
Query: 798 NNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETP 857
+ + F E E+ + K H+ AL I ++ + LS ++VDTG+S++V+ + + ++
Sbjct: 45 DGITFWESEITDLDKPHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSK 104
Query: 858 LRRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGA 917
L+ + F G +G I LP L F V+D A ++ +LGRPW H A
Sbjct: 105 LQGRKTPLTGFAGDTTFSIGTIQLPTIARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKA 164
Query: 918 VTSTLHQKLKFVKNGKLVTIHGEETYLVSQLSSFSC 953
V ST HQ +KF + ++G SQ SS C
Sbjct: 165 VPSTYHQCIKFPSYKGIAVVYG------SQRSSRKC 194
>At4g04050 putative transposon protein
Length = 681
Score = 70.5 bits (171), Expect = 1e-11
Identities = 41/131 (31%), Positives = 70/131 (53%)
Query: 798 NNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETP 857
+ + F E E + K H+ AL I ++ + LS++++DTG+S++V+ ++ + ++
Sbjct: 519 HQITFWESETTDLNKPHDDALVIRIDVGNYELSHIMIDTGSSVDVLFYDAFKRMGHLDSE 578
Query: 858 LRRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGA 917
L+ + F G LG I LP L +F V++ A ++ +LGRPW H A
Sbjct: 579 LQGRKTPLTGFAGDTTFSLGTIQLPTIARGVRRLTSFLVVNKKAPFNAILGRPWLHAMKA 638
Query: 918 VTSTLHQKLKF 928
V ST HQ +KF
Sbjct: 639 VPSTYHQCIKF 649
>At4g03800
Length = 637
Score = 69.7 bits (169), Expect = 2e-11
Identities = 47/147 (31%), Positives = 74/147 (49%), Gaps = 20/147 (13%)
Query: 797 CNNLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVM---PKSTLDQLCY 853
C+++ F E+E + H+ AL I+++ + +S +LVDTG+S++++ P S L
Sbjct: 182 CSSVSFDEEETRHIERPHDDALIITLDVANFKISRILVDTGSSVDLIFLGPPSPL----- 236
Query: 854 RETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTH 913
AF LG I LP+ G + ++ F V D A+Y+ +LG PW
Sbjct: 237 ------------VAFTSESAMSLGTIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIF 284
Query: 914 DAGAVTSTLHQKLKFVKNGKLVTIHGE 940
AV ST HQ LKF + + TI G+
Sbjct: 285 QMKAVPSTYHQWLKFPTSNGVETIWGD 311
>At1g20390 hypothetical protein
Length = 1791
Score = 66.2 bits (160), Expect = 2e-10
Identities = 43/176 (24%), Positives = 81/176 (45%), Gaps = 7/176 (3%)
Query: 802 FSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRS 861
F++ ++ HN L + + ++ VL+DTG+S++++ K L + + ++
Sbjct: 555 FTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPV 614
Query: 862 TFLVKAFDGSRKNVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTST 921
+ + FDG +G I LP+ +G + F V+ A Y+ +LG PW H A+ ST
Sbjct: 615 SKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPST 674
Query: 922 LHQKLKFVKNGKLVTIHG-------EETYLVSQLSSFSCIEAGSAEGTAFQGLTIE 970
HQ +KF + + T+ +Y S+L + ++ T G+ E
Sbjct: 675 YHQCVKFPTHNGIFTLRAPKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAE 730
Score = 42.4 bits (98), Expect = 0.003
Identities = 46/224 (20%), Positives = 91/224 (40%), Gaps = 13/224 (5%)
Query: 129 MEERIEELAKELRREIKA----NRGSSDTIKTHDLCLVPKVDIPKKFKVPEFDRYNGLTC 184
+ I E++ ++ R +R +T +T + V I K+ + + YNG
Sbjct: 124 LRRTISEMSSKIHRATSTAPELDRVLEETQQTPFTRRITNVSIRGAQKI-KLESYNGRND 182
Query: 185 PQNHIIKY---VRKMGNYSDN-DSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAF 240
P+ + + + + DN D+ F + L A W++ L + + +F +L ++F
Sbjct: 183 PKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAHNWFSRLKPNSIDSFHQLTSSF 242
Query: 241 KNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARITPALDEEETTQTFMKTLK 300
HY + S L S+SQ ES + R++ IT + +E +
Sbjct: 243 LKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFKLVVTNIT--VPDEAAIVALRNAVW 300
Query: 301 KD--YVERMIIAAPNNFSKMVTMGTRLEEAVREGIIVFEKVESS 342
D + + + + AP+ + +R E E +I+ K S+
Sbjct: 301 YDSRFRDDITLHAPSTLEDALHRASRFIELEEEKLILARKHNST 344
>At4g20490 putative protein
Length = 853
Score = 61.2 bits (147), Expect = 7e-09
Identities = 40/140 (28%), Positives = 65/140 (45%), Gaps = 14/140 (10%)
Query: 814 HNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVKAFDGSRK 873
HN AL I + + + +L+DTG+S+N+M TL ++ E+ + + +DG K
Sbjct: 477 HNDALVIKLVMEDFDVERILIDTGSSVNLMFLKTLFKMGISESKIMPKIRPMTGYDGEAK 536
Query: 874 NVLGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTST------------ 921
+GEI L + +G F V+D Y+ +LG PW + A+ ST
Sbjct: 537 MSIGEIKLQVQVGGITQKTKFVVIDSEPIYNAILGSPWIYSMKAIPSTNLHTIRQPEGLT 596
Query: 922 --LHQKLKFVKNGKLVTIHG 939
L + + K G L+ I G
Sbjct: 597 DMLRDREEAAKEGNLIAIKG 616
>At1g34080 putative protein
Length = 811
Score = 60.1 bits (144), Expect = 2e-08
Identities = 36/112 (32%), Positives = 63/112 (56%)
Query: 817 ALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVKAFDGSRKNVL 876
AL IS++ + + +L+DTG+S++++ TL ++ + ++ + + +F L
Sbjct: 476 ALVISLDVGNCEVQRILIDTGSSVDLIFLDTLVRMGISKKDIKGAPSPLVSFTIETSMSL 535
Query: 877 GEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKLKF 928
G I LP+T +I F V D A+Y+ +LG PW ++ AV ST HQ +KF
Sbjct: 536 GTITLPVTAQGVVKMIEFTVFDRPAAYNIILGTPWLYEMKAVPSTYHQCVKF 587
>At2g10670 pseudogene
Length = 929
Score = 50.4 bits (119), Expect = 1e-05
Identities = 70/304 (23%), Positives = 111/304 (36%), Gaps = 59/304 (19%)
Query: 177 DRYNGLTC--PQNHIIKYVRKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDV 230
++++GL P +H+ + R G N D + F SL + A W +L K+ +
Sbjct: 84 NKFHGLPMEDPLDHLDELERLCGLTKINGVSEDGFKLRLFPFSLGDKAHLWEKTLPKNSI 143
Query: 231 HTFDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARITPALDEEE 290
T+D+ AF + N+R R + + K++ESFCE +R++G T
Sbjct: 144 TTWDDCKMAFLAKFFSNSRTARLRNEISGFTLKQNESFCEAWERFKGYQ---TKCPHHGF 200
Query: 291 TTQTFMKTLKKDYVE--RMIIAAPNN---FSKMVTMGTRL-EEAVREGIIVFEKVESSAN 344
+ + + TL + + RM++ +N K + G L E + G E S
Sbjct: 201 SQASLLNTLYRGVLPKIRMLLDTASNGNFLKKDIEEGWELVENLAQSGGNYNEDYGRSIY 260
Query: 345 ASKRNGNGHHKK----------------------KETEVWMVSAGAGQSMATV------- 375
S + HH++ E E + V G A +
Sbjct: 261 TSSNTDDKHHREMKALNDKLDKIIQMQQKHVHFISEDEPFQVQEGENDQCAEIRYVHNQG 320
Query: 376 --APINAAQLPPLYPYAQYSQHPFFPPFYHQYPLPSGQPQVPVNAIVQQMQQQPPTQQQQ 433
P AA P P+ Y+Q F P + G Q QQ PP
Sbjct: 321 PSLPSAAAAAEPTKPFVPYNQSLGFVP---KQQFQGG----------YQQQQPPPGFTPH 367
Query: 434 QQQA 437
QQQA
Sbjct: 368 QQQA 371
Score = 33.9 bits (76), Expect = 1.2
Identities = 34/117 (29%), Positives = 55/117 (46%), Gaps = 17/117 (14%)
Query: 830 SNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVKAFDGSRKNVLGEI-DLPMTIG-- 886
+N L D GAS+++MP S + +L + T ++ D + K G + D+P+ I
Sbjct: 589 NNCLCDLGASVSLMPLSVVKKLGFVHYKPCDLTLILA--DRTSKTPFGLLEDVPVMINGV 646
Query: 887 --PENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKLKFVKNGKLVTIHGEE 941
P +F++ MD + +LGRP+ GAV VK GK+ GE+
Sbjct: 647 EVPTDFVVL--EMDGESKDPLILGRPFLASVGAVID--------VKQGKINLNLGED 693
>At4g08050
Length = 1428
Score = 49.3 bits (116), Expect = 3e-05
Identities = 32/107 (29%), Positives = 52/107 (47%), Gaps = 6/107 (5%)
Query: 177 DRYNGLTC--PQNHIIKYVRKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDV 230
++++GL P +H+ ++ R N D + F SL + A W +LS D +
Sbjct: 52 NKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLSHDSI 111
Query: 231 HTFDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRG 277
T+D+ AF + + N R R + SQK ESFCE +R++G
Sbjct: 112 TTWDDYKKAFLSKFFSNARTARLRNEIYGFSQKTGESFCEAWERFKG 158
Score = 38.5 bits (88), Expect = 0.047
Identities = 33/102 (32%), Positives = 48/102 (46%), Gaps = 9/102 (8%)
Query: 830 SNVLVDTGASLNVMPKS---TLDQLCYRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIG 886
S+ L D GAS+N+MP S L+ + Y+ L L+ A SRK+ DLP+ I
Sbjct: 589 SDCLCDLGASVNLMPLSMARRLEFIQYKPCDLT----LILADRSSRKHFGMLKDLPVMIN 644
Query: 887 PENFLITFQVMDINASYS--CLLGRPWTHDAGAVTSTLHQKL 926
F V+D+ + +LGRP+ GAV K+
Sbjct: 645 GVEVPTDFVVLDMEVEHKDPLILGRPFLASVGAVIDVKEGKI 686
>At4g03770 hypothetical protein
Length = 464
Score = 49.3 bits (116), Expect = 3e-05
Identities = 40/148 (27%), Positives = 63/148 (42%), Gaps = 12/148 (8%)
Query: 168 PKKFKVPEFDRYNGLTCPQNH----IIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYT 223
P K ++ F NG + P+ H II R + D+ + H F + L A W+T
Sbjct: 231 PGKLRIEYF---NGSSDPKGHLKLFIISVARAKFRPEERDAGLCHLFVEHLKGPALNWFT 287
Query: 224 SLSKDDVHTFDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARIT 283
L + V +F EL+ F Y S L SLSQ+ +E ++ ++R A++
Sbjct: 288 RLKGNSVDSFQELSTLFLKQYSVLIDPSTSDADLWSLSQQPNEPLRDFLAKFRSTLAKV- 346
Query: 284 PALDEEETTQTFMKTLKKDYVERMIIAA 311
E + TLKK R ++A
Sbjct: 347 ----EGINDLAALSTLKKALCLRARLSA 370
>At2g07480 F9A16.15
Length = 1012
Score = 48.9 bits (115), Expect = 3e-05
Identities = 81/375 (21%), Positives = 138/375 (36%), Gaps = 69/375 (18%)
Query: 145 KANRGSSDTIKTHD--LCLVPKVDIPKKFKVP-------EFDRYNGLTC--PQNHIIKYV 193
++N G+ D + H +VP FK+ + ++++GL P +H+ +
Sbjct: 6 QSNIGAGDAPRNHHQRAGIVPPPIQNNNFKIKSDLISMIQGNKFHGLPMEDPLDHLDNFD 65
Query: 194 RKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKNHYGFNTR 249
R N +S + F SL + A W +L + T+D+ AF + N+R
Sbjct: 66 RLCSLTKINGVSEESFKLRLFPFSLGDKAHLWEKTLPVKSIDTWDDCKKAFLAKFFSNSR 125
Query: 250 LKPSREFLRSLSQKKDESFCEYAQRWRGAAARITPALDEEETTQTFMKTLKKDYVERMII 309
R + +QK ESF E +R++G + P + + ++ L K RM++
Sbjct: 126 KARLRSEISGFNQKNSESFSEAWERFKGYTTQ-CPHHESLPPQYSILRCLPK---IRMLL 181
Query: 310 AAPNN---FSKMVTMGTRLEEAVREGIIVFEKVESSANASKRNGNGHHKKK--------- 357
+N +K V G L E + + + + N + HKK+
Sbjct: 182 DTASNGNILNKDVAEGWELVENLAQSHRNYNEDYDRTNRGSSDSEDKHKKEIKTLNDRID 241
Query: 358 --------------ETEVWMVSAGAGQSMATVAPINAA--------QLPPLYPYAQYSQH 395
E E+ + G ++ V+ + P +P Y +
Sbjct: 242 KLVLAQQRNVYYITEEELTQLQDGENLTIEEVSYLQNQGGYNKGFNNYKPPHPNLSYRSN 301
Query: 396 PFFPPFYHQYPLPSGQPQ-----VPVNAIVQQMQQQPP---TQQQQQQQARPTFPPIPML 447
P YP P QP VP N Q Q P TQQ QQ A+ +
Sbjct: 302 NVANPQDQVYP-PQNQPTQAKPFVPYNQGYNQKQNFGPPGFTQQPQQPSAQDS------- 353
Query: 448 YFELLPTLLLRGHCT 462
+ LP L++GH +
Sbjct: 354 EMKTLPQQLVQGHAS 368
>At1g38440 hypothetical protein
Length = 263
Score = 48.5 bits (114), Expect = 5e-05
Identities = 33/103 (32%), Positives = 47/103 (45%), Gaps = 12/103 (11%)
Query: 175 EFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFD 234
EFDR LT K+ S+ D + F SL + A W +L D + T+D
Sbjct: 21 EFDRLYNLT-----------KINGVSE-DGFKLRLFPFSLGDKAHIWEKNLPHDSITTWD 68
Query: 235 ELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRG 277
+ AF + + N R R + SQK ESFCE +R++G
Sbjct: 69 DCKKAFLSKFFSNARTARLRNEISGFSQKTSESFCEAWERFKG 111
>At3g32890 Athila ORF 1, putative
Length = 755
Score = 47.8 bits (112), Expect = 8e-05
Identities = 33/103 (32%), Positives = 47/103 (45%), Gaps = 12/103 (11%)
Query: 175 EFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFD 234
EFDR LT K+ S+ D + F SL + A W +L D + T+D
Sbjct: 21 EFDRLYNLT-----------KINGVSE-DGFKLRLFPFSLGDKAHIWEKNLPHDSITTWD 68
Query: 235 ELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRG 277
+ AF + + N R R + SQK ESFCE +R++G
Sbjct: 69 DCKKAFLSKFFSNARTARLRNEISGFSQKTRESFCEAWERFKG 111
Score = 33.5 bits (75), Expect = 1.5
Identities = 53/240 (22%), Positives = 101/240 (42%), Gaps = 20/240 (8%)
Query: 692 KAATPTTPPIDKATLTDVSPITKDVSRPSQSIEDSNLDEILRIIKRSDYKIVDQLLQTPS 751
+A+T +T A + + K PS ++ + + K K +D++
Sbjct: 414 EASTQSTSEEKAAIIERMVKRFKPTPLPSSALPWTFRKAWMEKYKSVASKQLDEIEAVMP 473
Query: 752 KISVLSLLLSSEAHRNTLLKVLEQAYVDHEVTVDRFGDIVGNITSCNNLWFSEDEMPEAG 811
I VL+L+ + H++ +LE+ + H+ D D + S + ++++ + G
Sbjct: 474 LIEVLNLI--PDPHKDVRNLILERIKMYHDS--DDESDATPSRASDKRI--VQEKLEDPG 527
Query: 812 KYHNLALHISVNCKSDML--SNVLVDTGASLNVMPKSTLDQLCYRETPLRRSTFLVKAFD 869
+ ++ C L S+ L D GAS+++MP S +L + + T ++ D
Sbjct: 528 SF-------TLPCSIGELAFSDCLCDLGASVSLMPLSVARRLEFIQYKPCDLTLILA--D 578
Query: 870 GSRKNVLGEI-DLPMTIGPENFLITFQVMDINASYS--CLLGRPWTHDAGAVTSTLHQKL 926
S + G + DLP+ I F V+D+ + +LGRP GAV K+
Sbjct: 579 RSFRKPFGMLKDLPVMINGVEVPTDFVVLDMEVEHKDPLILGRPLLASVGAVIDVREGKI 638
>At2g06160 putative Athila retroelement ORF1 protein
Length = 750
Score = 47.4 bits (111), Expect = 1e-04
Identities = 38/162 (23%), Positives = 70/162 (42%), Gaps = 3/162 (1%)
Query: 198 NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKNHYGFNTRLKPSREFL 257
N D F SL + W +L ++ + T+D+ AF + N+R R +
Sbjct: 79 NGVSEDGFKFRLFPFSLGDKTHLWEKTLPQNSITTWDDCKKAFFAKFFSNSRTARLRNEI 138
Query: 258 RSLSQKKDESFCEYAQRWRGAAARIT-PALDEEETTQTFMKTLKKDYVERMIIAAPNNF- 315
+QK++ESFCE +R++G + P + T + + + A+ NF
Sbjct: 139 SGFTQKQNESFCEAWERFKGYQTKCPHPGFKQASLLSTLYRGVLPKLRMLLDTASNGNFL 198
Query: 316 SKMVTMGTRLEEAVREGIIVF-EKVESSANASKRNGNGHHKK 356
+K V G L E + + + E + S + S + + HH++
Sbjct: 199 NKDVEKGWELVENLAQSDGNYNENYDKSISTSSESDDKHHRE 240
>At1g37180 hypothetical protein
Length = 661
Score = 46.2 bits (108), Expect = 2e-04
Identities = 36/139 (25%), Positives = 64/139 (45%), Gaps = 11/139 (7%)
Query: 137 AKELRREIKANRGSSDTIKTHDLCLVPKVDIPKKFKVPEFDRYNGLTCPQNHIIKY---V 193
A E+ + I+A R + T + L ++ + FK+P YNG P H+ + V
Sbjct: 284 APEVEKVIEATRRTPFTPRISKL----RIREFRDFKLPV---YNGKGDPNEHLTSFQVIV 336
Query: 194 RKMG-NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKNHYGFNTRLKP 252
R++ + D+ + F ++L A W+T L + + F +L+ AF YG+ +
Sbjct: 337 RRVPLEPYEEDAGLCKLFSENLSGPALTWFTQLEEGSIDNFKQLSTAFIKQYGYFIKSDI 396
Query: 253 SREFLRSLSQKKDESFCEY 271
+ L +LSQ D+ Y
Sbjct: 397 TEAHLWNLSQSADDPLRTY 415
>At4g07640 putative athila transposon protein
Length = 866
Score = 45.8 bits (107), Expect = 3e-04
Identities = 25/75 (33%), Positives = 37/75 (49%)
Query: 203 DSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKNHYGFNTRLKPSREFLRSLSQ 262
D + F SL + A W +L D + T+D+ AF + + N R R + SQ
Sbjct: 84 DGFKLRLFPFSLGDKAHIWEKNLPHDSIITWDDCKKAFLSKFFSNARTARLRNEISGFSQ 143
Query: 263 KKDESFCEYAQRWRG 277
K ESFCE +R++G
Sbjct: 144 KTGESFCEAWERFKG 158
>At3g31520 hypothetical protein
Length = 528
Score = 45.4 bits (106), Expect = 4e-04
Identities = 32/119 (26%), Positives = 53/119 (43%), Gaps = 7/119 (5%)
Query: 168 PKKFKVPEFDRYNGLTCPQNH----IIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYT 223
P K ++ F NG + P+ H II R + D+ + H F + L A W++
Sbjct: 239 PGKLRIEYF---NGSSDPKRHLKSFIISVARTKFRPEERDAGLCHLFVEHLKGPALCWFS 295
Query: 224 SLSKDDVHTFDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARI 282
L + + +F EL+ F Y L S L SLSQ+ +E ++ + R A++
Sbjct: 296 RLEGNSMDSFQELSTLFLKQYSVLIDLGTSDADLWSLSQQPNEPLRDFLAKLRSTLAKV 354
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 43.9 bits (102), Expect = 0.001
Identities = 25/66 (37%), Positives = 36/66 (53%), Gaps = 1/66 (1%)
Query: 876 LGEIDLPMTIGPENFLITFQVMDINASYSCLLGRPWTHDAGAVTSTLHQKLKF-VKNGKL 934
LG + LP+T ++ F V D A+Y+ +LG PW ++ V ST HQ +KF GK+
Sbjct: 353 LGTVVLPVTAQGVVKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGKM 412
Query: 935 VTIHGE 940
I E
Sbjct: 413 TGISTE 418
>At4g07360
Length = 675
Score = 43.5 bits (101), Expect = 0.001
Identities = 37/142 (26%), Positives = 62/142 (43%), Gaps = 19/142 (13%)
Query: 175 EFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFD 234
EFDR LT K+ S+ D + F SL + A W +L D + T+D
Sbjct: 10 EFDRLCNLT-----------KINGVSE-DGFKLRLFPFSLGDKAHIWEKNLLHDSITTWD 57
Query: 235 ELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWR------GAAARITPALDE 288
+ AF + + N R R + SQK ES CE +R++ + D
Sbjct: 58 DCKKAFLSKFFSNARTARLRNEISGFSQKTGESLCEAWERFKDGNYNEDCDRTVKGTADS 117
Query: 289 EETTQTFMKTLKKDYVERMIIA 310
++ + +KTL D ++R++++
Sbjct: 118 DDKHRKEIKTL-NDKLDRILLS 138
Score = 36.2 bits (82), Expect = 0.23
Identities = 56/236 (23%), Positives = 97/236 (40%), Gaps = 20/236 (8%)
Query: 702 DKATLTDVSPITKDVSRPSQSIEDSNLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLS 761
+ +T +VS + + S S+ + S +E II+R +V + TP L
Sbjct: 300 EASTQVEVSIVKFNHSAGSRHLTQSTSEEKAAIIER----MVKRFKPTPLPSRALPWTFR 355
Query: 762 S---EAHRNTLLKVLEQAYVDHEVTVDRFGDIVGNITSCNNLWFSEDEMPEAGKYHNLAL 818
E +++ K L++ V + ++ I NL +M + AL
Sbjct: 356 KAWMEKYKSVAAKQLDEIEVVMPL-MEVLNLIPDPHKDVRNLILERIKMYHDSDDESDAL 414
Query: 819 HISVNCKS---DMLSNVLVDTGASLNVMPKST---LDQLCYRETPLRRSTFLVKAFDGSR 872
H+ + + S+ L D GAS+++MP S L+ + Y+ L L+ A SR
Sbjct: 415 HLELLIRGLFKRTFSDCLCDLGASVSLMPLSVARRLEFIRYKPCDLT----LILADRSSR 470
Query: 873 KNVLGEIDLPMTIGPENFLITFQVMDINASY--SCLLGRPWTHDAGAVTSTLHQKL 926
+ D+P+ I F V+D+ + S +LGRP+ GAV K+
Sbjct: 471 RPFSMLKDVPVMINGVEVPTDFVVLDMEVEHKDSLILGRPFLASVGAVIDVREGKI 526
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 43.5 bits (101), Expect = 0.001
Identities = 26/111 (23%), Positives = 50/111 (44%), Gaps = 4/111 (3%)
Query: 177 DRYNGLTCPQNHIIKYVRKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHT 232
+ YNGL P+ ++ ++ G N +D D F ++L A W+T L +
Sbjct: 163 ESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSENLCGQALMWFTQLEPGSISN 222
Query: 233 FDELAAAFKNHYGFNTRLKPSREFLRSLSQKKDESFCEYAQRWRGAAARIT 283
F+EL+ F Y S L +LSQ +E+ + +++ ++++
Sbjct: 223 FNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFITKFKYVLSKLS 273
Score = 35.0 bits (79), Expect = 0.52
Identities = 23/88 (26%), Positives = 47/88 (53%), Gaps = 4/88 (4%)
Query: 799 NLWFSEDEMPEAGKYHNLALHISVNCKSDMLSNVLVDTGASLNVMPKSTLDQLCYRETPL 858
++ F E E + H+ AL ++++ + +S +L+DTG+S++++ STL+++ +
Sbjct: 408 SILFDEKETQHLERSHDDALVVTLDVANFEVSRILIDTGSSVDLIFLSTLERMGISRADV 467
Query: 859 RRSTFLVKAFDGSRKNVLGEIDLPMTIG 886
R V+ K V E+D + IG
Sbjct: 468 NRRKLGVE----RAKAVNDEVDKLLKIG 491
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.340 0.147 0.493
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,469,320
Number of Sequences: 26719
Number of extensions: 2026528
Number of successful extensions: 8483
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 8083
Number of HSP's gapped (non-prelim): 318
length of query: 2338
length of database: 11,318,596
effective HSP length: 115
effective length of query: 2223
effective length of database: 8,245,911
effective search space: 18330660153
effective search space used: 18330660153
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC135504.2


