

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135504.12 - phase: 0
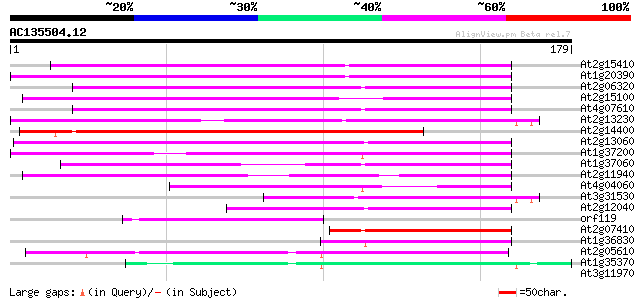
(179 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g15410 putative retroelement pol polyprotein 124 4e-29
At1g20390 hypothetical protein 119 7e-28
At2g06320 putative retroelement pol polyprotein 111 2e-25
At2g15100 putative retroelement pol polyprotein 111 2e-25
At4g07610 108 2e-24
At2g13230 pseudogene 108 2e-24
At2g14400 putative retroelement pol polyprotein 108 2e-24
At2g13060 pseudogene 106 8e-24
At1g37200 hypothetical protein 102 1e-22
At1g37060 Athila retroelment ORF 1, putative 97 4e-21
At2g11940 putative retroelement gag/pol polyprotein 88 2e-18
At4g04060 putative transposon protein 68 3e-12
At3g31530 hypothetical protein 62 2e-10
At2g12040 T10J7.2 59 1e-09
orf119 -mitochondrial genome- 52 2e-07
At2g07410 putative retroelement pol polyprotein 49 1e-06
At1g36830 hypothetical protein 46 1e-05
At2g05610 putative retroelement pol polyprotein 40 5e-04
At1g35370 hypothetical protein 40 9e-04
At3g11970 hypothetical protein 39 0.001
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 124 bits (310), Expect = 4e-29
Identities = 57/147 (38%), Positives = 88/147 (59%), Gaps = 1/147 (0%)
Query: 14 LIKCLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDAEDYVNLCDKCQ 73
L+ CL E Q+V+ ++H+G G H ++LA K+ + G YWPTM D E +V C+KCQ
Sbjct: 1422 LLSCLDDEEAQQVMREIHEGAGGNHSGGRALALKIRKHGQYWPTMNADCEKFVARCEKCQ 1481
Query: 74 RHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINYSTKWIEVEPLA 133
H P L + + P+PF W +D++ PFP A+ Q KY++V +Y TKW E E A
Sbjct: 1482 GHAPFIHIPSEVLQTAMPPYPFMRWAMDIIRPFP-ASRQKKYILVMTDYFTKWFEAESYA 1540
Query: 134 KIKAKNVLRFFKRNILARFGVPALVIS 160
+I++K V F +NI+ G+P +++
Sbjct: 1541 RIQSKEVQNFVWKNIICHHGLPYEIVT 1567
>At1g20390 hypothetical protein
Length = 1791
Score = 119 bits (299), Expect = 7e-28
Identities = 56/160 (35%), Positives = 93/160 (58%), Gaps = 1/160 (0%)
Query: 1 MEGTLYKRGFITPLIKCLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLK 60
M+ L K ++ CL E E++ + H+G G H ++LA K+ + G+YWPTM+
Sbjct: 1413 MKEHLLKVSAFGAMLNCLHGTEINEIMKETHEGAAGNHSGGRALALKLKKLGFYWPTMIS 1472
Query: 61 DAEDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAI 120
D + + C++CQRH P L + V+P+PF W +D++GP P A+ Q ++++V
Sbjct: 1473 DCKTFTAKCEQCQRHAPTIHQPTELLRAGVAPYPFMRWAMDIVGPMP-ASRQKRFILVMT 1531
Query: 121 NYSTKWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVIS 160
+Y TKW+E E A I+A +V F + I+ R G+P +I+
Sbjct: 1532 DYFTKWVEAESYATIRANDVQNFVWKFIICRHGLPYEIIT 1571
>At2g06320 putative retroelement pol polyprotein
Length = 466
Score = 111 bits (278), Expect = 2e-25
Identities = 55/140 (39%), Positives = 81/140 (57%), Gaps = 1/140 (0%)
Query: 21 NETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDAEDYVNLCDKCQRHGDMHL 80
+E + +L HD G H K+L+AG++WPTM KDA+++V+ CD CQR G++
Sbjct: 94 DEVEGILLHCHDSAYGGHFATFKTVSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISR 153
Query: 81 APPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINYSTKWIEVEPLAKIKAKNV 140
+ ++ F WGID +GPFP + G KY++V ++Y +KW+E AK V
Sbjct: 154 RNEMPQNPILEVDIFDVWGIDFMGPFPSSYGN-KYILVVVDYVSKWVEAIASPTNDAKVV 212
Query: 141 LRFFKRNILARFGVPALVIS 160
L+ FK I RFGV +VIS
Sbjct: 213 LKLFKTIIFPRFGVSWVVIS 232
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 111 bits (277), Expect = 2e-25
Identities = 54/156 (34%), Positives = 87/156 (55%), Gaps = 14/156 (8%)
Query: 5 LYKRGFITPLIKCLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDAED 64
LY+R F P C+ +T+ V+ +VHDG CG H +SLA KV + GYYWPT++ D E
Sbjct: 968 LYRRIFSAPDAVCIFGEQTRTVMKEVHDGTCGNHTGGRSLAFKVRKYGYYWPTLVADCEA 1027
Query: 65 YVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINYST 124
Y C++CQ+H + L P LT++ +P+PF W +D++GP ++ ST
Sbjct: 1028 YARKCEQCQKHAPLILQPAELLTTVSAPYPFMKWLMDIVGP--------------LHVST 1073
Query: 125 KWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVIS 160
+ +E + I V F ++I+ R G+P +++
Sbjct: 1074 RGVEAAAYSNITHVQVWNFIWKDIICRHGLPYEIVT 1109
>At4g07610
Length = 276
Score = 108 bits (270), Expect = 2e-24
Identities = 54/140 (38%), Positives = 81/140 (57%), Gaps = 1/140 (0%)
Query: 21 NETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDAEDYVNLCDKCQRHGDMHL 80
+E + +L H G + K+L+AG++WPTM K+A+++V CD C R G++
Sbjct: 79 DEVEGILLHCHCSTYGGNFTTFKTVSKILQAGFWWPTMFKEAQEFVLKCDSCHRKGNISK 138
Query: 81 APPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINYSTKWIEVEPLAKIKAKNV 140
+ ++ F WGID +GPFP + G KY++VA++Y +KWIE AK V
Sbjct: 139 RNEMPQNPILEVELFDVWGIDFIGPFPSSYGN-KYILVAVDYVSKWIEATASPTNDAKVV 197
Query: 141 LRFFKRNILARFGVPALVIS 160
L+ FK I +RFGVP +VIS
Sbjct: 198 LKLFKTIIFSRFGVPRVVIS 217
>At2g13230 pseudogene
Length = 353
Score = 108 bits (270), Expect = 2e-24
Identities = 63/181 (34%), Positives = 98/181 (53%), Gaps = 20/181 (11%)
Query: 1 MEGTLYKRGFITPLIKCLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLK 60
+E L+K PL+ C+ ++V+ +VH G CG + ++LA K+ R GY+WPTM+K
Sbjct: 125 VEEKLFKWRLSGPLMTCVEKEAARKVMKEVHGGSCGNNSGGRALAIKIKRLGYFWPTMIK 184
Query: 61 DAEDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAI 120
D C+KCQR+ P L+S+ SP+P W +D++GP A+ Q K ++V
Sbjct: 185 D-------CEKCQRNAPTIHLPAELLSSIASPYPLMRWSMDIIGPM-HASKQKKLVLVLT 236
Query: 121 NYSTKWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVIS------VLSRF------WGI 168
+Y +K E E A IK V F ++IL R GVP +++ + +RF WGI
Sbjct: 237 DYFSKLREAESYASIKEAQVESFMWKHILCRHGVPYEIVTDNGSQFISTRFQGFCDKWGI 296
Query: 169 K 169
+
Sbjct: 297 R 297
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 108 bits (269), Expect = 2e-24
Identities = 48/130 (36%), Positives = 80/130 (60%), Gaps = 2/130 (1%)
Query: 4 TLYKRGFITP-LIKCLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDA 62
+LY+RG P L+ GP + V+++VH+G+CG H +++A K+ + YYWPTM+ D
Sbjct: 952 SLYRRGVSDPYLLSIFGP-VVEIVMSEVHEGLCGSHSSGRAMAFKIKKMCYYWPTMITDC 1010
Query: 63 EDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINY 122
Y C +CQ H + P +S+ +P+PF W +D++GP ++T ++YL+V +Y
Sbjct: 1011 VKYAQRCKRCQLHAPLIHQPSELFSSISAPYPFMRWSMDIIGPLHRSTRGVQYLLVLTDY 1070
Query: 123 STKWIEVEPL 132
+KWIE E +
Sbjct: 1071 FSKWIEAEAI 1080
>At2g13060 pseudogene
Length = 930
Score = 106 bits (264), Expect = 8e-24
Identities = 54/159 (33%), Positives = 82/159 (50%), Gaps = 1/159 (0%)
Query: 2 EGTLYKRGFITPLIKCLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKD 61
E LYK + +C+ E +L+ H G H KVL+A ++WPTM +D
Sbjct: 174 EPYLYKHSYDGIYRRCIAATEVPSILSHCHSSSYGGHFATFKTVSKVLQADFWWPTMFRD 233
Query: 62 AEDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAIN 121
+ +++ CD CQR G + + + F WGID +GPFP + L Y++V ++
Sbjct: 234 TQKFISQCDPCQRRGKISKRNEMPPNFRLEVEVFDRWGIDFMGPFPPSNKNL-YILVDVD 292
Query: 122 YSTKWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVIS 160
Y +KW+E K + V++ FK I RFGVP +VIS
Sbjct: 293 YVSKWVEAIASLKNDSAVVMKLFKSIIFPRFGVPRIVIS 331
>At1g37200 hypothetical protein
Length = 1564
Score = 102 bits (254), Expect = 1e-22
Identities = 56/162 (34%), Positives = 86/162 (52%), Gaps = 12/162 (7%)
Query: 1 MEGTLYKRGFITPLIKCLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLK 60
MEG L KR P + C +T++++ +H+G CG H ++L ML
Sbjct: 1281 MEGRLMKRSVAGPYMVCTYGQQTKDLMKSMHEGQCGSHYSGRTLRC----------IMLA 1330
Query: 61 DAEDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATG--QLKYLVV 118
D + CDKCQRH PP E++S+ SP+PF W +D++ P + G +LK L+V
Sbjct: 1331 DCIAHSLRCDKCQRHAPTLHQPPEEMSSISSPYPFMKWSMDVVRPMEASGGKKRLKNLLV 1390
Query: 119 AINYSTKWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVIS 160
+YSTKWIE + ++ K V F NI+ R G+P +++
Sbjct: 1391 LTDYSTKWIEGKAFQQVTEKQVEVFLLENIVYRHGIPYEIVT 1432
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 97.4 bits (241), Expect = 4e-21
Identities = 51/144 (35%), Positives = 76/144 (52%), Gaps = 21/144 (14%)
Query: 17 CLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDAEDYVNLCDKCQRHG 76
C+ +E + +L H G H K+L+AG++WP+M KDA+++++ CD +
Sbjct: 1386 CVSEDEIEGILLHCHGFAYGGHFATFKTMSKILQAGFWWPSMFKDAQEFISKCDSFEN-- 1443
Query: 77 DMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINYSTKWIEVEPLAKIK 136
F WGID +GPFP + G KY++VAI+Y +KW+E
Sbjct: 1444 ------------------FDVWGIDFMGPFPSSYGN-KYILVAIDYVSKWVEAIASHTND 1484
Query: 137 AKNVLRFFKRNILARFGVPALVIS 160
A+ VL+ FK I RFGVP +VIS
Sbjct: 1485 ARVVLKLFKTIIFPRFGVPRIVIS 1508
>At2g11940 putative retroelement gag/pol polyprotein
Length = 1212
Score = 88.2 bits (217), Expect = 2e-18
Identities = 46/156 (29%), Positives = 79/156 (50%), Gaps = 19/156 (12%)
Query: 5 LYKRGFITPLIKCLGPNETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDAED 64
LY+R P C+ +T+ V+ ++HDG CG H +SLA K+ + GY+WPTM+ D E
Sbjct: 911 LYRRIISAPDAICIFGEQTRTVIKEIHDGTCGNHSGGRSLAFKLKKYGYFWPTMVADCEA 970
Query: 65 YVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINYST 124
Y + + G +SP PF W +D++GP +T +++L++
Sbjct: 971 YAHRSRRTFHFG-------------ISPLPFMKWSMDIVGPLHVSTRGVRFLLL------ 1011
Query: 125 KWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVIS 160
+W+E + I V F + I+ R G+P +++
Sbjct: 1012 EWVEAAAYSYITQVQVRHFIWKEIICRHGLPYEIVT 1047
>At4g04060 putative transposon protein
Length = 375
Score = 67.8 bits (164), Expect = 3e-12
Identities = 39/111 (35%), Positives = 56/111 (50%), Gaps = 19/111 (17%)
Query: 52 GYYWPTMLKDAEDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATG 111
GY+WPTML + + CDKCQRH P E++S+ SP+PF W +D++GP + G
Sbjct: 195 GYFWPTMLANCIAHSLRCDKCQRHAPTLHQLPKEMSSISSPYPFMKWSMDVVGPMEASGG 254
Query: 112 --QLKYLVVAINYSTKWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVIS 160
+LK L+V K V F NI+ R G+P +I+
Sbjct: 255 KKKLKNLLVT-----------------EKQVEDFLWENIVCRHGIPYEIIT 288
>At3g31530 hypothetical protein
Length = 831
Score = 62.0 bits (149), Expect = 2e-10
Identities = 35/100 (35%), Positives = 55/100 (55%), Gaps = 13/100 (13%)
Query: 82 PPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINYSTKWIEVEPLAKIKAKNVL 141
P L+S+ SP+PF W +D++GP + Q K ++V +Y +KWIE E A IK V
Sbjct: 648 PAELLSSIASPYPFMRWSMDIIGPMHPSK-QKKLVLVLTDYFSKWIEAESYASIKDAQVE 706
Query: 142 RFFKRNILARFGVPALVIS------VLSRF------WGIK 169
F ++IL R G+P +++ + +RF WGI+
Sbjct: 707 NFVLKHILCRHGIPYEIVTDNGSQFISTRFQGFCDKWGIR 746
>At2g12040 T10J7.2
Length = 976
Score = 58.9 bits (141), Expect = 1e-09
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 1/91 (1%)
Query: 70 DKCQRHGDMHLAPPVELTSLVSPWPFAWWGIDLLGPFPKATGQLKYLVVAINYSTKWIEV 129
D CQR G + + ++ F WGI+ +GP P + L Y++V ++Y +KW+E
Sbjct: 544 DPCQRRGKISNCNEMPQKFILEVKVFDCWGINFMGPIPPSNKNL-YILVVVDYVSKWVEA 602
Query: 130 EPLAKIKAKNVLRFFKRNILARFGVPALVIS 160
K + V++ FK I FGVP +VIS
Sbjct: 603 IASPKNDSAVVMKLFKCIIFPHFGVPRIVIS 633
>orf119 -mitochondrial genome-
Length = 119
Score = 51.6 bits (122), Expect = 2e-07
Identities = 27/64 (42%), Positives = 35/64 (54%), Gaps = 2/64 (3%)
Query: 37 QHLDVKSLARKVLRAGYYWPTMLKDAEDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFA 96
QHL K R VL+AG+YWPT KDA +V+ CD CQR G+ + ++ F
Sbjct: 26 QHL--KQWPRFVLQAGFYWPTTFKDAHGFVSSCDACQRKGNFTKRNEMPQHFILEVEVFD 83
Query: 97 WWGI 100
WGI
Sbjct: 84 VWGI 87
>At2g07410 putative retroelement pol polyprotein
Length = 411
Score = 48.9 bits (115), Expect = 1e-06
Identities = 26/58 (44%), Positives = 36/58 (61%), Gaps = 1/58 (1%)
Query: 103 LGPFPKATGQLKYLVVAINYSTKWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVIS 160
+G FP + G KY++VA++Y +KW+E AK VL+ FK I RFGV +VIS
Sbjct: 1 MGSFPSSYGN-KYILVAVDYVSKWVEAIVSPTNDAKVVLKLFKTIIFTRFGVHMVVIS 57
>At1g36830 hypothetical protein
Length = 243
Score = 45.8 bits (107), Expect = 1e-05
Identities = 23/63 (36%), Positives = 38/63 (59%), Gaps = 2/63 (3%)
Query: 100 IDLLGPFPKATGQ--LKYLVVAINYSTKWIEVEPLAKIKAKNVLRFFKRNILARFGVPAL 157
+D++GP + G+ LK L+V +YSTKWIE + ++ K V F NI+ + G+P
Sbjct: 1 MDVVGPMEASGGKKKLKNLLVLTDYSTKWIEAKAFQQVTEKQVEDFLWENIVCQHGIPYE 60
Query: 158 VIS 160
+I+
Sbjct: 61 IIT 63
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 40.4 bits (93), Expect = 5e-04
Identities = 35/160 (21%), Positives = 66/160 (40%), Gaps = 9/160 (5%)
Query: 6 YKRGFITPLIKCLGPNET---QEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDA 62
+ +G + K + PN + +L +H G H K + + ++ +YW +M+KD
Sbjct: 452 WSQGVLRRKNKIVVPNNSGIKDTILRWLHCSGMGGHSG-KEVTHQRVKGLFYWKSMVKDI 510
Query: 63 EDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAWW---GIDLLGPFPKATGQLKYLVVA 119
+ ++ C CQ+ + A P L L P P W +D + P + G+ +VV
Sbjct: 511 QAFIRSCGTCQQCKSDNAASPGLLQPL--PIPDRIWSDVSMDFIDGLPLSNGKTVIMVVV 568
Query: 120 INYSTKWIEVEPLAKIKAKNVLRFFKRNILARFGVPALVI 159
S + A V + + N+ G P+ ++
Sbjct: 569 DRLSKAAHFIALAHPYSAMTVAQAYLDNVFKLHGCPSSIV 608
>At1g35370 hypothetical protein
Length = 1447
Score = 39.7 bits (91), Expect = 9e-04
Identities = 40/150 (26%), Positives = 58/150 (38%), Gaps = 20/150 (13%)
Query: 38 HLDVKSLARKVLRAGYYWPTMLKDAEDYVNLCDKCQRHGDMHLAPPVELTSLVSPWPFAW 97
H VKSL +YW M+KD + ++ C CQ+ + A P L L P P
Sbjct: 1074 HQRVKSL--------FYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPL--PIPDKI 1123
Query: 98 W---GIDLLGPFPKATGQLKYLVVAINYSTKWIEVEPLAKIKAKNVLRFFKRNILARFGV 154
W +D + P + G+ +VV S V A V + F N+ G
Sbjct: 1124 WCDVSMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQAFLDNVYKHHGC 1183
Query: 155 PALVIS-----VLSRFWGIKSLIRLEPFNL 179
P ++S S FW K +L+ L
Sbjct: 1184 PTSIVSDRDVLFTSDFW--KEFFKLQGVEL 1211
>At3g11970 hypothetical protein
Length = 1499
Score = 39.3 bits (90), Expect = 0.001
Identities = 36/154 (23%), Positives = 58/154 (37%), Gaps = 11/154 (7%)
Query: 21 NETQEVLADVHDGICGQHLDVKSLARKVLRAGYYWPTMLKDAEDYVNLCDKCQRHGDMHL 80
N +L +H G H + + + ++ +YW M+KD + Y+ C CQ+
Sbjct: 1079 NIKNTILLWLHGSGVGGHSG-RDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPA 1137
Query: 81 APPVELTSLVSPWPFAWW---GIDLLGPFPKATGQLKYLVVAINYSTKWIEVEPLAKIKA 137
A P L L P P W +D + P + G+ +VV S + A
Sbjct: 1138 ASPGLLQPL--PIPDTIWSEVSMDFIEGLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSA 1195
Query: 138 KNVLRFFKRNILARFGVPALVIS-----VLSRFW 166
V + N+ G P ++S S FW
Sbjct: 1196 LTVAHAYLDNVFKLHGCPTSIVSDRDVVFTSEFW 1229
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.143 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,064,605
Number of Sequences: 26719
Number of extensions: 161131
Number of successful extensions: 505
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 446
Number of HSP's gapped (non-prelim): 51
length of query: 179
length of database: 11,318,596
effective HSP length: 93
effective length of query: 86
effective length of database: 8,833,729
effective search space: 759700694
effective search space used: 759700694
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 56 (26.2 bits)
Medicago: description of AC135504.12


