

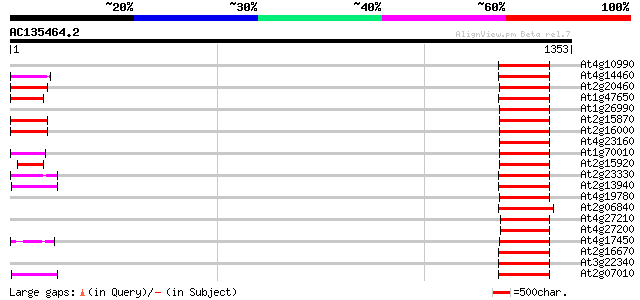
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135464.2 - phase: 0 /pseudo
(1353 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g10990 putative retrotransposon polyprotein 158 2e-38
At4g14460 retrovirus-related like polyprotein 148 2e-35
At2g20460 putative retroelement pol polyprotein 144 3e-34
At1g47650 hypothetical protein 140 4e-33
At1g26990 polyprotein, putative 140 5e-33
At2g15870 putative retroelement pol polyprotein 136 9e-32
At2g16000 putative retroelement pol polyprotein 135 2e-31
At4g23160 putative protein 134 5e-31
At1g70010 hypothetical protein 133 6e-31
At2g15920 putative retroelement pol polyprotein 132 1e-30
At2g23330 putative retroelement pol polyprotein 129 1e-29
At2g13940 putative retroelement pol polyprotein 125 1e-28
At4g19780 putative LTR retrotransposon (fragment) 124 3e-28
At2g06840 putative retroelement pol polyprotein 124 4e-28
At4g27210 putative protein 122 1e-27
At4g27200 putative protein 122 1e-27
At4g17450 retrotransposon like protein 121 2e-27
At2g16670 putative retroelement pol polyprotein 121 2e-27
At3g22340 hypothetical protein 119 9e-27
At2g07010 putative retroelement pol polyprotein 119 1e-26
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 158 bits (399), Expect = 2e-38
Identities = 71/122 (58%), Positives = 96/122 (78%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
+PG GL + S L L G+SDADWG C D+RRS+TG+C ++G SL++WKSKKQ +VSRSS
Sbjct: 866 NPGQGLMYSASSELCLNGFSDADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSS 925
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+E+EYR+L +TCE+ L LL DL+VT + L CD++SALH+A+NP+FHERTKH++I
Sbjct: 926 TESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEI 985
Query: 1300 DC 1301
DC
Sbjct: 986 DC 987
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 148 bits (374), Expect = 2e-35
Identities = 66/124 (53%), Positives = 92/124 (73%)
Query: 1178 ETSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSR 1237
+ +PG GL + D + L G+SDADW C DTRRSI+G+C ++G SL+SWKSKKQ + SR
Sbjct: 1309 KANPGQGLMYSADYEICLNGFSDADWAACKDTRRSISGFCIYLGTSLISWKSKKQAVASR 1368
Query: 1238 SSSEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHL 1297
SS+E+EYR++ +TCE+ L LL DL++ + L CD++SALH + NP+FHERTKH+
Sbjct: 1369 SSTESEYRSMAQATCEIIWLQQLLKDLHIPLTCPAKLFCDNKSALHSSLNPVFHERTKHI 1428
Query: 1298 DIDC 1301
+IDC
Sbjct: 1429 EIDC 1432
Score = 87.0 bits (214), Expect = 7e-17
Identities = 41/97 (42%), Positives = 58/97 (59%), Gaps = 1/97 (1%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V +WLMNSV I QSL+Y + WN+L +RF + D RI +++ L Q
Sbjct: 96 RCNDIVSTWLMNSVDKKIGQSLLYIATVQGIWNNLLSRFKQDDAPRIFDIEQKLSKIEQG 155
Query: 62 SLSIIEYFTELKSYWEELEVFCPIPNCTCNVRCSCEA 98
S+ I Y+T L + WEE + +P CTC RC C+A
Sbjct: 156 SMDISTYYTALLTLWEEHRNYVELPVCTCG-RCECDA 191
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 144 bits (363), Expect = 3e-34
Identities = 71/120 (59%), Positives = 90/120 (74%), Gaps = 1/120 (0%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ + L L GY+DADWG CPD+RRS TG+ F+G SL+SW+SKKQ VSRSS+E
Sbjct: 1290 GQGLFYSAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAE 1349
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL ++CE+ L LL L V S VP L DS +A++IA+NP+FHERTKH++IDC
Sbjct: 1350 AEYRALALASCEMAWLSTLLLALRVH-SGVPILYSDSTAAVYIATNPVFHERTKHIEIDC 1408
Score = 77.0 bits (188), Expect = 7e-14
Identities = 35/90 (38%), Positives = 58/90 (63%), Gaps = 1/90 (1%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V SWL+NSV+P I +S++ + A D W DL RF+ + R L +++ RQ
Sbjct: 127 RCNSMVKSWLLNSVSPQIYRSILRLNDATDIWRDLFDRFNLTNLPRTYNLTQEIQDLRQG 186
Query: 62 SLSIIEYFTELKSYWEELEVFCPIPN-CTC 90
++S+ EY+T LK+ W++L+ + + CTC
Sbjct: 187 TMSLSEYYTLLKTLWDQLDSTEALDDPCTC 216
>At1g47650 hypothetical protein
Length = 1409
Score = 140 bits (354), Expect = 4e-33
Identities = 68/122 (55%), Positives = 89/122 (72%), Gaps = 1/122 (0%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
S G GLF+ + L L G++D+DW CPD+RRS TG+ F+G SL+SW+SKKQ +VSRSS
Sbjct: 1072 SVGQGLFYSASADLTLKGFADSDWASCPDSRRSTTGFSMFVGDSLISWRSKKQHVVSRSS 1131
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYRAL TCEL L LL L + VP L DS +A++IA+NP+FHERTKH+++
Sbjct: 1132 AEAEYRALALVTCELVWLHTLLMSLKAS-YLVPVLYSDSTTAIYIATNPVFHERTKHIEL 1190
Query: 1300 DC 1301
DC
Sbjct: 1191 DC 1192
Score = 79.7 bits (195), Expect = 1e-14
Identities = 34/79 (43%), Positives = 54/79 (68%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RC ++V SWL+NSV+P I +S++ + A D W DL +RF+ + R L +++ FRQ
Sbjct: 126 RCKSMVKSWLLNSVSPQIYRSILRMNDASDIWRDLHSRFNVTNLPRTYNLTQEIQDFRQG 185
Query: 62 SLSIIEYFTELKSYWEELE 80
+LS+ EY+T LK+ W++LE
Sbjct: 186 TLSLSEYYTRLKTLWDQLE 204
>At1g26990 polyprotein, putative
Length = 1436
Score = 140 bits (353), Expect = 5e-33
Identities = 65/120 (54%), Positives = 88/120 (73%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ D++ L G+SD+DW CPDTRR +TG+ F+G SLVSW+SKKQ +VS SS+E
Sbjct: 1264 GQGLFYGADTNFDLRGFSDSDWQTCPDTRRCVTGFAIFVGNSLVSWRSKKQDVVSMSSAE 1323
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRA+ +T EL L Y+LT + + L CD+++ALHIA+N +FHERTKH++ DC
Sbjct: 1324 AEYRAMSVATKELIWLGYILTAFKIPFTHPAYLYCDNEAALHIANNSVFHERTKHIENDC 1383
>At2g15870 putative retroelement pol polyprotein
Length = 1264
Score = 136 bits (342), Expect = 9e-32
Identities = 66/120 (55%), Positives = 86/120 (71%), Gaps = 1/120 (0%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ S L L G++D+DW CPD+R S TG+ F+G SL+S +SKKQ +VSRSS+E
Sbjct: 1093 GQGLFYSASSDLTLKGFADSDWASCPDSRHSTTGFTMFVGDSLISLRSKKQHVVSRSSAE 1152
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL +TCEL L LL L + +P L DS +A++IA NP+FHERTKH++IDC
Sbjct: 1153 AEYRALALATCELVWLHTLLASL-TAATTIPILFSDSTAAIYIAINPVFHERTKHIEIDC 1211
Score = 76.6 bits (187), Expect = 9e-14
Identities = 34/91 (37%), Positives = 57/91 (62%), Gaps = 1/91 (1%)
Query: 1 IRCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQ 60
I CN +V SWL+NS++P I +S++ + D W DL +RF+ + R L +++ RQ
Sbjct: 96 ISCNTMVKSWLLNSMSPQIYRSILRMNDVSDIWRDLNSRFNMTNLPRTYNLTQEIQDLRQ 155
Query: 61 NSLSIIEYFTELKSYWEELEVFCPIPN-CTC 90
++LS+ EY+T LK+ W++L + + CTC
Sbjct: 156 STLSLSEYYTRLKTLWDQLNSTEELDDPCTC 186
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 135 bits (339), Expect = 2e-31
Identities = 65/120 (54%), Positives = 85/120 (70%), Gaps = 1/120 (0%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ S L L G++D+DW C D+RRS T + F+G SL+SW+SKKQ VSRSS+E
Sbjct: 1283 GQGLFYSASSDLTLKGFADSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAE 1342
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL +TCE+ L LL L + VP L DS +A++IA+NP+FHERTKH+ +DC
Sbjct: 1343 AEYRALALATCEMVWLFTLLVSLQAS-PPVPILYSDSTAAIYIATNPVFHERTKHIKLDC 1401
Score = 82.0 bits (201), Expect = 2e-15
Identities = 37/90 (41%), Positives = 59/90 (65%), Gaps = 1/90 (1%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V SWL+NSV+P I +S++ + A D W DL +RF+ + R L +++ FRQ
Sbjct: 123 RCNSMVKSWLLNSVSPQIYRSILRMNDASDIWRDLNSRFNVTNLPRTYNLTQEIQDFRQG 182
Query: 62 SLSIIEYFTELKSYWEELEVFCPIPN-CTC 90
+LS+ EY+T LK+ W++L+ + CTC
Sbjct: 183 TLSLSEYYTRLKTLWDQLDSTEALDEPCTC 212
>At4g23160 putative protein
Length = 1240
Score = 134 bits (336), Expect = 5e-31
Identities = 63/120 (52%), Positives = 84/120 (69%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ + + L +SDA + C DTRRS GYC F+G SL+SWKSKKQQ+VS+SS+E
Sbjct: 429 GQGLFYSSQAEMQLQVFSDASFQSCKDTRRSTNGYCMFLGTSLISWKSKKQQVVSKSSAE 488
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL +T E+ L +L + SK L CD+ +A+HIA+N +FHERTKH++ DC
Sbjct: 489 AEYRALSFATDEMMWLAQFFRELQLPLSKPTLLFCDNTAAIHIATNAVFHERTKHIESDC 548
>At1g70010 hypothetical protein
Length = 1315
Score = 133 bits (335), Expect = 6e-31
Identities = 63/120 (52%), Positives = 86/120 (71%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ S L L Y++AD+ C D+RRS +GYC F+G SL+ WKS+KQ +VS+SS+E
Sbjct: 1144 GQGLFYSATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAE 1203
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYR+L +T EL L L +L V SK L CD+++A+HIA+N +FHERTKH++ DC
Sbjct: 1204 AEYRSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDC 1263
Score = 75.1 bits (183), Expect = 3e-13
Identities = 33/85 (38%), Positives = 51/85 (59%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V SWL+NSV+ I S++Y +A W DL RF ++ R+ L++ +HS RQ
Sbjct: 33 RCNSMVKSWLLNSVSKEIYTSILYFPTAAAIWKDLYTRFHKSSLPRLYKLRQQIHSLRQG 92
Query: 62 SLSIIEYFTELKSYWEELEVFCPIP 86
+L + Y T ++ WEEL +P
Sbjct: 93 NLDLSSYHTRTQTLWEELTSLQAVP 117
>At2g15920 putative retroelement pol polyprotein
Length = 1100
Score = 132 bits (333), Expect = 1e-30
Identities = 67/120 (55%), Positives = 86/120 (70%), Gaps = 1/120 (0%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ S L L G++D+DW C D+RRS TG+ F+G SL+S +SKKQ++VSRSS+E
Sbjct: 925 GQGLFYSASSDLTLKGFADSDWVSCADSRRSTTGFTMFVGDSLISLRSKKQRVVSRSSAE 984
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AEYRAL +TCEL L LL L S VP L +S +A++IA NP+FHERTKH +IDC
Sbjct: 985 AEYRALALATCELVSLHTLLVSLTAATS-VPILFSNSTTAIYIAINPVFHERTKHTEIDC 1043
Score = 53.1 bits (126), Expect = 1e-06
Identities = 22/62 (35%), Positives = 39/62 (62%)
Query: 19 ITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQNSLSIIEYFTELKSYWEE 78
I +S++ + A D W DL +RF+ + R L +++ RQ +LS+ EY+T LK+ W++
Sbjct: 84 ICRSILRMNDASDIWRDLNSRFNMTNLPRTYNLTQEIQDLRQGTLSLSEYYTRLKTLWDQ 143
Query: 79 LE 80
L+
Sbjct: 144 LD 145
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 129 bits (324), Expect = 1e-29
Identities = 64/122 (52%), Positives = 85/122 (69%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
SP G+F DSSL + Y D+D+ CP TRRS++ Y ++G S +SWK+KKQ VS SS
Sbjct: 1323 SPAQGIFLRSDSSLIINAYCDSDYNACPLTRRSLSAYVVYLGDSPISWKTKKQDTVSYSS 1382
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYRA+ + EL+ L LL DL V S L CDS++A+HIA+NP+FHERTKH++
Sbjct: 1383 AEAEYRAMAYTLKELKWLKALLKDLGVHHSSPMKLHCDSEAAIHIAANPVFHERTKHIES 1442
Query: 1300 DC 1301
DC
Sbjct: 1443 DC 1444
Score = 57.4 bits (137), Expect = 6e-08
Identities = 29/114 (25%), Positives = 55/114 (47%), Gaps = 6/114 (5%)
Query: 1 IRCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQ 60
I N+++ W+ S+ P+I ++ + A W +L+ RFS + VR + L+ ++ + Q
Sbjct: 81 IAINSMIVGWIRTSIDPTIRSTVGFVSEASQLWENLRRRFSVGNGVRKTLLKDEIAACTQ 140
Query: 61 NSLSIIEYFTELKSYWEELEVFCPIPNCTCNVRCSCEAMRNSRRGSDTPLSHRF 114
+ ++ Y+ L WEEL+ N C CEA + + + H+F
Sbjct: 141 DGQPVLAYYGRLIKLWEELQ------NYKSGRECKCEAASDIEKEREDDRVHKF 188
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 125 bits (315), Expect = 1e-28
Identities = 62/122 (50%), Positives = 82/122 (66%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
SPG G+ D L L Y D+DW CP TRRSI+ Y +G S +SWK+KKQ VS SS
Sbjct: 1328 SPGQGILLNADPDLTLEVYCDSDWQSCPLTRRSISAYVVLLGGSPISWKTKKQDTVSHSS 1387
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYRA+ + E++ L LL +L + S L CDS++A+HIA+NP+FHERTKH++
Sbjct: 1388 AEAEYRAMSYALKEIKWLRKLLKELGIEQSTPARLYCDSKAAIHIAANPVFHERTKHIES 1447
Query: 1300 DC 1301
DC
Sbjct: 1448 DC 1449
Score = 77.0 bits (188), Expect = 7e-14
Identities = 34/111 (30%), Positives = 60/111 (53%), Gaps = 1/111 (0%)
Query: 4 NNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQNSL 63
N+++ W+ S+ P + ++ + A W DL+ RFS ++VRI ++ L S RQ+
Sbjct: 95 NSMIVGWIRTSIEPKVKATVTFISDAHLLWKDLKQRFSVGNKVRIHQIRAQLSSCRQDGQ 154
Query: 64 SIIEYFTELKSYWEELEVFCPIPNCTCNVRCSCEAMRNSRRGSDTPLSHRF 114
++IEY+ L + WEE ++ P+ CTC + C C A + + H+F
Sbjct: 155 AVIEYYGRLSNLWEEYNIYKPVTVCTCGL-CRCGATSEPTKEREEEKIHQF 204
>At4g19780 putative LTR retrotransposon (fragment)
Length = 306
Score = 124 bits (312), Expect = 3e-28
Identities = 61/120 (50%), Positives = 79/120 (65%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+P D L GY+D+D+G C D+RR + YC FIG SLVSWKSKKQ VS S++E
Sbjct: 133 GQGLFYPADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIGDSLVSWKSKKQDTVSMSTAE 192
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AE+RA+ T E+ L LL D V L D+ +ALHI +N +FHERTK +++DC
Sbjct: 193 AEFRAMSQGTKEMIWLSRLLDDFKVPFIPPAYLYSDNTAALHIVNNSVFHERTKFVELDC 252
>At2g06840 putative retroelement pol polyprotein
Length = 1102
Score = 124 bits (311), Expect = 4e-28
Identities = 62/132 (46%), Positives = 88/132 (65%), Gaps = 1/132 (0%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
SP G+ D +L L Y D+D+ CP TRRS++ Y ++G + +SWK+KKQ VS SS
Sbjct: 933 SPDQGILLRSDRALSLTAYCDSDYNPCPRTRRSLSAYVLYLGDTPISWKTKKQDTVSSSS 992
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYRA+ + E++ L L+T L V ++ L CDSQ+A+HIA+NP+FHERTKH++
Sbjct: 993 AEAEYRAMAYTLKEIKWLKALMTTLGVDHTQPILLFCDSQAAIHIAANPVFHERTKHIEK 1052
Query: 1300 DC-QCSEGETSK 1310
DC Q + T K
Sbjct: 1053 DCHQVRDAVTDK 1064
>At4g27210 putative protein
Length = 1318
Score = 122 bits (306), Expect = 1e-27
Identities = 59/117 (50%), Positives = 77/117 (65%)
Query: 1184 GLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSEAE 1243
G+ + RDS L YSD+DWG C TRRS+ G C F+G +LVSW SKK VSRSS+EAE
Sbjct: 1055 GISYSRDSPTLLQAYSDSDWGNCKQTRRSVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAE 1114
Query: 1244 YRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDID 1300
Y++L + E+ L LL +L + P L CD+ SA+++ +NP FH RTKH DID
Sbjct: 1115 YKSLSDAASEILWLSTLLRELRIPLPDTPELFCDNLSAVYLTANPAFHARTKHFDID 1171
>At4g27200 putative protein
Length = 819
Score = 122 bits (306), Expect = 1e-27
Identities = 59/117 (50%), Positives = 77/117 (65%)
Query: 1184 GLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSEAE 1243
G+ + RDS L YSD+DWG C TRRS+ G C F+G +LVSW SKK VSRSS+EAE
Sbjct: 556 GISYSRDSPTLLQAYSDSDWGNCKQTRRSVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAE 615
Query: 1244 YRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDID 1300
Y++L + E+ L LL +L + P L CD+ SA+++ +NP FH RTKH DID
Sbjct: 616 YKSLSDAASEILWLSTLLRELRIPLPDTPELFCDNLSAVYLTANPAFHARTKHFDID 672
>At4g17450 retrotransposon like protein
Length = 1433
Score = 121 bits (304), Expect = 2e-27
Identities = 59/120 (49%), Positives = 77/120 (64%)
Query: 1182 GTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSSSE 1241
G GLF+ D L GY+D+D+G C D+RR + YC FIG LVSWKSKKQ VS S++E
Sbjct: 1260 GQGLFYSADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAE 1319
Query: 1242 AEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDIDC 1301
AE+RA+ T E+ L L D V L CD+ +ALHI +N +FHERTK +++DC
Sbjct: 1320 AEFRAMSQGTKEMIWLSRLFDDFKVPFIPPAYLYCDNTAALHIVNNSVFHERTKFVELDC 1379
Score = 55.8 bits (133), Expect = 2e-07
Identities = 32/108 (29%), Positives = 56/108 (51%), Gaps = 20/108 (18%)
Query: 2 RCNNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQN 61
RCN++V +WL+N VT + WNDL +RF ++ R L++ +H+ +Q
Sbjct: 129 RCNSMVKTWLLNVVT--------------EMWNDLFSRFRVSNLPRKYQLEQSIHTLKQG 174
Query: 62 SLSIIEYFTELKSYWEELEVFCPIPNCTCNVR-CSCEAMRNSRRGSDT 108
+L + Y+T+ K+ WE+L VR C+CE ++ ++T
Sbjct: 175 NLDLSTYYTKKKTLWEQL-----ANTRVLTVRKCNCEHVKELLEEAET 217
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 121 bits (304), Expect = 2e-27
Identities = 60/123 (48%), Positives = 82/123 (65%)
Query: 1178 ETSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSR 1237
+ PG G+F R + G+ D+DW G P +RRS+TGY G S +SWK+KKQ VS+
Sbjct: 1158 KADPGQGVFLRRSGDFQITGWCDSDWAGDPMSRRSVTGYFVQFGDSPISWKTKKQDTVSK 1217
Query: 1238 SSSEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHL 1297
SS+EAEYRA+ EL L LL L V+ + + CDS+SA++IA+NP+FHERTKH+
Sbjct: 1218 SSAEAEYRAMSFLASELLWLKQLLFSLGVSHVQPMIMCCDSKSAIYIATNPVFHERTKHI 1277
Query: 1298 DID 1300
+ID
Sbjct: 1278 EID 1280
>At3g22340 hypothetical protein
Length = 220
Score = 119 bits (299), Expect = 9e-27
Identities = 58/122 (47%), Positives = 81/122 (65%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
+PG G+ D+ L L + DAD CP TRRS+T + +G S +SWK++KQ VSRSS
Sbjct: 22 NPGQGILLRADAELTLTAWCDADHATCPLTRRSLTAWFIQLGGSPISWKTRKQDTVSRSS 81
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYR++ + E+ L LL L V CS L CD+QSA+++++NP+FHERTKH+
Sbjct: 82 AEAEYRSMADTVSEILWLRQLLITLGVDCSSPIPLFCDNQSAIYLSANPVFHERTKHVGS 141
Query: 1300 DC 1301
DC
Sbjct: 142 DC 143
>At2g07010 putative retroelement pol polyprotein
Length = 1413
Score = 119 bits (298), Expect = 1e-26
Identities = 58/122 (47%), Positives = 82/122 (66%)
Query: 1180 SPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIG*SLVSWKSKKQQIVSRSS 1239
SPG G+ + L L Y D+D+ CP TRRS++ Y +G S +SWK+KKQ VS SS
Sbjct: 1240 SPGQGILLSANKDLTLEVYCDSDFQSCPLTRRSLSAYVVLLGGSPISWKTKKQDTVSHSS 1299
Query: 1240 SEAEYRALVASTCELQ*LVYLLTDLNVTCSKVPGLLCDSQSALHIASNPIFHERTKHLDI 1299
+EAEYRA+ + E++ L LL +L +T + L CDS++A+ IA+NP+FHERTKH++
Sbjct: 1300 AEAEYRAMSVALKEIKWLNKLLKELGITLAAPTRLFCDSKAAISIAANPVFHERTKHIER 1359
Query: 1300 DC 1301
DC
Sbjct: 1360 DC 1361
Score = 68.2 bits (165), Expect = 3e-11
Identities = 29/111 (26%), Positives = 59/111 (53%), Gaps = 1/111 (0%)
Query: 4 NNLVHSWLMNSVTPSITQSLIYTDSAVDAWNDLQARFSRADRVRISTLQRDLHSFRQNSL 63
N+++ W+ S+ P + ++ + A W++L+ RFS ++V + ++ L + RQ+
Sbjct: 90 NSMIVGWIRASIEPKVKSTVTFICDAHQLWSELKQRFSVGNKVHVHQIKTQLAACRQDGQ 149
Query: 64 SIIEYFTELKSYWEELEVFCPIPNCTCNVRCSCEAMRNSRRGSDTPLSHRF 114
+I+Y+ L WEE +++ PI C C + C+C A + + H+F
Sbjct: 150 PVIDYYGRLCKLWEEFQIYKPITVCKCGL-CTCGATLEPSKEREEEKIHQF 199
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.360 0.159 0.618
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,084,675
Number of Sequences: 26719
Number of extensions: 968973
Number of successful extensions: 4039
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 92
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 3857
Number of HSP's gapped (non-prelim): 159
length of query: 1353
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1242
effective length of database: 8,352,787
effective search space: 10374161454
effective search space used: 10374161454
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC135464.2


