

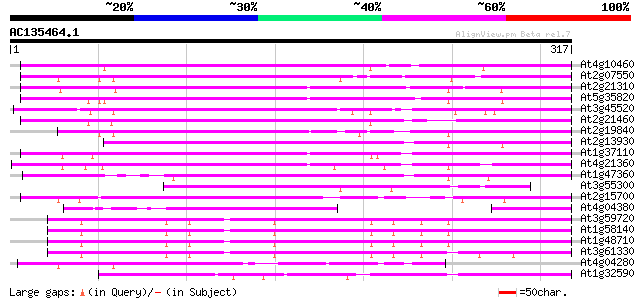
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135464.1 - phase: 0 /pseudo
(317 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g10460 putative retrotransposon 207 5e-54
At2g07550 putative retroelement pol polyprotein 204 7e-53
At2g21310 putative retroelement pol polyprotein 201 3e-52
At5g35820 copia-like retrotransposable element 196 2e-50
At3g45520 copia-like polyprotein 195 3e-50
At2g21460 putative retroelement pol polyprotein 191 6e-49
At2g19840 copia-like retroelement pol polyprotein 164 8e-41
At2g13930 putative retroelement pol polyprotein 153 1e-37
At1g37110 147 1e-35
At4g21360 putative transposable element 145 3e-35
At1g47360 polyprotein, putative 137 6e-33
At3g55300 putative protein 106 2e-23
At2g15700 copia-like retroelement pol polyprotein 100 1e-21
At4g04380 putative polyprotein 96 3e-20
At3g59720 copia-type reverse transcriptase-like protein 82 4e-16
At1g58140 hypothetical protein 82 4e-16
At1g48710 hypothetical protein 82 4e-16
At3g61330 copia-type polyprotein 81 7e-16
At4g04280 putative transposon protein 79 2e-15
At1g32590 hypothetical protein, 5' partial 78 7e-15
>At4g10460 putative retrotransposon
Length = 1230
Score = 207 bits (528), Expect = 5e-54
Identities = 135/343 (39%), Positives = 188/343 (54%), Gaps = 37/343 (10%)
Query: 7 DIEKFTGSNDFGL*KVKMQAILTQQKCVEALKGEAEMPATLTQEEK-------------R 53
++EKF G D+ L K K+ A + AL+ + L EE+
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 54 EMVDKAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQLYSFKMM 113
E KA+S IVL + D+VLR +E TA SML + LYM+K+L +R LKQ+LYS+KM
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 114 ESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILYGK-EGTAT 172
E+ S+ + EF +++ DL N V + DED+A+LLL SLPK F+ KDT+ YG T +
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 173 LEEVQAALRTKELTKFKDMKVDEG-SEGLNV-----TRGRNEHRGKGKGKSRSKSRSKGF 226
++EV AA+ +KEL + K G +EGL V TRG +E + KG K RS+SRSKG+
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKG-NKGRSRSRSKGW 245
Query: 227 DKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEE------------GYESAGAL 274
C++C ++GHFK CP+KG + A S E GY + AL
Sbjct: 246 K----GCWICGEEGHFKTSCPNKGKQQNKGKDQASGSKGEAATIKGNTSEGSGYYVSEAL 301
Query: 275 VVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
T WV+D+GC+YHM +KE+FE L GG VR+GN
Sbjct: 302 HSTDVNLGNEWVMDTGCNYHMTHKKEWFEELSEDAGGTVRMGN 344
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 204 bits (518), Expect = 7e-53
Identities = 133/342 (38%), Positives = 197/342 (56%), Gaps = 38/342 (11%)
Query: 7 DIEKFTGSNDFGL*KVKMQA---ILTQQKCVEALKGEAEMPATLTQ-----EEKREMVD- 57
++EKF G D+ + K K+ A IL ++ + E + L + EEK E +
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 58 ------KAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQLYSFK 111
KA+SAIVL + D+VLR + +E+TAA+ML + LYM+K+L +R KQ+LYSFK
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 112 MMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILYGK-EGT 170
M E+ S+ + EF +I+ DL N+ V + DED+A+LLL +LPK+F+ KDT+ Y +
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 171 ATLEEVQAALRTKEL---TKFKDMKVDEGSEGLNVTRGRNEHRGKGKGKSRSKSRSKGFD 227
TL+EV AA+ +KEL + K +KV +EGL V + +NE++GKG+ K + K + KG
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKVQ--AEGLYV-KDKNENKGKGEQKGKGKGK-KGKS 242
Query: 228 KSKYKCFLCHKQGHFKKDCPD------------KGGDGSPSVQVAEASNEEGYESAGALV 275
K K C+ C ++GHF+ CP+ KG +AEA+ GY + AL
Sbjct: 243 KKKPGCWTCGEEGHFRSSCPNQNKPQFKQSQVVKGESSGGKGNLAEAA---GYYVSEALS 299
Query: 276 VTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
T E W+LD+GCSYHM ++E+F GG VR+GN
Sbjct: 300 STEVHLEDEWILDTGCSYHMTYKREWFHEFNEDAGGSVRMGN 341
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 201 bits (512), Expect = 3e-52
Identities = 126/335 (37%), Positives = 197/335 (58%), Gaps = 28/335 (8%)
Query: 7 DIEKFTGSNDFGL*KVKMQAI---------LTQQKCVEALKGEAEMPATLTQEEKREMVD 57
++EK G D+ L K K+ A L + + +E + AE + LT+ E + + +
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 58 K---AKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQLYSFKMME 114
K A+S ++L LG+ VLR V +E TAA M+ + L+M KSL +R LKQ+LY +KM +
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 115 SKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILYGKEGTATLE 174
S +I E + +F K++ DL N++V++ DED+A++LL SLPK F+ KDT+ YGK T L+
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGKT-TLALD 185
Query: 175 EVQAALRTKELTKFKDMK-VDEGSEGLNV-TRGRNEHRGKGKGKSRSKSRSKGFDKSKYK 232
E+ A+R+K L K + S+ L V RGR+E R K +++S+SRSK + K
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSERNKSQSRSK--SREKKV 243
Query: 233 CFLCHKQGHFKKDC---PDKGGDGSPSVQVAEASNEEGYES-AGALVV------TSWEPE 282
C++C K+GHFKK C +K G+ S + E+SN G + A AL V + E +
Sbjct: 244 CWVCGKEGHFKKQCYVWKEKNKKGNNS-EKGESSNVIGQAADAAALAVREESNADNQEVD 302
Query: 283 KSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
W++D+GCS+HM PR+++F + G V++ N
Sbjct: 303 NEWIMDTGCSFHMTPRRDWFVEFDESQTGRVKMAN 337
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 196 bits (497), Expect = 2e-50
Identities = 129/342 (37%), Positives = 194/342 (56%), Gaps = 35/342 (10%)
Query: 7 DIEKFTGSNDFGL*KVKMQAILTQQKCVEALKGEAEM----------------PATLTQ- 49
++EKF G D+ L K K+ A + +E L E E P T T
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 50 -EEK--REMVDKAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQ 106
E+K +E KA+S I+L LG+ VLR V ++ TAA M+ + L+M KSL +R LKQ+
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 107 LYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILYG 166
LY +KM E+ ++ E + +F K++ DL N++V + DED+A++LL SLP+ F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 167 KEGTATLEEVQAALRTKELTKFKDMK-VDEGSEGLNV-TRGRNEHRGKGKGKSRSKSRSK 224
K T LEE+ +A+R+K L K + S+GL V RGR+E RGKG K++S+S+SK
Sbjct: 187 KT-TLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKNKSRSKSK 245
Query: 225 GFDKSKYKCFLCHKQGHFKKDC---PDKGGDGSPSVQVAEASNEEGYESAGALVVT---- 277
G K+ C++C K+GHFKK C ++ GS S + ++ A ALVV+
Sbjct: 246 GAGKT---CWICGKEGHFKKQCYVWKERNKQGSTSERGEASTVTARVTDAAALVVSRALL 302
Query: 278 --SWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
+ +W+LD+GCS+HM RK++ G VR+GN
Sbjct: 303 GFAEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGN 344
>At3g45520 copia-like polyprotein
Length = 1363
Score = 195 bits (495), Expect = 3e-50
Identities = 136/355 (38%), Positives = 198/355 (55%), Gaps = 49/355 (13%)
Query: 3 GSKWDIEKFTGSNDFGL*KVKMQAILTQQKCVEALKGEAEMP---------ATLTQEEKR 53
G++ ++EKF G D+ + K K+ A + L+ E+E P + ++E+R
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLR-ESETPMGKERDSEKSDEDEKEER 61
Query: 54 EMVD-------KAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQ 106
E ++ KA+S IVL + D+VLR + +E +AA+ML + LYM+K+L +R LKQ+
Sbjct: 62 EKMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQK 121
Query: 107 LYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILYG 166
LYSFKM E+ SI + EF I+ DL N+ V + DED+A+LLL SLPK F+ KDT+ Y
Sbjct: 122 LYSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYS 181
Query: 167 KEGTA-TLEEVQAALRTKELTKFKDMK--VDEGSEGLNV-----TRGRNEHRGKGKGKSR 218
T +L+EV AA+ ++EL +F +K + +EGL V RGR+E + KGKGK R
Sbjct: 182 SGKTVLSLDEVAAAIYSREL-EFGSVKKSIKGQAEGLYVKDKAENRGRSEQKDKGKGK-R 239
Query: 219 SKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKG-------GDGSPSVQVAEASNEEG---- 267
SKS KSK C++C + GH K CP+K G + + EG
Sbjct: 240 SKS------KSKRGCWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSGGKGNLVEGSVNF 293
Query: 268 YESAG-----ALVVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
ESAG AL T E W++D+GC YHM ++E+ E + GG VR+GN
Sbjct: 294 VESAGMFVSEALSSTDIHLEDEWIMDTGCIYHMTHKREWLEDFDEEAGGSVRMGN 348
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 191 bits (484), Expect = 6e-49
Identities = 131/333 (39%), Positives = 184/333 (54%), Gaps = 42/333 (12%)
Query: 7 DIEKFTGSNDFGL*KVKMQAILTQQKCVEALKGEAEM-----PATLTQEEKREMV----- 56
++EKF G D+ + K K+ A L ALK E ++ LT+EE++E V
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 57 -----DKAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQLYSFK 111
KA+SAIVL + D+VLR + +E +AA+ML + LYM+K+L +R KQ+LYSFK
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 112 MMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILYG-KEGT 170
M E+ SI + EF +I+ DL N V + DED+A+LLL SLPK F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 171 ATLEEVQAALRTKELTKFKDMKVDEG-SEGLNV-----TRGRNEHRGKGKGKSRSKSRSK 224
+L+EV AA+ +KEL + K +G +EGL V TRGR E RG +S+S+S
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKEKTETRGRTEQRGNNNNNKKSRSKS- 245
Query: 225 GFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYESAGALVVTSWEPEKS 284
+SK C++C G S + S G + AL T E
Sbjct: 246 ---RSKKGCWIC----------------GESSNGSSNYSEANGLYVSEALSSTDIHLEDE 286
Query: 285 WVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
WV+D+GCSYHM ++E+FE L GG VR+GN
Sbjct: 287 WVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGN 319
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 164 bits (414), Expect = 8e-41
Identities = 102/307 (33%), Positives = 173/307 (56%), Gaps = 32/307 (10%)
Query: 28 LTQQKCVEALKGEAEM-PATLTQ--EEKREMVD---KAKSAIVLCLGDKVLRDVAREATA 81
L ++ + LK E E PA Q EE++ +D KA I + +GDKVLR++ TA
Sbjct: 7 LVERAPLPPLKEEDESDPAKKKQRIEEEKARIDQDEKAMDMIFINVGDKVLRNIENSKTA 66
Query: 82 ASMLAKWESLYMTKSLAHRQLLKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMED 141
A A + LY+ KSL +R L+ ++Y+++M +SK++ E + EF K++ DL N+++ + D
Sbjct: 67 AEAWATLDKLYLVKSLPNRVYLQLKVYNYRMQDSKTLEENVDEFQKMISDLNNLQIQVPD 126
Query: 142 EDKALLLLCSLPKSFEHFKDTILYGKEGTATLEEVQAALRTKELTKFKDMKVDEG----- 196
E +A+L+L +LP S++ K+T+ YG+EG L++V +A ++KEL +++ G
Sbjct: 127 EVQAILILSALPDSYDMLKETLKYGREG-IKLDDVISAAKSKEL----ELRDSSGGSRPV 181
Query: 197 SEGLNVTRGRNEHRGKGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDC------PDKG 250
EGL V RG+++ RG KS + K C++C K+GHFK+ C
Sbjct: 182 GEGLYV-RGKSQARGSDGPKS---------TEGKKVCWICGKEGHFKRQCYKWLEKNKAN 231
Query: 251 GDGSPSVQVAEASNEEGYESAGALVVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEG 310
G G ++ +A + G ++ + + ++ W++D+GCS+HM PRKEY + +
Sbjct: 232 GAGETALVKDDAQDLVGLVASEVNMSEGKDDQEEWIMDTGCSFHMTPRKEYLMDFVEAKS 291
Query: 311 GVVRLGN 317
G VR+ N
Sbjct: 292 GKVRMAN 298
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 153 bits (387), Expect = 1e-37
Identities = 99/279 (35%), Positives = 149/279 (52%), Gaps = 20/279 (7%)
Query: 54 EMVDKAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQLYSFKMM 113
E DKAK+ I L + DKVLR + TAA + L+M +SL HR + Y+FKM
Sbjct: 44 ERCDKAKNVIFLNVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQ 103
Query: 114 ESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILY-GKEGTAT 172
E+K I E + +F KI+ DL ++++++ DE +A+LLL SLP ++ +T+ Y
Sbjct: 104 ENKKIDENIDDFLKIVADLNHLQIDVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLR 163
Query: 173 LEEVQAALRTKELTKFKDMK-VDEGSEGLNVTRGRNEHRG-KGKGKSRSKSRSKGFDKSK 230
L++V A R KE ++ + V EG G+N ++G KGK +SRSKS K
Sbjct: 164 LDDVMVAARDKERELSQNNRPVVEGHFARGRPDGKNNNQGNKGKNRSRSKSAD-----GK 218
Query: 231 YKCFLCHKQGHFKKDC------PDKGGDGSPSVQVAEASNEEGYESAGALVVT------S 278
C++C K+GHFKK C GS + + + A + E + A L+ T +
Sbjct: 219 RVCWICGKEGHFKKQCYKWIERNKSKQQGSDNGESSLAKSTEAFNPAMVLLATDETLVVT 278
Query: 279 WEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
WVLD+GCS+HM PRK++F+ G V++GN
Sbjct: 279 DSIANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKMGN 317
>At1g37110
Length = 1356
Score = 147 bits (370), Expect = 1e-35
Identities = 105/339 (30%), Positives = 171/339 (49%), Gaps = 35/339 (10%)
Query: 7 DIEKFTGSNDFGL*KVKMQAIL------------TQQKCVEALKGEAEMPA--------- 45
+I+ F G DF L K+++QA L + K V K EA+ +
Sbjct: 9 EIKVFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSGTK 68
Query: 46 TLTQEEKREMVDKAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQ 105
+ K E ++AK+ I+ + D VL V AT A + A YM SL +R +
Sbjct: 69 EVPDPVKIEQSEQAKNIIINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIYTQL 128
Query: 106 QLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILY 165
+LYSFKM+ + +I + + EF +I+ +L ++E+ +++E +A+L+L SLP S K T+ Y
Sbjct: 129 KLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLKHTLKY 188
Query: 166 GKEGTATLEEVQAALRTKELTKFKDMKVDEGSEGLNVT--RGR-----NEHRGKGKGKSR 218
G + T T+++V ++ ++ E + + +D+G + T RGR N+ G+GKG+SR
Sbjct: 189 GNK-TLTVQDVTSSAKSLERELAEAVDLDKGQAAVLYTTERGRPLVRNNQKGGQGKGRSR 247
Query: 219 SKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYESAGALVVTS 278
S S K+K C+ C K+GH KKDC + Q E + AL V
Sbjct: 248 SNS------KTKVPCWYCKKEGHVKKDCYSRKKKMESEGQGEAGVITEKLVFSEALSVNE 301
Query: 279 WEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
+ W+LDSGC+ HM R+++F + K + LG+
Sbjct: 302 QMVKDLWILDSGCTSHMTSRRDWFISFQEKGNTTILLGD 340
>At4g21360 putative transposable element
Length = 1308
Score = 145 bits (366), Expect = 3e-35
Identities = 110/352 (31%), Positives = 179/352 (50%), Gaps = 49/352 (13%)
Query: 2 MGSKWDIEKFTGSNDFGL*KVKMQAIL------------TQQKCVEALKGEA-------- 41
M +K +I+ F G DF L K++++A L T K + +K E
Sbjct: 3 MSTKVEIKTFNGDRDFSLWKIRIEAQLGVLGLKPALSDFTLTKTILVVKSEKKESESEDD 62
Query: 42 EMPATLTQEE----KREMVDKAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSL 97
E + T+E K E D+AK+ I+ + D VL V TAA + A L+M SL
Sbjct: 63 ETDSKKTEEVPDPIKFEQSDQAKNFIINHITDTVLLKVQHCVTAAELWATLNKLFMETSL 122
Query: 98 AHRQLLKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFE 157
+R + +LYSFKM+++ SI + EF +I+ +L ++++ + +E +A+L+L SLP S+
Sbjct: 123 PNRIYTQLRLYSFKMVDNLSIDQNTDEFLRIVAELGSLQIQVGEEVQAILILNSLPPSYI 182
Query: 158 HFKDTILYGKEGTATLEEVQAALRT--KELTKFKDMKVDEGSEGL-NVTRGRNEHR---G 211
K T+ YG + T ++++V ++ ++ +EL++ K+ S L RGR + + G
Sbjct: 183 QLKHTLKYGNK-TLSVQDVVSSAKSLERELSEQKETIRAPASTALYTAERGRPQTKNTQG 241
Query: 212 KGKGKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDC------PDKGGDGSPSVQVAEASNE 265
+GKG+ RS S KS+ C+ C K+GH KKDC + G G V +
Sbjct: 242 QGKGRGRSNS------KSRLTCWFCKKEGHVKKDCYAGKRKLENEGQGKAGVITEKLVYS 295
Query: 266 EGYESAGALVVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
E AL + E + WV+DSGC+YHM R ++F E ++ LG+
Sbjct: 296 E------ALSMYDQEAKDKWVIDSGCTYHMTSRMDWFSEFNENETTMILLGD 341
>At1g47360 polyprotein, putative
Length = 1182
Score = 137 bits (346), Expect = 6e-33
Identities = 102/328 (31%), Positives = 157/328 (47%), Gaps = 44/328 (13%)
Query: 8 IEKFTGSNDFGL*KVKMQAILTQQKCVEALKGEAEMPATLTQEEKREMVDKAKSAIVLCL 67
I F S DF L K ++ A L+ + + G+ P T +EE E I LC
Sbjct: 6 IAVFDESGDFSLWKTRIMAHLSVIGLKDVVIGKTITPLTAEEEEDPE------KKIELC- 58
Query: 68 GDKVLRDVAREATAASMLAKWESL---YMTKSLAHRQLLKQQLYSFKMMESKSISEQLTE 124
A+EA WE+L +M +SL HR + Y+FKM E+K I E + +
Sbjct: 59 ------QTAQEA--------WETLDRLFMIRSLPHRVYTQLSFYTFKMQENKKIDENIDD 104
Query: 125 FNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILY-GKEGTATLEEVQAALRTK 183
F KI+ DL ++++ + DE +A+LLL SLP ++ +T+ Y L++V R K
Sbjct: 105 FLKIVADLNHLQIEVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRLDDVMVGARDK 164
Query: 184 ELTKFKDMK-VDEGSEGLNVTRGRNEHRG-KGKGKSRSKSRSKGFDKSKYKCFLCHKQGH 241
E ++ + V EG G+N ++G KGK +SRSKS K C++C K+GH
Sbjct: 165 ERELSQNNRPVAEGHYARGRPEGKNNNQGNKGKNRSRSKSAD-----GKRVCWICGKEGH 219
Query: 242 FKKDC---PDKGGDGSPSVQVAEASNEEGYE---------SAGALVVTSWEPEKSWVLDS 289
FKK C ++ V E+S + E + G ++ + W LD+
Sbjct: 220 FKKQCYKWLERNKSKQQGSDVGESSLAKSSEIFDPAMVIMATGETLLVTGRDADEWFLDT 279
Query: 290 GCSYHMCPRKEYFETLILKEGGVVRLGN 317
GCS+HM PR++ F+ G V++GN
Sbjct: 280 GCSFHMTPRRDLFKDFKELSSGFVKMGN 307
>At3g55300 putative protein
Length = 262
Score = 106 bits (264), Expect = 2e-23
Identities = 65/215 (30%), Positives = 112/215 (51%), Gaps = 14/215 (6%)
Query: 88 WESLYMTKSLAHRQLLKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALL 147
+E Y TK+LA+ LK + SFKM+E+KSI E L K++ DLA++++N DED+AL
Sbjct: 36 YEGNYQTKTLANMIYLKHKFTSFKMVENKSIEENLDTSLKLVADLASLDINTSDEDQALQ 95
Query: 148 LLCSLPKSFEHFKDTILYGK-EGTATLEEVQAALRTKEL----TKFKDMKVDEGSEGLNV 202
+ LP ++ DT+ YG + T TL V ++ +KE+ ++ + L
Sbjct: 96 FMAGLPPQYDSLVDTLKYGSGKDTLTLNAVISSAYSKEIELKEKGLLNVSKPDSEALLGD 155
Query: 203 TRGRNEHRGKGK---GKSRSKSRSKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQV 259
+GR+ ++G + G S + + + CF+C K+ H+K++CP + ++
Sbjct: 156 FKGRSNNKGNKRPCTGSSSGNGKFQSRPSNNTSCFICGKEDHWKRECPQR----KKHAEL 211
Query: 260 AEASNEEGYESAGALVVTSWEPEKSWVLDSGCSYH 294
A + E L V++ K WV+DSG ++H
Sbjct: 212 ANIATEP--PQPLVLTVSTQSTCKEWVMDSGRTFH 244
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 100 bits (249), Expect = 1e-21
Identities = 94/339 (27%), Positives = 161/339 (46%), Gaps = 58/339 (17%)
Query: 7 DIEKFTGSNDFGL*KVKMQA---------------------ILTQQKCVEALK---GEAE 42
++++F G+ DF L KV+M A ++ ++ V G+ E
Sbjct: 8 EVDRFDGTGDFSLWKVRMLAHYGVLGLKGILNDEQLLRDPPVIEEEAAVAGRDYHVGDFE 67
Query: 43 MPATLTQEEKREMVDKAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQL 102
+P+ + K E +KAK IVL +G++VLR + TAA+M + + LYM SL +R
Sbjct: 68 LPSNVDLI-KFEKSEKAKDLIVLNVGNQVLRKIKNCETAAAMWSTLKRLYMETSLPNRIY 126
Query: 103 LKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDT 162
L+ + Y++KM +S+SI + +F K++ DL NI V++ +E +A+LLL SL E+ +
Sbjct: 127 LQLKFYTYKMTDSRSIDGNVDDFLKLVTDLNNIGVDVTEEVQAILLLSSLYDRKENSQKV 186
Query: 163 ILYGKEGTATLEEVQAALRTKELTKFKDMKVDEGSEGLNVTRGRNEHRGKGKGKSRSKSR 222
EG +E + + +K ++ + G +V R + + + +G+S K+
Sbjct: 187 ESSQSEGLYVEQEAD---------QRRGLKKEKANHGEDVLRAKAD---QNQGQSTIKTT 234
Query: 223 SKGFDKSKYKCFLCHKQGHFKKDCPDKGGDGS--PSVQVAEASNEEGYESAGALVVTS-- 278
KKD G GS S+ +S E LV+T+
Sbjct: 235 RV-------------VSSVVKKDI----GRGSVLKSLHKPSSSANIATEPKQPLVLTASP 277
Query: 279 WEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
+ ++ WV+DSGCS+H+ K+ L +GG V +GN
Sbjct: 278 QDTKEEWVMDSGCSFHITLDKDSLFDLQEFDGGKVLMGN 316
>At4g04380 putative polyprotein
Length = 778
Score = 95.9 bits (237), Expect = 3e-20
Identities = 66/156 (42%), Positives = 92/156 (58%), Gaps = 19/156 (12%)
Query: 31 QKCVEALKGEAEMPATLTQEEKREMVDKAKSAIVLCLGDKVLRDVAREATAASMLAKWES 90
+K E K E E +E+KR K +S IVL + D+VL EA+ +
Sbjct: 33 EKSDEEEKEECEKVEAF-EEKKR----KTRSTIVLSVSDRVL-----EAS--------DR 74
Query: 91 LYMTKSLAHRQLLKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLC 150
LYM+K+L ++ LKQ+LY FKM E+ S+ + EF I+ DL N+ V + DED+ +LLL
Sbjct: 75 LYMSKALPNQIYLKQKLYRFKMSENLSMEGNIDEFLHIVADLENLNVLVSDEDQTILLLM 134
Query: 151 SLPKSFEHFKDTILYGKEGTA-TLEEVQAALRTKEL 185
SLPKSF+ KDT+ Y T TL+EV AA+ +KEL
Sbjct: 135 SLPKSFDQLKDTLQYSSGKTVLTLDEVTAAIYSKEL 170
Score = 42.7 bits (99), Expect = 3e-04
Identities = 19/45 (42%), Positives = 27/45 (59%)
Query: 273 ALVVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
AL T E W++D+GCSYHM ++E+ E + G VR+GN
Sbjct: 187 ALSSTDIHLEDEWIMDTGCSYHMTHKREWLEDFDEEAGCSVRMGN 231
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 82.0 bits (201), Expect = 4e-16
Identities = 96/348 (27%), Positives = 145/348 (41%), Gaps = 55/348 (15%)
Query: 22 VKMQAILTQQKCVEAL-KG--EAEMPATLTQEEKREMVDKAK-SAIVLCLGDKVLRDVAR 77
++M+AIL E + KG E E +L+Q +K + D K LCL + L +
Sbjct: 23 LRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQGLDEDTF 82
Query: 78 EATAASMLAK--WESLYMTKSLAHR------QLLKQQLYSFKMMESKSISEQLTEFNKIL 129
E + AK WE L + A + Q L+ + + +M E + +S+ F+++L
Sbjct: 83 EKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY---FSRVL 139
Query: 130 DDLANIEVNMEDEDKALLL---LCSLPKSFEHFKDTILYGKEGTA-TLEEVQAALRTKEL 185
N++ N E D ++ L SL FEH I K+ A T+E++ +L+ E
Sbjct: 140 TVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEE 199
Query: 186 TKFKDMKVDEGSEGLNVT----------RGRNEHRGKGKG-------------------K 216
K K + E + +T RG + RG+G+G +
Sbjct: 200 KKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGE 259
Query: 217 SRSKSRSKGFDKSKY-----KCFLCHKQGHFKKDC--PDKGGDGSPSVQVAEASNEEGYE 269
+ S+ R KG KS+Y KC+ C K GH+ +C P + V E EE
Sbjct: 260 NSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFKEKANYVEEKIQEEDML 319
Query: 270 SAGALVVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
+ E W LDSG S HMC RK F L G V LG+
Sbjct: 320 LMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGD 367
>At1g58140 hypothetical protein
Length = 1320
Score = 82.0 bits (201), Expect = 4e-16
Identities = 96/348 (27%), Positives = 145/348 (41%), Gaps = 55/348 (15%)
Query: 22 VKMQAILTQQKCVEAL-KG--EAEMPATLTQEEKREMVDKAK-SAIVLCLGDKVLRDVAR 77
++M+AIL E + KG E E +L+Q +K + D K LCL + L +
Sbjct: 23 LRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQGLDEDTF 82
Query: 78 EATAASMLAK--WESLYMTKSLAHR------QLLKQQLYSFKMMESKSISEQLTEFNKIL 129
E + AK WE L + A + Q L+ + + +M E + +S+ F+++L
Sbjct: 83 EKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY---FSRVL 139
Query: 130 DDLANIEVNMEDEDKALLL---LCSLPKSFEHFKDTILYGKEGTA-TLEEVQAALRTKEL 185
N++ N E D ++ L SL FEH I K+ A T+E++ +L+ E
Sbjct: 140 TVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEE 199
Query: 186 TKFKDMKVDEGSEGLNVT----------RGRNEHRGKGKG-------------------K 216
K K + E + +T RG + RG+G+G +
Sbjct: 200 KKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGE 259
Query: 217 SRSKSRSKGFDKSKY-----KCFLCHKQGHFKKDC--PDKGGDGSPSVQVAEASNEEGYE 269
+ S+ R KG KS+Y KC+ C K GH+ +C P + V E EE
Sbjct: 260 NSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEEKANYVEEKIQEEDML 319
Query: 270 SAGALVVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
+ E W LDSG S HMC RK F L G V LG+
Sbjct: 320 LMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGD 367
>At1g48710 hypothetical protein
Length = 1352
Score = 82.0 bits (201), Expect = 4e-16
Identities = 96/348 (27%), Positives = 145/348 (41%), Gaps = 55/348 (15%)
Query: 22 VKMQAILTQQKCVEAL-KG--EAEMPATLTQEEKREMVDKAK-SAIVLCLGDKVLRDVAR 77
++M+AIL E + KG E E +L+Q +K + D K LCL + L +
Sbjct: 23 LRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQGLDEDTF 82
Query: 78 EATAASMLAK--WESLYMTKSLAHR------QLLKQQLYSFKMMESKSISEQLTEFNKIL 129
E + AK WE L + A + Q L+ + + +M E + +S+ F+++L
Sbjct: 83 EKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY---FSRVL 139
Query: 130 DDLANIEVNMEDEDKALLL---LCSLPKSFEHFKDTILYGKEGTA-TLEEVQAALRTKEL 185
N++ N E D ++ L SL FEH I K+ A T+E++ +L+ E
Sbjct: 140 TVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEE 199
Query: 186 TKFKDMKVDEGSEGLNVT----------RGRNEHRGKGKG-------------------K 216
K K + E + +T RG + RG+G+G +
Sbjct: 200 KKKKKEDIIEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGE 259
Query: 217 SRSKSRSKGFDKSKY-----KCFLCHKQGHFKKDC--PDKGGDGSPSVQVAEASNEEGYE 269
+ S+ R KG KS+Y KC+ C K GH+ +C P + V E EE
Sbjct: 260 NSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEEKANYVEEKIQEEDML 319
Query: 270 SAGALVVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
+ E W LDSG S HMC RK F L G V LG+
Sbjct: 320 LMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGD 367
>At3g61330 copia-type polyprotein
Length = 1352
Score = 81.3 bits (199), Expect = 7e-16
Identities = 97/354 (27%), Positives = 153/354 (42%), Gaps = 67/354 (18%)
Query: 22 VKMQAILTQQKCVEAL-KG--EAEMPATLTQEEKREMVDKAK-SAIVLCLGDKVLRDVAR 77
++M+AIL E + KG E E +L+Q +K + D K LCL + L +
Sbjct: 23 LRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQGLDEDTF 82
Query: 78 EATAASMLAK--WESLYMTKSLAHR------QLLKQQLYSFKMMESKSISEQLTEFNKIL 129
E + AK WE L + A + Q L+ + + +M E + +S+ F+++L
Sbjct: 83 EKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDY---FSRVL 139
Query: 130 DDLANIEVNMEDEDKALLL---LCSLPKSFEHFKDTILYGKEGTA-TLEEVQAALRTKEL 185
N++ N E D ++ L SL FEH I K+ A T+E++ +L+ E
Sbjct: 140 TVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYEE 199
Query: 186 TKFKDMKVDEGSEGLNVT----------RGRNEHRGKGKG-------------------K 216
K K + E + +T RG + RG+G+G +
Sbjct: 200 KKKKKEDIAEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGE 259
Query: 217 SRSKSRSKGFDKSKY-----KCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASN---EEGY 268
+ S+ R KG KS+Y KC+ C K GH+ +C +PS + E EE
Sbjct: 260 NSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECK------APSNKKFEEKAHYVEEKI 313
Query: 269 ESAGALVVTSWEPEK-----SWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
+ L++ S++ ++ W LDSG S HMC RK F L G V LG+
Sbjct: 314 QEEDMLLMASYKKDEQKENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGD 367
>At4g04280 putative transposon protein
Length = 1104
Score = 79.3 bits (194), Expect = 2e-15
Identities = 71/266 (26%), Positives = 121/266 (44%), Gaps = 46/266 (17%)
Query: 5 KWDIEKFTGSNDFGL*KVKMQA---ILTQQKCVEALKGEAEMPATLTQEEKREMVD---- 57
K +I+ F G DF K++++A +L + + K +P +E++ E D
Sbjct: 2 KVEIKTFNGDRDFSFWKIRIEAQLGVLGLKNSLTDFKLTKTVPVAKKEEKESEYEDDASD 61
Query: 58 -----------------KAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHR 100
+AK+ I+ + D VL V TAA + A L+M SL +R
Sbjct: 62 IKQATEEPDPIKFEQSEQAKNFIINHITDTVLLKVQHCKTAAEIWATLNKLFMETSLLNR 121
Query: 101 QLLKQQLYSFKMMESKSISEQLTEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFK 160
+ +LYSFKM+++ SI + + EF +IL D ++ N+ LP S+ K
Sbjct: 122 IYTQLKLYSFKMVDTLSIDQNVDEFLRILADC--LDQNL------------LPSSYIQLK 167
Query: 161 DTILYGKEGTATLEEVQAALRTKELTKFKDMKVDEGSEGLNVTRGRNEHRGKGKGKSRSK 220
T+ +A E + + T+E K M + G R +++ +G+GK+R +
Sbjct: 168 HTLNTDVVSSAKSLERKLS-ETQESNKNVSMALYTTDRGRPQVRNQDK---QGQGKNRGR 223
Query: 221 SRSKGFDKSKYKCFLCHKQGHFKKDC 246
S S K++ C+ C K+ H K+DC
Sbjct: 224 SNS----KTRVTCWFCKKEAHVKRDC 245
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 77.8 bits (190), Expect = 7e-15
Identities = 68/286 (23%), Positives = 123/286 (42%), Gaps = 34/286 (11%)
Query: 51 EKREMVDKAKSAIVLCLGDKVLRDVAREATAASMLAKWESLYMTKSLAHRQLLKQQLYSF 110
EK K K+ + + +L+ + ++ T+ + + Y L++ SF
Sbjct: 20 EKTVKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRSF 79
Query: 111 KMMESKSISEQLTEF----NKILDDLANIEVNMEDE-----------DKALLLLCSLPKS 155
+++E K I E +T + +I +D+ N+ +M D +K ++C++ +S
Sbjct: 80 EVLEMK-IGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEES 138
Query: 156 FEHFKDTILYGKEGTATLEEVQAALRTKELTKFK---DMKVDEGSEGLNVTRGRNEHRGK 212
+ K+ + G + + + E + E K + D G RGR +
Sbjct: 139 -NNIKELTVDGLQSSLMVHEQNLSRHDVEERVLKAETQWRPDGG-------RGRGGSPSR 190
Query: 213 GKGKSRSKSRSKGF-DKSKYKCFLCHKQGHFKKDCPDKGGDGSPSVQVAEASNEEGYESA 271
G+G+ + R +G+ ++ +CF CHK GH+K +CP S + EE
Sbjct: 191 GRGRGGYQGRGRGYVNRDTVECFKCHKMGHYKAECP------SWEKEANYVEMEEDLLLM 244
Query: 272 GALVVTSWEPEKSWVLDSGCSYHMCPRKEYFETLILKEGGVVRLGN 317
+ E ++ W LDSGCS HMC +E+F L VRLG+
Sbjct: 245 AHVEQIGDEEKQIWFLDSGCSNHMCGTREWFLELDSGFKQNVRLGD 290
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,009,710
Number of Sequences: 26719
Number of extensions: 293922
Number of successful extensions: 1653
Number of sequences better than 10.0: 145
Number of HSP's better than 10.0 without gapping: 88
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 1321
Number of HSP's gapped (non-prelim): 243
length of query: 317
length of database: 11,318,596
effective HSP length: 99
effective length of query: 218
effective length of database: 8,673,415
effective search space: 1890804470
effective search space used: 1890804470
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135464.1


