

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.4 + phase: 0
(631 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
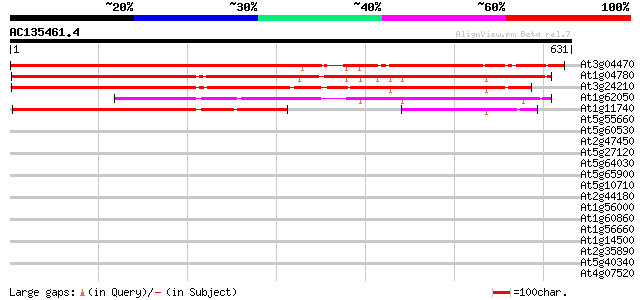
Score E
Sequences producing significant alignments: (bits) Value
At3g04470 unknown protein 845 0.0
At1g04780 unknown protein 645 0.0
At3g24210 hypothetical protein 596 e-170
At1g62050 unknown protein 358 5e-99
At1g11740 unknown protein 336 3e-92
At5g55660 putative protein 37 0.044
At5g60530 late embryonic abundant protein - like 36 0.074
At2g47450 CAO chloroplast signal recognition particle chromo pro... 35 0.13
At5g27120 SAR DNA-binding protein - like 35 0.17
At5g64030 ankyrin-like protein 34 0.22
At5g65900 ATP-dependent RNA helicase-like 33 0.48
At5g10710 unknown protein 33 0.48
At2g44180 methionine aminopeptidase-like protein 33 0.48
At1g56000 hypothetical protein 33 0.48
At1g60860 GCN4-complementing like protein 33 0.63
At1g56660 hypothetical protein 33 0.63
At1g14500 hypothetical protein 33 0.63
At2g35890 putative calcium-dependent protein kinase 32 0.82
At5g40340 unknown protein 32 1.1
At4g07520 hypothetical protein 32 1.1
>At3g04470 unknown protein
Length = 640
Score = 845 bits (2183), Expect = 0.0
Identities = 442/664 (66%), Positives = 526/664 (78%), Gaps = 72/664 (10%)
Query: 1 MEDISKYKHSPAHVAVLNRDHASLRRLISTIPTLPKPGEVTTESESLSAEIQADQISSVI 60
MED SKY HSPAH+AV+ RDHA+LRR++S +P L K GEVTTE+ES+ +E +AD +S+VI
Sbjct: 1 MEDYSKYTHSPAHLAVVLRDHAALRRIVSDLPRLAKAGEVTTEAESMESESRADSVSAVI 60
Query: 61 DRRDVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVI 120
DRRDVPGRETPLHL VRLRDP++AEILMSAGADWSLQNE GWS+LQEAVC REE+IA++I
Sbjct: 61 DRRDVPGRETPLHLAVRLRDPVSAEILMSAGADWSLQNENGWSALQEAVCTREEAIAMII 120
Query: 121 ARYYQPLAWSKWCRRLPRVIGSASRIRDFYMEISFHFESSVIPFIGRIAPSDTYRIWKRG 180
AR+YQPLAW+KWCRRLPR+I SASRIRDFYMEI+FHFESSVIPFIGRIAPSDTYRIWKRG
Sbjct: 121 ARHYQPLAWAKWCRRLPRIIASASRIRDFYMEITFHFESSVIPFIGRIAPSDTYRIWKRG 180
Query: 181 ANLRADMTLAGFDGFRIQRSDQSFLFLGEGYASEDGNVSLPRGSLIALAHKEKEITNALE 240
+NLRADMTLAGFDGF+IQRSDQ+FLFLG+GY+SEDG +SL GSLI L+HKEKE+TNALE
Sbjct: 181 SNLRADMTLAGFDGFKIQRSDQTFLFLGDGYSSEDGKMSLSPGSLIVLSHKEKEMTNALE 240
Query: 241 GAGTQPTESEIANEVSMMFQTNMYRPGIDVTQAELVPNLNWRRQEKTEVVGDWKAKVYDM 300
GAG QPT++E+A+EV++M QTNMYRPGIDVTQAELV +LNWRRQE+TE+VG+WKAKVYDM
Sbjct: 241 GAGAQPTDAEVAHEVALMSQTNMYRPGIDVTQAELVSHLNWRRQERTEMVGNWKAKVYDM 300
Query: 301 LNVMVSVKSRRVPGALNDEEVFANGED------------YDDVLTAEERVQLDSALRMGN 348
L+VMVSVKSRRVPGA+ DEE+FA E+ ++DVLT EER+QL+SAL+ GN
Sbjct: 301 LHVMVSVKSRRVPGAMTDEELFAVDEERTAVTNGAETDGFEDVLTPEERLQLNSALQTGN 360
Query: 349 SDGVCQDEEPGAGGGGFDGRGNLYENCEV-----NGVVKEKKGWFGWNK----------- 392
SD + +DEE CEV NG +K+KKGWFGWNK
Sbjct: 361 SDAI-EDEE-----------------CEVTDQQENGALKDKKGWFGWNKKGSNTEDTKLK 402
Query: 393 --------NPNQRSSDQQ-KLQPEFPNDDHGEMKSKKGKDQNLKKKKKKG-ASNESKSES 442
+ NQ+ Q+ + + N+DHG+ +KKGK+ KKKKKKG A +E K ES
Sbjct: 403 KGSKSAPEDGNQKGKSQKSSMVSDHANEDHGD--AKKGKE---KKKKKKGVAGDEVKRES 457
Query: 443 EFKKGVRPVLWLTQDFPLKTDELLPLLDILANKVKAIRRLRELLTTKLPPGTFPVKVAIP 502
E+KKG+RPVLWLT DFPL TDELLPLLDILANKVKA+RRLRELLTTKLP GTFPVK+AIP
Sbjct: 458 EYKKGLRPVLWLTPDFPLTTDELLPLLDILANKVKAVRRLRELLTTKLPLGTFPVKLAIP 517
Query: 503 IVPTIRVIVTFTKFEELQ-TPEEFSTPLSSPVSFQDAKYKESEGST-SWVSWMKGSRGMP 560
I+PT+RV+VTFTKFEELQ EEFSTP SSPV F DAK SE S+ SW+SWM+ +
Sbjct: 518 IIPTVRVVVTFTKFEELQAAEEEFSTPPSSPV-FHDAKSSSSENSSPSWISWMRSGK--- 573
Query: 561 SSDSDSHIHSHRYKDEVDPFIIPSDYKWVDANERKRRMKAKRAKIKKNKKQTVAKGRDGA 620
SSD+D S+RYKDE DPF+IPSDYKW+D+ E+KRRMKAK+AK + KKQ K +
Sbjct: 574 SSDND----SNRYKDEADPFLIPSDYKWIDSAEKKRRMKAKKAK-SRRKKQAATKAAGAS 628
Query: 621 HQGS 624
G+
Sbjct: 629 DSGT 632
>At1g04780 unknown protein
Length = 664
Score = 645 bits (1664), Expect = 0.0
Identities = 350/663 (52%), Positives = 454/663 (67%), Gaps = 66/663 (9%)
Query: 3 DISKYKHSPAHVAVLNRDHASLRRLISTIPTLPKPGEVTTESESLSAEIQADQISSVIDR 62
D++KY HSP H AV+ RD+A L++L+S +P + P EV E+ S++ E +AD I++VIDR
Sbjct: 7 DVTKYGHSPVHHAVVTRDYAGLKKLLSALPKMRDPSEVQNEAASVAEETKADSIAAVIDR 66
Query: 63 RDVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVIAR 122
RDV R+T LHL V+L D +AE+LM+AGADWSLQNE GWS+LQEA+C REE IA++I R
Sbjct: 67 RDVVNRDTALHLAVKLGDETSAEMLMAAGADWSLQNEHGWSALQEAICGREERIAMIIVR 126
Query: 123 YYQPLAWSKWCRRLPRVIGSASRIRDFYMEISFHFESSVIPFIGRIAPSDTYRIWKRGAN 182
+YQPLAW+KWCRRLPR++ + R+RDFYMEI+FHFESSVIPFI R+APSDTY+IWKRGAN
Sbjct: 127 HYQPLAWAKWCRRLPRLVATMHRMRDFYMEITFHFESSVIPFISRVAPSDTYKIWKRGAN 186
Query: 183 LRADMTLAGFDGFRIQRSDQSFLFLGEGYASEDGNVSLPRGSLIALAHKEKEITNALEGA 242
LRADMTLAGFDGFRIQRSDQ+ LFLG+G SEDG V P GSL+ ++HK+KEI NAL+GA
Sbjct: 187 LRADMTLAGFDGFRIQRSDQTILFLGDG--SEDGKV--PSGSLLMISHKDKEIMNALDGA 242
Query: 243 GTQPTESEIANEVSMMFQTNMYRPGIDVTQAELVPNLNWRRQEKTEVVGDWKAKVYDMLN 302
G +E E+ EV+ M +T+++RPGIDVTQA L P L WRRQEKTE+VG WKAKVYDM N
Sbjct: 243 GAAASEEEVRQEVAAMSKTSIFRPGIDVTQAVLFPQLTWRRQEKTEMVGQWKAKVYDMHN 302
Query: 303 VMVSVKSRRVPGALNDEEVFAN--------GEDYDDVLTAEERVQLDSALRMGNSDGVCQ 354
V+VS+KSRRVPGA+ DEE+F+N ED D+LT +E+ QL+ AL++ +
Sbjct: 303 VVVSIKSRRVPGAMTDEELFSNTNQENDTESEDLGDILTEDEKRQLELALKLDS-----P 357
Query: 355 DEEPGAGGGGFDGRGN--LYENCEV-----NGVVK-EKKGWF-GWNKN----------PN 395
+E + N +E+ E+ NG K EKKGWF GW K P
Sbjct: 358 EESSNGESSRISQKQNSCSFEDREIPVTDGNGYCKQEKKGWFSGWRKREEGHRRSSVPPR 417
Query: 396 QRSSDQQKLQPEFPNDD---HGEMKSKKGKDQNLK---KKKKKGASNESKS--------- 440
+K+ +DD G + K G+ ++ + + +G + SK+
Sbjct: 418 NSLCVDEKVSDLLGDDDSPSRGGRQIKPGRHSTVETVVRNENRGLRDSSKASTSEGSGSS 477
Query: 441 -------ESEFKKGVRPVLWLTQDFPLKTDELLPLLDILANKVKAIRRLRELLTTKLPPG 493
E+E+KKG+RPVLWL++ FPL+T ELLPLLDILANKVKAIRRLREL+TTKLP G
Sbjct: 478 KRKEGNKENEYKKGLRPVLWLSERFPLQTKELLPLLDILANKVKAIRRLRELMTTKLPSG 537
Query: 494 TFPVKVAIPIVPTIRVIVTFTKFEELQTPE-EFSTPLSSPVSF-----QDAKYKESEGST 547
TFPVKVAIP++PTIRV+VTFTKFEEL+ E EF TP SSP S ++ S S+
Sbjct: 538 TFPVKVAIPVIPTIRVLVTFTKFEELEAIEDEFVTPPSSPTSSVKNSPREETQSSSNPSS 597
Query: 548 SWVSWMKGSRGMPSSDSDS-HIHSHRYKDEVDPFIIPSDYKWVDANERKRRMKAKRAKIK 606
SW W+K PS+ S S + + +++ DPF IP Y W+ A E+K++++ K K K
Sbjct: 598 SWFQWIKTPSQRPSTSSSSGGFNIGKAENDQDPFAIPRGYNWITAEEKKKKVQEKN-KAK 656
Query: 607 KNK 609
K K
Sbjct: 657 KGK 659
>At3g24210 hypothetical protein
Length = 614
Score = 596 bits (1537), Expect = e-170
Identities = 318/601 (52%), Positives = 420/601 (68%), Gaps = 33/601 (5%)
Query: 3 DISKYKHSPAHVAVLNRDHASLRRLISTIPTLPKPGEVTTESESLSAEIQADQISSVIDR 62
D+SKY HS H AV ++D+ LR ++S++P P E+ TE+ SLS E ++D IS+VIDR
Sbjct: 5 DVSKYIHSSVHKAVASKDYVGLRAILSSLPKPKDPCEIQTEAASLSEEAKSDTISAVIDR 64
Query: 63 RDVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVIAR 122
RDVP R+TPLHL V+L D + E+LM AGADW+LQNE GW++LQEAVC+R+E+IA++I R
Sbjct: 65 RDVPSRDTPLHLAVKLCDSTSVEMLMVAGADWTLQNEDGWNALQEAVCSRQEAIAMIIVR 124
Query: 123 YYQPLAWSKWCRRLPRVIGSASRIRDFYMEISFHFESSVIPFIGRIAPSDTYRIWKRGAN 182
+YQPLAW+KWCRRLPR+I + S+++DFY+E+SFHFESSV+PF+ ++APSDTY++WK G+N
Sbjct: 125 HYQPLAWAKWCRRLPRLIATMSKMKDFYLEMSFHFESSVVPFVSKVAPSDTYKVWKVGSN 184
Query: 183 LRADMTLAGFDGFRIQRSDQSFLFLGEGYASEDGNVSLPRGSLIALAHKEKEITNALEGA 242
LRADMT+AGFDGF+IQRSDQS LFLG+G SEDG V+ RGSL + HK+KE+ NAL+GA
Sbjct: 185 LRADMTMAGFDGFKIQRSDQSILFLGDG--SEDGEVA--RGSLYMVNHKDKEVMNALDGA 240
Query: 243 GTQPTESEIANEVSMMFQTNMYRPGIDVTQAELVPNLNWRRQEKTEVVGDWKAKVYDMLN 302
P+E E+ EV+ M ++N++RPGIDVTQA L P NWRRQEK+E+VG WKAKVY+M N
Sbjct: 241 CGVPSEEEVRKEVAAMCKSNIFRPGIDVTQAVLSPQTNWRRQEKSEMVGPWKAKVYEMNN 300
Query: 303 VMVSVKSRRVPGALNDEEVFANGEDYDDVLTAEERVQLDSALRMGNSDGVCQDEEPGAGG 362
V+VSVKSR+VPG+L V +D DVLT EE+ QL+ AL++ D P G
Sbjct: 301 VVVSVKSRKVPGSL---AVEGGDDDICDVLTDEEQKQLEDALKLDLPD------FPNVNG 351
Query: 363 GGFDGRGNLYENCEVNGVVKEKKGWF-GWNKNPNQRSSDQQKLQPEFPNDDHGEMKSKKG 421
+ ++ E N EKKGW GW K ++ Q P + ++ + G
Sbjct: 352 ENGFIVHHQEDHVEANKKA-EKKGWLGGWRKKETKQEKQIQYAPPRSSLCVNEKVSNLLG 410
Query: 422 KDQNLK-----------KKKKKGASNESKSESEFKKGVRPVLWLTQDFPLKTDELLPLLD 470
+ +K +K K S++++ ESE+KKG+RPVLWL+ +FPL+T+ELLPLLD
Sbjct: 411 ESNQIKPGRHSVDNEHYRKPKALVSDKTRHESEYKKGLRPVLWLSPNFPLQTEELLPLLD 470
Query: 471 ILANKVKAIRRLRELLTTKLPPGTFPVKVAIPIVPTIRVIVTFTKFEELQTPEEFSTPLS 530
ILANKVKAIRRLRELLTTKLP GTFPVKVAIP+VPTI+V+VTFTKFEE++ +EF TPLS
Sbjct: 471 ILANKVKAIRRLRELLTTKLPSGTFPVKVAIPVVPTIKVLVTFTKFEEIEPVDEFKTPLS 530
Query: 531 SPVS----FQDAKYKESEGSTSWVSWMKGSRGMPSSDSDSHIHSHRYKDEVDPFIIPSDY 586
SP S A + S+S SW + SR S+ I SH +D + Y
Sbjct: 531 SPTSSGYESPAAATTDFTNSSSSSSWFRRSR---SNKDGGSISSHLLHSSLDLILNTIKY 587
Query: 587 K 587
+
Sbjct: 588 R 588
>At1g62050 unknown protein
Length = 497
Score = 358 bits (919), Expect = 5e-99
Identities = 220/524 (41%), Positives = 294/524 (55%), Gaps = 68/524 (12%)
Query: 119 VIARYYQPLAWSKWCRRLPRVIGSASRIRDFYMEISFHFESSVIPFIGRIAPSDTYRIWK 178
V+ R++ LAW KW RRLPR+I R+RDFYMEISFHFESSVIPF+G+IAPSDTY+IWK
Sbjct: 3 VLLRHHHRLAWCKWRRRLPRLIAVLRRMRDFYMEISFHFESSVIPFVGKIAPSDTYKIWK 62
Query: 179 RGANLRADMTLAGFDGFRIQRSDQSFLFLGEGYASEDGNVSLPRGSLIALAHKEKEITNA 238
R NLRAD TLAGFDG +IQR+DQSFLFLG+G D ++ + GSL+ L +++I +A
Sbjct: 63 RDGNLRADTTLAGFDGLKIQRADQSFLFLGDG----DESLDISPGSLLVLNRDDRKILDA 118
Query: 239 LEGAGTQPTESEIANEVSMMFQTNMYRPGIDVTQAELVPNLNWRRQEKTEVVGDWKAKVY 298
E AG ++S+IA S Q+++YRPG+DVT+AELV NWRRQEK E VG+WKA+VY
Sbjct: 119 FESAGAPISDSDIAGFCS---QSSLYRPGMDVTKAELVGRTNWRRQEKMENVGEWKARVY 175
Query: 299 DMLNVMVSVKSRRVPGALNDEEVFANGEDYDD--VLTAEERVQLDSALRMGNSDGVCQDE 356
++ V S +SR++ E+V D DD L AE L R
Sbjct: 176 EIQKVTFSFRSRKIVTGDGSEQVLPLELDEDDEGFLVAENPSFLAPPPRSQRRH------ 229
Query: 357 EPGAGGGGFDGRGNLYENCEVNGVVKEKKGWFGWNKN------------PNQRSSDQQKL 404
+ VKE + W + P Q S ++ +
Sbjct: 230 ---------------------SSFVKEDRDWISVGRKSVDVYPSAAPPPPRQSQSPRRSV 268
Query: 405 -QPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKS-----------ESEFKKGVRPVL 452
Q + P +++ + + ++ A + S E EF K + P +
Sbjct: 269 SQFQPPRRSMAQVQPPRRSVAQAQPPRRSVALPSTVSPVNPPPSPQIKEKEFVKSLHPSV 328
Query: 453 WLTQDFPLKTDELLPLLDILANKVKAIRRLRELLTTKLPPGTFPVKVAIPIVPTIRVIVT 512
WLT+ FPLKT+ELLPLLDILANKVKA+ R+R+LLTTK PPGTFPVK++IP+VPT++V++T
Sbjct: 329 WLTEQFPLKTEELLPLLDILANKVKAVARMRDLLTTKFPPGTFPVKLSIPVVPTVKVVIT 388
Query: 513 FTKFEELQTPEEFSTPLSSPVSFQDAKYKESEGSTSWVSWMKGSRGMPSS--DSDSHIHS 570
FTKF EL E F TPLSSP A E + S + S PS+ + S
Sbjct: 389 FTKFVELPPAETFYTPLSSPRFLYSATAGEEDHSDGEKPDARNSISRPSTWLRLATGKGS 448
Query: 571 HRYKDE-----VDPFIIPSDYKWVDANERKRRMKAKRAKIKKNK 609
HR +E DPF IP YKW + + +K+KK+K
Sbjct: 449 HRRVEEEEQQTPDPFAIPIGYKWTSMSSSNSK-SGNSSKMKKSK 491
>At1g11740 unknown protein
Length = 624
Score = 336 bits (861), Expect = 3e-92
Identities = 174/309 (56%), Positives = 220/309 (70%), Gaps = 7/309 (2%)
Query: 4 ISKYKHSPAHVAVLNRDHASLRRLISTIPTLPKPGEVTTESESLSAEIQADQISSVIDRR 63
I Y HSP H AV+ DHA L RL+S++P L P ++ TES+S+S E A++IS+VIDRR
Sbjct: 14 IDDYAHSPVHYAVVVGDHAGLTRLVSSLPKLTDPEQIHTESDSMSQERVAEKISTVIDRR 73
Query: 64 DVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVIARY 123
DVP ETPLHL VRL D AA+ + SAGAD +LQN GW+SL EA+C R I I R
Sbjct: 74 DVPFGETPLHLAVRLGDVFAAKTISSAGADITLQNAAGWNSLHEALCRRNSEITEAILRD 133
Query: 124 YQPLAWSKWCRRLPRVIGSASRIRDFYMEISFHFESSVIPFIGRIAPSDTYRIWKRGANL 183
+ AW KW RRLP +I SR+RDFYMEISFHFESSVIPF+G+IAPSDTY+IWKRG +L
Sbjct: 134 HHRSAWCKWRRRLPHLIAVLSRMRDFYMEISFHFESSVIPFVGKIAPSDTYKIWKRGGDL 193
Query: 184 RADMTLAGFDGFRIQRSDQSFLFLGEGYASEDGNVSLPRGSLIALAHKEKEITNALEGAG 243
RAD +LAGFDGF+I+R++QSFLFL +G D + + G+L+ L +K I NA E A
Sbjct: 194 RADTSLAGFDGFKIRRANQSFLFLSDG----DEFLDVSPGTLLVLNRDDKTILNAFENAK 249
Query: 244 TQPTESEIANEVSMMFQTNMYRPGIDVTQAELVPNLNWRRQEKTEVVGDWKAKVYDMLNV 303
++S+IA Q+++YRPG+DVT+A+LV NWRRQ K E VG+WKAK YD+ NV
Sbjct: 250 DPISDSDIA---GFCGQSSLYRPGMDVTKAKLVEITNWRRQAKIETVGEWKAKAYDVENV 306
Query: 304 MVSVKSRRV 312
S KSR+V
Sbjct: 307 SFSFKSRKV 315
Score = 144 bits (363), Expect = 1e-34
Identities = 77/168 (45%), Positives = 101/168 (59%), Gaps = 17/168 (10%)
Query: 441 ESEFKKGVRPVLWLTQDFPLKTDELLPLLDILANKVKAIRRLRELLTTKLPPGTFPVKVA 500
E E K +RP +WLT FPLKT+ELLPLLDILAN VKA+RR+RELLTTK P GTFPVK++
Sbjct: 365 EKEIVKSLRPSVWLTDQFPLKTEELLPLLDILANHVKAVRRMRELLTTKFPAGTFPVKLS 424
Query: 501 IPIVPTIRVIVTFTKFEELQTPEEFSTPLSSPVSF---------------QDAKYKESEG 545
IP++PT++V++TF+KF L +EF TPLSSP D + S
Sbjct: 425 IPVIPTVKVVMTFSKFVALPPLDEFYTPLSSPKHLLAAMEDQHDVHDDEKLDRRISSSRP 484
Query: 546 STSWVSWMKGSRGMPSSDSDSHIHSHRYKDEVDPFIIPSDYKWVDANE 593
S S SW++ + S + + VDPF IP+ YKW ++
Sbjct: 485 SFSTPSWLRLNINATGKSSRRRLVDE--EQVVDPFTIPTGYKWTSNSD 530
>At5g55660 putative protein
Length = 759
Score = 36.6 bits (83), Expect = 0.044
Identities = 41/173 (23%), Positives = 72/173 (40%), Gaps = 24/173 (13%)
Query: 319 EEVFANGEDYDDVLTAEERVQLDS-----ALRMGNSDGVCQDEEPGAGGGGFDGRGNLYE 373
EE G+ DDV AE+ +D AL+ N + ++EE G + N +
Sbjct: 165 EEKLVGGDKGDDVDEAEKVENVDEDDKEEALKEKNEAELAEEEETNKGEEVKEA--NKED 222
Query: 374 NCEVNGVVKE------KKGWFGWNKNPNQRSSDQQKLQPEFPNDDHGEMKSKKGKDQNLK 427
+ E + V E K N++ + D+++ + E NDD K+ K +++K
Sbjct: 223 DVEADTKVAEPEVEDKKTESKDENEDKEEEKEDEKEDEKEESNDD------KEDKKEDIK 276
Query: 428 KKKKKGASNESKS-----ESEFKKGVRPVLWLTQDFPLKTDELLPLLDILANK 475
K K+G K+ E KK + P D P++ + + L + +K
Sbjct: 277 KSNKRGKGKTEKTRGKTKSDEEKKDIEPKTPFFSDRPVRERKSVERLVAVVDK 329
>At5g60530 late embryonic abundant protein - like
Length = 439
Score = 35.8 bits (81), Expect = 0.074
Identities = 23/94 (24%), Positives = 43/94 (45%), Gaps = 2/94 (2%)
Query: 356 EEPGAGGGGFDGRGNLYENCEVNGVVKEKKGWFGWNKNPNQRSSDQQKLQPEFPNDDHGE 415
+ G G G +G+GN + + G + G K+ Q D++K + + +
Sbjct: 25 QSDGGKGNGNNGKGNEVQVDKGKGDNGKSNG--NGPKDKEQEKKDKEKAAKDKKEKEKKD 82
Query: 416 MKSKKGKDQNLKKKKKKGASNESKSESEFKKGVR 449
+ K+ KD+ K+K+KK + K + E K+ R
Sbjct: 83 KEEKEKKDKERKEKEKKDKLEKEKKDKERKEKER 116
>At2g47450 CAO chloroplast signal recognition particle chromo
protein
Length = 373
Score = 35.0 bits (79), Expect = 0.13
Identities = 22/62 (35%), Positives = 33/62 (52%), Gaps = 5/62 (8%)
Query: 47 LSAEIQADQISSVIDRRDVPGRETPLHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQ 106
L AE AD +D RD+ G T LH+ P E L+ GAD +++E+G ++L+
Sbjct: 178 LLAEAGAD-----LDHRDMRGGLTALHMAAGYVRPEVVEALVELGADIEVEDERGLTALE 232
Query: 107 EA 108
A
Sbjct: 233 LA 234
>At5g27120 SAR DNA-binding protein - like
Length = 533
Score = 34.7 bits (78), Expect = 0.17
Identities = 36/154 (23%), Positives = 62/154 (39%), Gaps = 36/154 (23%)
Query: 323 ANGEDYDDVLTAEERVQLDSALR------MGNSDGVCQ--------DEEPGAGGGGFDGR 368
A G+ D+ + E R++L++ LR +G G + D++ G GG
Sbjct: 372 ALGDSQDNTMGVENRLKLEARLRTLEGKDLGRLSGSAKGKPKIEVYDKDKKKGSGGLITP 431
Query: 369 GNLYENC-----------EVNGVVKEKKGWFGWNKNPNQRSSDQQKLQPEFPNDDHGEMK 417
Y NGV KEKK +K +++ D+++ + E P+ K
Sbjct: 432 AKTYNTAADSLLQTPTVDSENGV-KEKK-----DKKKKKKADDEEEAKTEEPSKKKSNKK 485
Query: 418 SKKGKDQNL-----KKKKKKGASNESKSESEFKK 446
+ + + K+KKKK E ++E KK
Sbjct: 486 KTEAEPETAEEPAKKEKKKKRKHEEEETEMPAKK 519
>At5g64030 ankyrin-like protein
Length = 829
Score = 34.3 bits (77), Expect = 0.22
Identities = 24/109 (22%), Positives = 45/109 (41%), Gaps = 13/109 (11%)
Query: 346 MGNSDGVCQDEEPGAGGGGFDGRGNLYENCEVNGVVKEKKGWF---------GWNKNPNQ 396
+ N DG+ + P A +G G +E+ V ++KKG +K NQ
Sbjct: 54 LDNKDGIKKQMTPPAE----EGNGQKFEDAPVETPNEDKKGDGDASLPKEDESSSKQDNQ 109
Query: 397 RSSDQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKSESEFK 445
++K + EF + +++ G+DQ K + G + + + K
Sbjct: 110 EEKKEEKTKEEFTPSSETKSETEGGEDQKDDSKSENGGGGDLDEKKDLK 158
>At5g65900 ATP-dependent RNA helicase-like
Length = 633
Score = 33.1 bits (74), Expect = 0.48
Identities = 26/104 (25%), Positives = 46/104 (44%), Gaps = 3/104 (2%)
Query: 392 KNPNQRSSDQQKLQPEFP-NDDHGEMKSKKGKDQNLKKKKKKGASNESKSESEFKKGVRP 450
+ P ++ +KLQ ND+ E+ +++ + + KKK++K +S+ E K
Sbjct: 84 EEPKKKKKKNKKLQQRGDTNDEEDEVIAEEEEPKKKKKKQRKDTEAKSEEEEVEDKEEEK 143
Query: 451 VLWLTQDFPLKTDELLPLLDILANKVK--AIRRLRELLTTKLPP 492
L T KT E L L D +K R+ ++ +PP
Sbjct: 144 KLEETSIMTNKTFESLSLSDNTYKSIKEMGFARMTQIQAKAIPP 187
>At5g10710 unknown protein
Length = 312
Score = 33.1 bits (74), Expect = 0.48
Identities = 27/96 (28%), Positives = 42/96 (43%), Gaps = 11/96 (11%)
Query: 268 IDVTQAELVPNLNWRRQEKTEVVGDWKAKVYDMLNVMVSVKSRRVPGALNDEEVFANGED 327
+D T+A L L R+ + D + D LN R +EVF +GE+
Sbjct: 14 LDTTRARLSNLLKRHRELSDRLTRDSDKTMLDRLNKEFEAARRS-----QSQEVFLDGEE 68
Query: 328 YDDVL--TAEERVQLDSALRMGNSDG----VCQDEE 357
++D L T ERV +++ + N + VC EE
Sbjct: 69 WNDGLLATLRERVHMEADRKADNGNAGFSLVCHPEE 104
>At2g44180 methionine aminopeptidase-like protein
Length = 441
Score = 33.1 bits (74), Expect = 0.48
Identities = 41/168 (24%), Positives = 66/168 (38%), Gaps = 29/168 (17%)
Query: 374 NCEVNGVVKEKKGWFGWNKNPNQRSSDQQKLQP-----EFPNDDHGEMKSKKGKDQNLKK 428
N EV + KE N N +Q SSD K E D++ E + + + KK
Sbjct: 5 NPEVATMGKENTEAESSNGNESQLSSDLTKSLDLAEVKEDEKDNNQEEEDGLKAEASTKK 64
Query: 429 KKKKGASNESKSESE------------FKKGVRPV----------LWLTQDFPLKTDELL 466
KKKK S + KS + F G P LW T + E L
Sbjct: 65 KKKKSKSKKKKSSLQQTDPPSIPVLELFPSGDFPQGEIQQYNDDNLWRTTSEEKREMERL 124
Query: 467 --PLLDILANKVKAIRRLRELLTTKLPPGTFPVKVAIPIVPTIRVIVT 512
P+ + L + R++R+ + + L PG + + + T+R +++
Sbjct: 125 QKPIYNSLRQAAEVHRQVRKYMRSILKPGMLMIDLCETLENTVRKLIS 172
>At1g56000 hypothetical protein
Length = 416
Score = 33.1 bits (74), Expect = 0.48
Identities = 42/157 (26%), Positives = 64/157 (40%), Gaps = 35/157 (22%)
Query: 432 KGASNESKSESEFKKGVRPVLWLTQDFP-LKTD---ELLPLLDILANKVKAIRRLRELLT 487
K NES +S F G+ + WL ++ P L TD E L D + K I R
Sbjct: 160 KALCNESGVKSMFGTGIAKMEWLEEEIPWLLTDSKGENLGRFDGVVASDKNIVSPRFTQV 219
Query: 488 TKLPPGTFPVKVA-----------IPIVPTIRVIVTFTKFEELQTPEEFSTPLSSPVSFQ 536
T LPP P+ ++ IP++P +++ F + PLSS + +
Sbjct: 220 TGLPP---PLDLSLVPELATKLQNIPVLPCFSLMLAFKE------------PLSS-IPVK 263
Query: 537 DAKYKESEGSTSWVSWMKGSRGMPSSDSDSHIHSHRY 573
+K SE +SW P +DS ++S Y
Sbjct: 264 GLSFKNSE----ILSWAHCESTKPGRSTDSVLNSECY 296
>At1g60860 GCN4-complementing like protein
Length = 776
Score = 32.7 bits (73), Expect = 0.63
Identities = 15/51 (29%), Positives = 28/51 (54%)
Query: 72 LHLTVRLRDPIAAEILMSAGADWSLQNEQGWSSLQEAVCNREESIALVIAR 122
LH+ + DPI E+L+ GAD ++++ G + L + + + A V+ R
Sbjct: 688 LHVACQSGDPILLELLLQFGADINMRDYHGRTPLHHCIASGNNAFAKVLLR 738
>At1g56660 hypothetical protein
Length = 522
Score = 32.7 bits (73), Expect = 0.63
Identities = 17/66 (25%), Positives = 37/66 (55%), Gaps = 2/66 (3%)
Query: 381 VKEKKGWFGWNKNPNQRSSDQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKS 440
++E+ G K ++ ++++K +P+ + + +SK +D+ +K KK+KG + +
Sbjct: 169 LEEEDGKKNKKKEKDESGTEEKKKKPK--KEKKQKEESKSNEDKKVKGKKEKGEKGDLEK 226
Query: 441 ESEFKK 446
E E KK
Sbjct: 227 EDEEKK 232
>At1g14500 hypothetical protein
Length = 436
Score = 32.7 bits (73), Expect = 0.63
Identities = 23/88 (26%), Positives = 41/88 (46%), Gaps = 14/88 (15%)
Query: 1 MEDISKYKHSPAHVAVLNRDHASLRRLISTIPTLPKPGEVTTESESLSAEIQADQISSVI 60
++D+S H+ H+AV+N L+ L + + + +TES+ L
Sbjct: 132 IQDVSVNGHNALHLAVMNDRFEILQVLTGWLQRMSQKDSASTESDFL------------- 178
Query: 61 DRRDVPGRETPLHLTVRLRDPIAAEILM 88
+R+D+ TPLHL D A ++L+
Sbjct: 179 NRKDL-AHNTPLHLAAYKEDHQAVKLLL 205
>At2g35890 putative calcium-dependent protein kinase
Length = 520
Score = 32.3 bits (72), Expect = 0.82
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query: 397 RSSDQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKSESEFKK 446
+ D+ KL ND HG+ K +GK++ +K+ +G S+ E+KK
Sbjct: 48 KDDDKTKL-----NDTHGDPKLLEGKEKPAQKQTSQGQGGRKCSDEEYKK 92
>At5g40340 unknown protein
Length = 1008
Score = 32.0 bits (71), Expect = 1.1
Identities = 14/46 (30%), Positives = 29/46 (62%)
Query: 399 SDQQKLQPEFPNDDHGEMKSKKGKDQNLKKKKKKGASNESKSESEF 444
S +++ + + P D E+ ++ K + KKKK++G S + ++E+EF
Sbjct: 816 STKKERKRKKPKHDEEEVPNETEKPEKKKKKKREGKSKKKETETEF 861
>At4g07520 hypothetical protein
Length = 734
Score = 32.0 bits (71), Expect = 1.1
Identities = 29/97 (29%), Positives = 42/97 (42%), Gaps = 11/97 (11%)
Query: 356 EEPGAGGGGFDGRGNLYENCEVNGVVKEKKGWFGWNKNPNQRSSDQQKLQPEFPNDD--- 412
E G G + G + E +GV E +G+ P+ SS P+ P D
Sbjct: 592 ETDGVETDGVETGGVETHDVETDGV--EPEGFVLEFVEPDDPSSAMVLYVPDVPIGDVQV 649
Query: 413 HGEMKSKKGKDQNLKKKKK------KGASNESKSESE 443
E ++ KG D+ LKKK K KG N+ K+E +
Sbjct: 650 ERETEADKGDDETLKKKTKKKGRKGKGKCNDKKNEKK 686
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,031,373
Number of Sequences: 26719
Number of extensions: 697394
Number of successful extensions: 2420
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 45
Number of HSP's that attempted gapping in prelim test: 2274
Number of HSP's gapped (non-prelim): 148
length of query: 631
length of database: 11,318,596
effective HSP length: 105
effective length of query: 526
effective length of database: 8,513,101
effective search space: 4477891126
effective search space used: 4477891126
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC135461.4


