

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.15 + phase: 0 /pseudo
(1184 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
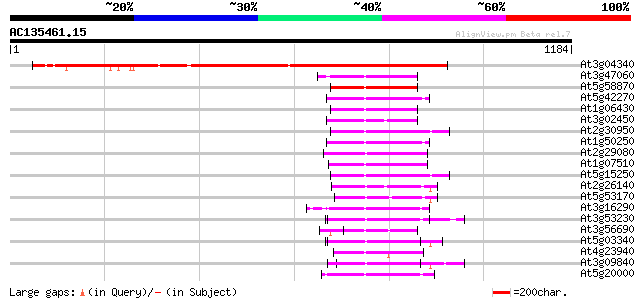
At3g04340 unknown protein 839 0.0
At3g47060 FtsH metalloprotease - like protein 155 2e-37
At5g58870 cell division protein - like 153 6e-37
At5g42270 cell division protein FtsH 146 6e-35
At1g06430 cell division protease FtsH, putative 145 1e-34
At3g02450 cell division protein FtsH-like protein 145 2e-34
At2g30950 zinc dependent protease (VAR2) 144 2e-34
At1g50250 putative chloroplast FtsH protease 144 2e-34
At2g29080 AAA-type like ATPase 142 1e-33
At1g07510 unknown protein 140 4e-33
At5g15250 FtsH-like protein Pftf precursor - like 138 2e-32
At2g26140 FtsH like protease 137 3e-32
At5g53170 cell division protein FtsH protease-like 136 6e-32
At3g16290 putative FtsH-like metalloprotease 124 2e-28
At3g53230 CDC48 - like protein 124 3e-28
At3g56690 calmodulin-binding protein 123 7e-28
At5g03340 transitional endoplasmic reticulum ATPase 122 1e-27
At4g23940 cell division protein - like 122 1e-27
At3g09840 putative transitional endoplasmic reticulum ATPase 121 3e-27
At5g20000 26S proteasome AAA-ATPase subunit RPT6a - like protein 117 5e-26
>At3g04340 unknown protein
Length = 1320
Score = 839 bits (2168), Expect = 0.0
Identities = 469/952 (49%), Positives = 638/952 (66%), Gaps = 87/952 (9%)
Query: 48 QNNHTVLSQPTNSTLVLSQCC--LTKQLILRALFCFAVGVSTFGTFQIAPAFALPTIPWV 105
+ N VLS NS +TK L+ ALFC A+G+S +FQ APA A+P + V
Sbjct: 87 EGNELVLSSEYNSAKTRESVIQFVTKPLVY-ALFCIAIGLSPIRSFQ-APALAVPFVSDV 144
Query: 106 QFLSKNKENKNQ--------HEYSDCTQKVLDTVPSLLRTIEEVRKGNGDFEDVKRALEF 157
+ K + + + HE+SD T+++L+TV LL+TIE VRK NG+ +V AL+
Sbjct: 145 IWKKKKERVREKEVVLKAVDHEFSDYTRRLLETVSVLLKTIEIVRKENGEVAEVGAALDA 204
Query: 158 VKLKKYEMEKEILERMHPVLMDLKEELRLLQIKEGEISWQMAEVNREHRKLMG------- 210
VK++K +++KEI+ ++ + L++E LL + +I + + ++ KL+
Sbjct: 205 VKVEKEKLQKEIMSGLYRDMRRLRKERDLLMKRADKIVDEALSLKKQSEKLLRKGAREKM 264
Query: 211 -----------------WE-MDMKDNVVNEVEKKVL----------DKRMVELEKKWNEI 242
WE +D D+++ + E L ++ VEL K +N
Sbjct: 265 EKLEESVDIMESEYNKIWERIDEIDDIILKKETTTLSFGVRELIFIERECVELVKSFNRE 324
Query: 243 LVKIDEMEDV-------ISRKETVA-------------LSYGVLEI----CFIQRECENL 278
L + E V +SR E + VLE+ F R+ +
Sbjct: 325 LNQ-KSFESVPESSITKLSRSEIKQELVNAQRKHLEQMILPNVLELEEVDPFFDRDSVDF 383
Query: 279 VERFKQEIKQ-KKIGSSFASSVNKLSKSVIQEDLETVQRKQIEQTILPSIVDVDDLGPFF 337
R K+ +++ KK+ + + K K +E L +K E + + + F
Sbjct: 384 SLRIKKRLEESKKLQRDLQNRIRKRMKKFGEEKLFV--QKTPEGEAVKGFPEAEVKWMFG 441
Query: 338 HQDSV---DFAQHLERSLKDSREQQK-NLEAQIRKDMQYDKEKRSVVYSPEEEERILLDR 393
++ V HL K +E+ K +L+ ++ +D+ + K+ Y + +E++LLDR
Sbjct: 442 EKEVVVPKAIQLHLRHGWKKWQEEAKADLKQKLLEDVDFGKQ-----YIAQRQEQVLLDR 496
Query: 394 DRVVSKTWYNEEKNRWEMDPVAVPHAVSKKLIEHVRIRYDGRAMYIALKGEDKEFYVDIK 453
DRVVSKTWYNE+K+RWEMDP+AVP+AVS+KLI+ RIR+D MY+ALKG+DKEFYVDIK
Sbjct: 497 DRVVSKTWYNEDKSRWEMDPMAVPYAVSRKLIDSARIRHDYAVMYVALKGDDKEFYVDIK 556
Query: 454 EFERLFEYIGGFDVLYRKMLACGIPTAVHLMWIPLSELSVHQRISVILRFPLRFLSGRWN 513
E+E LFE GGFD LY KMLACGIPT+VHLMWIP+SELS+ Q+ ++ R R +
Sbjct: 557 EYEMLFEKFGGFDALYLKMLACGIPTSVHLMWIPMSELSLQQQFLLVTRVVSRVFNALRK 616
Query: 514 SETVLTTTNLIFDNIKEMTDDIMTVIGFPIVEYILPNPVRVKLGMAWPEE--ETMNTPWY 571
++ V + + + I+ + DDIM + FP++E+I+P +R++LGMAWPEE +T+ + WY
Sbjct: 617 TQVVSNAKDTVLEKIRNINDDIMMAVVFPVIEFIIPYQLRLRLGMAWPEEIEQTVGSTWY 676
Query: 572 LNWQLNAEARVQSRRADGDFRWIMLFIARAAISGFVLINVFQFMRRKIPRLLGYGPIQKN 631
L WQ AE +SR + DF+W + F+ R++I GFVL +VF+F++RK+PRLLGYGP +++
Sbjct: 677 LQWQSEAEMNFKSRNTE-DFQWFLWFLIRSSIYGFVLYHVFRFLKRKVPRLLGYGPFRRD 735
Query: 632 PNRRKLEQMAYYFDERKGRMRDRRREGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEE 691
PN RK ++ YF RK R++ +R+ G+DPIKTAF+ MKRVK PPIPL NF+SI+SM+EE
Sbjct: 736 PNVRKFWRVKSYFTYRKRRIKQKRKAGIDPIKTAFDRMKRVKNPPIPLKNFASIESMREE 795
Query: 692 ISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEA 751
I+EVVAFLQNP+AFQEMGARAPRGVLIVGERGTGKTSLA+AIAAEA+VPVV ++AQ+LEA
Sbjct: 796 INEVVAFLQNPKAFQEMGARAPRGVLIVGERGTGKTSLALAIAAEARVPVVNVEAQELEA 855
Query: 752 GMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVEL 811
G+WVGQSA+NVRELFQTARDLAPVI+FVEDFDLFAGVRGKF+HT+ QDHE+FINQLLVEL
Sbjct: 856 GLWVGQSAANVRELFQTARDLAPVIIFVEDFDLFAGVRGKFVHTKQQDHESFINQLLVEL 915
Query: 812 DGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQ 871
DGFEKQDGVVLMATTRN KQIDEAL+RPGRMDR+FHLQ PT+ ERE IL++AA+ETMD +
Sbjct: 916 DGFEKQDGVVLMATTRNHKQIDEALRRPGRMDRVFHLQSPTEMERERILHNAAEETMDRE 975
Query: 872 LVEYVDWKKVAEKTALLRPIELKLVPIALEGSAFRSKVLDTDEIMSYCSFFA 923
LV+ VDW+KV+EKT LLRPIELKLVP+ALE SAFRSK LDTDE++SY S+FA
Sbjct: 976 LVDLVDWRKVSEKTTLLRPIELKLVPMALESSAFRSKFLDTDELLSYVSWFA 1027
>At3g47060 FtsH metalloprotease - like protein
Length = 802
Score = 155 bits (391), Expect = 2e-37
Identities = 86/213 (40%), Positives = 125/213 (58%), Gaps = 8/213 (3%)
Query: 649 GRMRDRRREGVDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEM 708
G++R R+ G D K + I + + +D KEE+ E+V FL+NP + +
Sbjct: 300 GQLRTRKAGGPDGGKVSGGG------ETITFADVAGVDEAKEELEEIVEFLRNPEKYVRL 353
Query: 709 GARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQT 768
GAR PRGVL+VG GTGKT LA A+A EA+VP + A + ++VG AS VR+LF
Sbjct: 354 GARPPRGVLLVGLPGTGKTLLAKAVAGEAEVPFISCSASEF-VELYVGMGASRVRDLFAR 412
Query: 769 ARDLAPVILFVEDFDLFAGVR-GKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTR 827
A+ AP I+F+++ D A R GKF N + E +NQLL E+DGF+ V+++ T
Sbjct: 413 AKKEAPSIIFIDEIDAVAKSRDGKFRMGSNDEREQTLNQLLTEMDGFDSNSAVIVLGATN 472
Query: 828 NLKQIDEALQRPGRMDRIFHLQRPTQAERENIL 860
+D AL+RPGR DR+ ++ P + RE+IL
Sbjct: 473 RADVLDPALRRPGRFDRVVTVETPDKIGRESIL 505
>At5g58870 cell division protein - like
Length = 806
Score = 153 bits (386), Expect = 6e-37
Identities = 79/185 (42%), Positives = 114/185 (60%), Gaps = 2/185 (1%)
Query: 677 IPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
I + + +D KEE+ E+V FL+NP + +GAR PRGVL+VG GTGKT LA A+A E
Sbjct: 326 ITFADVAGVDEAKEELEEIVEFLKNPDRYVRLGARPPRGVLLVGLPGTGKTLLAKAVAGE 385
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVR-GKFIHT 795
+ VP + A + ++VG AS VR+LF A+ AP I+F+++ D A R GKF
Sbjct: 386 SDVPFISCSASEF-VELYVGMGASRVRDLFARAKKEAPSIIFIDEIDAVAKSRDGKFRMV 444
Query: 796 ENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAE 855
N + E +NQLL E+DGF+ V+++ T +D AL+RPGR DR+ ++ P +
Sbjct: 445 SNDEREQTLNQLLTEMDGFDSSSAVIVLGATNRADVLDPALRRPGRFDRVVTVESPDKVG 504
Query: 856 RENIL 860
RE+IL
Sbjct: 505 RESIL 509
>At5g42270 cell division protein FtsH
Length = 704
Score = 146 bits (369), Expect = 6e-35
Identities = 87/218 (39%), Positives = 122/218 (55%), Gaps = 8/218 (3%)
Query: 670 KRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSL 729
+ V + + + + D K E+ EVV FL+NP + +GA+ P+G L+VG GTGKT L
Sbjct: 240 QEVPETGVTFGDVAGADQAKLELQEVVDFLKNPDKYTALGAKIPKGCLLVGPPGTGKTLL 299
Query: 730 AMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVR 789
A A+A EA VP A + ++VG AS VR+LF+ A+ AP I+F+++ D R
Sbjct: 300 ARAVAGEAGVPFFSCAASEF-VELFVGVGASRVRDLFEKAKSKAPCIVFIDEIDAVGRQR 358
Query: 790 GKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQ 849
G + N + E INQLL E+DGF GV+++A T +D AL RPGR DR +
Sbjct: 359 GAGMGGGNDEREQTINQLLTEMDGFSGNSGVIVLAATNRPDVLDSALLRPGRFDRQVTVD 418
Query: 850 RPTQAERENIL--YSAAKETMDDQLVEYVDWKKVAEKT 885
RP A R IL +S K D VD++KVA +T
Sbjct: 419 RPDVAGRVQILKVHSRGKAIGKD-----VDYEKVARRT 451
>At1g06430 cell division protease FtsH, putative
Length = 649
Score = 145 bits (366), Expect = 1e-34
Identities = 80/184 (43%), Positives = 109/184 (58%), Gaps = 1/184 (0%)
Query: 677 IPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
+ ++ + +D K++ EVV FL+ P F +GAR P+GVL+VG GTGKT LA AIA E
Sbjct: 217 VTFDDVAGVDEAKQDFMEVVEFLKKPERFTAVGARIPKGVLLVGPPGTGKTLLAKAIAGE 276
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTE 796
A VP I + M+VG AS VR+LF+ A++ AP I+FV++ D RG I
Sbjct: 277 AGVPFFSISGSEF-VEMFVGVGASRVRDLFKKAKENAPCIVFVDEIDAVGRQRGTGIGGG 335
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
N + E +NQLL E+DGFE GV+++A T +D AL RPGR DR + P R
Sbjct: 336 NDEREQTLNQLLTEMDGFEGNTGVIVVAATNRADILDSALLRPGRFDRQVSVDVPDVKGR 395
Query: 857 ENIL 860
+IL
Sbjct: 396 TDIL 399
>At3g02450 cell division protein FtsH-like protein
Length = 622
Score = 145 bits (365), Expect = 2e-34
Identities = 76/191 (39%), Positives = 116/191 (59%), Gaps = 4/191 (2%)
Query: 670 KRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSL 729
+R K P + ++ +DS K+E+ E+V+ LQ ++++GAR PRGVL+VG GTGKT L
Sbjct: 324 RRSKNPTVGFDDVEGVDSAKDELVEIVSCLQGSINYKKLGARLPRGVLLVGPPGTGKTLL 383
Query: 730 AMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVR 789
A A+A EA VP + A + ++VG+ A+ +R+LF AR +P I+F+++ D G R
Sbjct: 384 ARAVAGEAGVPFFSVSASEF-VELFVGRGAARIRDLFNAARKNSPSIIFIDELDAVGGKR 442
Query: 790 GKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQ 849
G+ + N + + +NQLL E+DGFE V+++A T + +D AL RPGR R +
Sbjct: 443 GR---SFNDERDQTLNQLLTEMDGFESDTKVIVIAATNRPEALDSALCRPGRFSRKVLVA 499
Query: 850 RPTQAERENIL 860
P Q R IL
Sbjct: 500 EPDQEGRRKIL 510
>At2g30950 zinc dependent protease (VAR2)
Length = 695
Score = 144 bits (364), Expect = 2e-34
Identities = 93/254 (36%), Positives = 134/254 (52%), Gaps = 5/254 (1%)
Query: 677 IPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
+ ++ + +D K++ EVV FL+ P F +GA+ P+GVL++G GTGKT LA AIA E
Sbjct: 224 VTFDDVAGVDEAKQDFMEVVEFLKKPERFTAVGAKIPKGVLLIGPPGTGKTLLAKAIAGE 283
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTE 796
A VP I + M+VG AS VR+LF+ A++ AP I+FV++ D RG I
Sbjct: 284 AGVPFFSISGSEF-VEMFVGVGASRVRDLFKKAKENAPCIVFVDEIDAVGRQRGTGIGGG 342
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
N + E +NQLL E+DGFE GV+++A T +D AL RPGR DR + P R
Sbjct: 343 NDEREQTLNQLLTEMDGFEGNTGVIVVAATNRADILDSALLRPGRFDRQVSVDVPDVKGR 402
Query: 857 ENIL--YSAAKETMDDQLVEYVDWKKVAEKTALLRPIELKLVPIALEGSAFRSKVLDTDE 914
+IL ++ K+ +D +E + + A L L L G R+ + +
Sbjct: 403 TDILKVHAGNKKFDNDVSLEIIAMRTPGFSGADL--ANLLNEAAILAGRRARTSISSKEI 460
Query: 915 IMSYCSFFAGMENT 928
S AGME T
Sbjct: 461 DDSIDRIVAGMEGT 474
>At1g50250 putative chloroplast FtsH protease
Length = 716
Score = 144 bits (364), Expect = 2e-34
Identities = 87/218 (39%), Positives = 121/218 (54%), Gaps = 8/218 (3%)
Query: 670 KRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSL 729
+ V + + + + D K E+ EVV FL+NP + +GA+ P+G L+VG GTGKT L
Sbjct: 252 QEVPETGVSFADVAGADQAKLELQEVVDFLKNPDKYTALGAKIPKGCLLVGPPGTGKTLL 311
Query: 730 AMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVR 789
A A+A EA VP A + ++VG AS VR+LF+ A+ AP I+F+++ D R
Sbjct: 312 ARAVAGEAGVPFFSCAASEF-VELFVGVGASRVRDLFEKAKSKAPCIVFIDEIDAVGRQR 370
Query: 790 GKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQ 849
G + N + E INQLL E+DGF GV+++A T +D AL RPGR DR +
Sbjct: 371 GAGMGGGNDEREQTINQLLTEMDGFSGNSGVIVLAATNRPDVLDSALLRPGRFDRQVTVD 430
Query: 850 RPTQAERENIL--YSAAKETMDDQLVEYVDWKKVAEKT 885
RP A R IL +S K D VD+ KVA +T
Sbjct: 431 RPDVAGRVKILQVHSRGKALGKD-----VDFDKVARRT 463
>At2g29080 AAA-type like ATPase
Length = 809
Score = 142 bits (358), Expect = 1e-33
Identities = 83/220 (37%), Positives = 117/220 (52%), Gaps = 1/220 (0%)
Query: 663 KTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGER 722
K + K I + + D K+EI E V FL+NP+ ++++GA+ P+G L+VG
Sbjct: 305 KATITRADKHSKNKIYFKDVAGCDEAKQEIMEFVHFLKNPKKYEDLGAKIPKGALLVGPP 364
Query: 723 GTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDF 782
GTGKT LA A A E+ VP + I M+VG S VR LFQ AR AP I+F+++
Sbjct: 365 GTGKTLLAKATAGESGVPFLSISGSDFME-MFVGVGPSRVRHLFQEARQAAPSIIFIDEI 423
Query: 783 DLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRM 842
D RG+ N + E+ +NQLLVE+DGF GVV++A T +D+AL RPGR
Sbjct: 424 DAIGRARGRGGLGGNDERESTLNQLLVEMDGFGTTAGVVVLAGTNRPDILDKALLRPGRF 483
Query: 843 DRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVA 882
DR + +P R+ I K+ D Y + A
Sbjct: 484 DRQITIDKPDIKGRDQIFKIYLKKIKLDHEPSYYSQRLAA 523
>At1g07510 unknown protein
Length = 813
Score = 140 bits (353), Expect = 4e-33
Identities = 82/210 (39%), Positives = 115/210 (54%), Gaps = 2/210 (0%)
Query: 674 KPPIPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAI 733
K I + + + K+EI E V FLQNP+ ++++GA+ P+G L+VG GTGKT LA A
Sbjct: 321 KNKIYFKDVAGCEEAKQEIMEFVHFLQNPKKYEDLGAKIPKGALLVGPPGTGKTLLAKAT 380
Query: 734 AAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGK-F 792
A E+ VP + I M+VG S VR LFQ AR AP I+F+++ D RG+
Sbjct: 381 AGESAVPFLSISGSDF-MEMFVGVGPSRVRNLFQEARQCAPSIIFIDEIDAIGRARGRGG 439
Query: 793 IHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPT 852
N + E+ +NQLLVE+DGF GVV++A T +D+AL RPGR DR + +P
Sbjct: 440 FSGGNDERESTLNQLLVEMDGFGTTAGVVVLAGTNRPDILDKALLRPGRFDRQITIDKPD 499
Query: 853 QAERENILYSAAKETMDDQLVEYVDWKKVA 882
R+ I K+ D Y + A
Sbjct: 500 IKGRDQIFQIYLKKIKLDHEPSYYSQRLAA 529
>At5g15250 FtsH-like protein Pftf precursor - like
Length = 687
Score = 138 bits (348), Expect = 2e-32
Identities = 89/254 (35%), Positives = 130/254 (51%), Gaps = 5/254 (1%)
Query: 677 IPLNNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAE 736
I + + +D K++ E+V FL+ P F +GA+ P+GVL+ G GTGKT LA AIA E
Sbjct: 220 ITFEDVAGVDEAKQDFEEIVEFLKTPEKFSALGAKIPKGVLLTGPPGTGKTLLAKAIAGE 279
Query: 737 AKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTE 796
A VP + + M+VG AS R+LF A+ +P I+F+++ D +RG I
Sbjct: 280 AGVPFFSLSGSEF-IEMFVGVGASRARDLFNKAKANSPCIVFIDEIDAVGRMRGTGIGGG 338
Query: 797 NQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAER 856
N + E +NQ+L E+DGF GV+++A T + +D AL RPGR DR + P R
Sbjct: 339 NDEREQTLNQILTEMDGFAGNTGVIVIAATNRPEILDSALLRPGRFDRQVSVGLPDIRGR 398
Query: 857 ENIL--YSAAKETMDDQLVEYVDWKKVAEKTALLRPIELKLVPIALEGSAFRSKVLDTDE 914
E IL +S +K+ D + + + A L L L G + K+ T+
Sbjct: 399 EEILKVHSRSKKLDKDVSLSVIAMRTPGFSGADL--ANLMNEAAILAGRRGKDKITLTEI 456
Query: 915 IMSYCSFFAGMENT 928
S AGME T
Sbjct: 457 DDSIDRIVAGMEGT 470
>At2g26140 FtsH like protease
Length = 717
Score = 137 bits (346), Expect = 3e-32
Identities = 81/230 (35%), Positives = 126/230 (54%), Gaps = 13/230 (5%)
Query: 680 NNFSSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKV 739
++ +D K E+ E+V +L++P+ F +G + P+GVL+VG GTGKT LA AIA EA V
Sbjct: 227 SDVKGVDEAKAELEEIVHYLRDPKRFTRLGGKLPKGVLLVGPPGTGKTMLARAIAGEAGV 286
Query: 740 PVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQD 799
P + E M+VG A VR+LF A+ +P I+F+++ D G R + Q
Sbjct: 287 PFFSCSGSEFEE-MFVGVGARRVRDLFSAAKKCSPCIIFIDEIDAIGGSRNP---KDQQY 342
Query: 800 HEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENI 859
+ +NQ+LVELDGF++ +G++++A T + +D+AL RPGR DR + P R I
Sbjct: 343 MKMTLNQMLVELDGFKQNEGIIVVAATNFPESLDKALVRPGRFDRHIVVPNPDVEGRRQI 402
Query: 860 LYSAAKETMDDQLVEYVDWKKVAEKTA------LLRPIELKLVPIALEGS 903
L S + + E VD +A T L + + + A++GS
Sbjct: 403 LESHMSKVLK---AEDVDLMIIARGTPGFSGADLANLVNVAALKAAMDGS 449
>At5g53170 cell division protein FtsH protease-like
Length = 806
Score = 136 bits (343), Expect = 6e-32
Identities = 88/227 (38%), Positives = 122/227 (52%), Gaps = 20/227 (8%)
Query: 686 DSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIK 745
D K+E+ EVV +L+NP F +G + P+G+L+ G GTGKT LA AIA EA VP
Sbjct: 368 DDAKQELEEVVEYLKNPSKFTRLGGKLPKGILLTGAPGTGKTLLAKAIAGEAGVPFFYRA 427
Query: 746 AQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFI-HTENQDHEAFI 804
+ E M+VG A VR LFQ A+ AP I+F+++ D R ++ HT+ H
Sbjct: 428 GSEFEE-MFVGVGARRVRSLFQAAKKKAPCIIFIDEIDAVGSTRKQWEGHTKKTLH---- 482
Query: 805 NQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENI--LYS 862
QLLVE+DGFE+ +G+++MA T +D AL RPGR DR + P RE I LY
Sbjct: 483 -QLLVEMDGFEQNEGIIVMAATNLPDILDPALTRPGRFDRHIVVPSPDVRGREEILELYL 541
Query: 863 AAKETMDDQLVEYVDWKKVAEKTA------LLRPIELKLVPIALEGS 903
K +D VD K +A T L + + + A+EG+
Sbjct: 542 QGKPMSED-----VDVKAIARGTPGFNGADLANLVNIAAIKAAVEGA 583
>At3g16290 putative FtsH-like metalloprotease
Length = 876
Score = 124 bits (312), Expect = 2e-28
Identities = 85/260 (32%), Positives = 133/260 (50%), Gaps = 16/260 (6%)
Query: 626 GPIQKNPNRRKLEQMAYYFDERKGRMRDRRREGVDPIKTAFEHMKRVKKPPIPLNNFSSI 685
G +KNP QMA F + R+R + + E+++R + + + +
Sbjct: 368 GTGEKNPYL----QMAMQFMKSGARVRRASNKRLP------EYLER--GVDVKFTDVAGL 415
Query: 686 DSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIK 745
++ E+ E+V F + ++ G + P G+L+ G G GKT LA A+A EA V I
Sbjct: 416 GKIRLELEEIVKFFTHGEMYRRRGVKIPGGILLCGPPGVGKTLLAKAVAGEAGVNFFSIS 475
Query: 746 AQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFIN 805
A Q ++VG AS VR L+Q AR+ AP ++F+++ D RG + Q+ +A +N
Sbjct: 476 ASQF-VEIYVGVGASRVRALYQEARENAPSVVFIDELDAVGRERGLIKGSGGQERDATLN 534
Query: 806 QLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAK 865
QLLV LDGFE + V+ +A+T +D AL RPGR DR + +P R IL A+
Sbjct: 535 QLLVSLDGFEGRGEVITIASTNRPDILDPALVRPGRFDRKIFIPKPGLIGRMEILQVHAR 594
Query: 866 ETMDDQLVEYVDWKKVAEKT 885
+ + E +D+ VA T
Sbjct: 595 K---KPMAEDLDYMAVASMT 611
>At3g53230 CDC48 - like protein
Length = 815
Score = 124 bits (311), Expect = 3e-28
Identities = 71/215 (33%), Positives = 116/215 (53%), Gaps = 5/215 (2%)
Query: 672 VKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLIVGERGTGKTSLA 730
V+ P + + ++++K E+ E V + +++P F++ G +GVL G G GKT LA
Sbjct: 473 VEVPNVSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLA 532
Query: 731 MAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRG 790
AIA E + + IK +L MW G+S +NVRE+F AR AP +LF ++ D A RG
Sbjct: 533 KAIANECQANFISIKGPELLT-MWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRG 591
Query: 791 KFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQR 850
+ + +NQLL E+DG + V ++ T ID AL RPGR+D++ ++
Sbjct: 592 NSVGDAGGAADRVLNQLLTEMDGMNAKKTVFIIGATNRPDIIDPALLRPGRLDQLIYIPL 651
Query: 851 PTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT 885
P + R I S +++ + + VD + +A+ T
Sbjct: 652 PDEESRYQIFKSCLRKS---PVAKDVDLRALAKYT 683
Score = 99.4 bits (246), Expect = 1e-20
Identities = 78/302 (25%), Positives = 143/302 (46%), Gaps = 25/302 (8%)
Query: 667 EHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF----LQNPRAFQEMGARAPRGVLIVGER 722
E +KR + + + + ++++++++ L++P+ F+ +G + P+G+L+ G
Sbjct: 192 EPIKREDEERLDEVGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPP 251
Query: 723 GTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDF 782
G+GKT +A A+A E I ++ + + G+S SN+R+ F+ A AP I+F+++
Sbjct: 252 GSGKTLIARAVANETGAFFFCINGPEIMSKL-AGESESNLRKAFEEAEKNAPSIIFIDEI 310
Query: 783 DLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRM 842
D A R K T + ++QLL +DG + + V++M T ID AL+R GR
Sbjct: 311 DSIAPKREK---THGEVERRIVSQLLTLMDGLKSRAHVIVMGATNRPNSIDPALRRFGRF 367
Query: 843 DRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKTALLRPIELKLVPIALEG 902
DR + P + R +L T + +L E VD ++V++ T +L +
Sbjct: 368 DREIDIGVPDEIGRLEVL---RIHTKNMKLAEDVDLERVSKDTHGYVGADLAALCTEAAL 424
Query: 903 SAFRSKV----LDTDEIMSYCSFFAGMENTHYLCLDFQFFDAPMAEANQNCQEVKQNVGE 958
R K+ LD +EI + + N H+ A N N +++ V E
Sbjct: 425 QCIREKMDVIDLDDEEIDAEILNSMAVSNDHF----------QTALGNSNPSALRETVVE 474
Query: 959 SP 960
P
Sbjct: 475 VP 476
>At3g56690 calmodulin-binding protein
Length = 1022
Score = 123 bits (308), Expect = 7e-28
Identities = 78/224 (34%), Positives = 121/224 (53%), Gaps = 23/224 (10%)
Query: 655 RREGVDPIKTAFEHMKRVKKP-----------PIPLNNFSSI---DSMKEEISEVVAFLQ 700
R++G + FE + K +P N+ + + +K ++ E V + Q
Sbjct: 685 RKQGEHTLSVGFEDFENAKTKIRPSAMREVILEVPKVNWEDVGGQNEVKNQLMEAVEWPQ 744
Query: 701 NPR-AFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSA 759
+ AF+ +G R P G+L+ G G KT +A A+A+EAK+ + +K +L + WVG+S
Sbjct: 745 KHQDAFKRIGTRPPSGILMFGPPGCSKTLMARAVASEAKLNFLAVKGPELFS-KWVGESE 803
Query: 760 SNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQD---HEAFINQLLVELDGFEK 816
VR LF AR AP I+F ++ D A +RGK EN + ++QLLVELDG +
Sbjct: 804 KAVRSLFAKARANAPSIIFFDEIDSLASIRGK----ENDGVSVSDRVMSQLLVELDGLHQ 859
Query: 817 QDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENIL 860
+ GV ++A T +ID AL RPGR DR+ ++ P + +RE IL
Sbjct: 860 RVGVTVIAATNRPDKIDSALLRPGRFDRLLYVGPPNETDREAIL 903
Score = 91.3 bits (225), Expect = 3e-18
Identities = 52/158 (32%), Positives = 86/158 (53%), Gaps = 4/158 (2%)
Query: 704 AFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVR 763
+ +G R +GVLI G GTGKTSLA A + V + ++ ++G+S +
Sbjct: 409 SLSSLGLRPTKGVLIHGPPGTGKTSLARTFARHSGVNFFSVNGPEI-ISQYLGESEKALD 467
Query: 764 ELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLM 823
E+F++A + P ++F++D D A R + +Q + LL +DG + DGVV++
Sbjct: 468 EVFRSASNATPAVVFIDDLDAIAPARKEGGEELSQ---RMVATLLNLMDGISRTDGVVVI 524
Query: 824 ATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILY 861
A T I+ AL+RPGR+DR + P+ +R +IL+
Sbjct: 525 AATNRPDSIEPALRRPGRLDREIEIGVPSSTQRSDILH 562
>At5g03340 transitional endoplasmic reticulum ATPase
Length = 810
Score = 122 bits (306), Expect = 1e-27
Identities = 66/197 (33%), Positives = 107/197 (53%), Gaps = 2/197 (1%)
Query: 672 VKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLIVGERGTGKTSLA 730
V+ P + + ++++K E+ E V + +++P F++ G +GVL G G GKT LA
Sbjct: 472 VEVPNVSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLA 531
Query: 731 MAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRG 790
AIA E + + +K +L MW G+S +NVRE+F AR AP +LF ++ D A RG
Sbjct: 532 KAIANECQANFISVKGPELLT-MWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRG 590
Query: 791 KFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQR 850
+ +NQLL E+DG + V ++ T ID AL RPGR+D++ ++
Sbjct: 591 NSAGDAGGAADRVLNQLLTEMDGMNAKKTVFIIGATNRPDIIDSALLRPGRLDQLIYIPL 650
Query: 851 PTQAERENILYSAAKET 867
P + R NI + +++
Sbjct: 651 PDEDSRLNIFKACLRKS 667
Score = 99.0 bits (245), Expect = 1e-20
Identities = 70/264 (26%), Positives = 134/264 (50%), Gaps = 25/264 (9%)
Query: 667 EHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF----LQNPRAFQEMGARAPRGVLIVGER 722
E +KR + + + + ++++++++ L++P+ F+ +G + P+G+L+ G
Sbjct: 191 EPVKREDEERLDEVGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPP 250
Query: 723 GTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDF 782
G+GKT +A A+A E I ++ + + G+S SN+R+ F+ A AP I+F+++
Sbjct: 251 GSGKTLIARAVANETGAFFFCINGPEIMSKL-AGESESNLRKAFEEAEKNAPSIIFIDEI 309
Query: 783 DLFAGVRGKFIHTENQDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRM 842
D A R K T + ++QLL +DG + + V++M T ID AL+R GR
Sbjct: 310 DSIAPKREK---TNGEVERRIVSQLLTLMDGLKSRAHVIVMGATNRPNSIDPALRRFGRF 366
Query: 843 DRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKT--------------ALL 888
DR + P + R +L T + +L E VD +++++ T A L
Sbjct: 367 DREIDIGVPDEIGRLEVL---RIHTKNMKLAEDVDLERISKDTHGYVGADLAALCTEAAL 423
Query: 889 RPIELKLVPIALEGSAFRSKVLDT 912
+ I K+ I LE + +++L++
Sbjct: 424 QCIREKMDVIDLEDDSIDAEILNS 447
>At4g23940 cell division protein - like
Length = 946
Score = 122 bits (306), Expect = 1e-27
Identities = 73/200 (36%), Positives = 111/200 (55%), Gaps = 12/200 (6%)
Query: 683 SSIDSMKEEISEVVAFLQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVV 742
+ ID +E+ E+V +L+NP F +MG + P GVL+ G G GKT +A AIA EA VP
Sbjct: 433 AGIDEAVDELQELVKYLKNPDLFDKMGIKPPHGVLLEGPPGCGKTLVAKAIAGEAGVPFY 492
Query: 743 EIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTEN----- 797
++ + + VG ++ +R+LF+ A+ P ++F+++ D A R + I EN
Sbjct: 493 QMAGSEF-VEVLVGVGSARIRDLFKRAKVNKPSVIFIDEIDALA-TRRQGIFKENSDQLY 550
Query: 798 ----QDHEAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQ 853
Q+ E +NQLL+ELDGF+ GV+ + T +D AL RPGR DR ++ P
Sbjct: 551 NAATQERETTLNQLLIELDGFDTGKGVIFLGATNRRDLLDPALLRPGRFDRKIRVRPPNA 610
Query: 854 AERENIL-YSAAKETMDDQL 872
R +IL A+K M D +
Sbjct: 611 KGRLDILKIHASKVKMSDSV 630
>At3g09840 putative transitional endoplasmic reticulum ATPase
Length = 809
Score = 121 bits (303), Expect = 3e-27
Identities = 68/198 (34%), Positives = 110/198 (55%), Gaps = 3/198 (1%)
Query: 672 VKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVLIVGERGTGKTSLA 730
V+ P + N+ ++++K E+ E V + +++P F++ G +GVL G G GKT LA
Sbjct: 472 VEVPNVSWNDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLA 531
Query: 731 MAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRG 790
AIA E + + +K +L MW G+S +NVRE+F AR AP +LF ++ D A RG
Sbjct: 532 KAIANECQANFISVKGPELLT-MWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRG 590
Query: 791 KFIHTENQDH-EAFINQLLVELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQ 849
+ + +NQLL E+DG + V ++ T ID AL RPGR+D++ ++
Sbjct: 591 GGSGGDGGGAADRVLNQLLTEMDGMNAKKTVFIIGATNRPDIIDSALLRPGRLDQLIYIP 650
Query: 850 RPTQAERENILYSAAKET 867
P + R NI +A +++
Sbjct: 651 LPDEDSRLNIFKAALRKS 668
Score = 99.0 bits (245), Expect = 1e-20
Identities = 78/286 (27%), Positives = 141/286 (49%), Gaps = 25/286 (8%)
Query: 691 EISEVVAF-LQNPRAFQEMGARAPRGVLIVGERGTGKTSLAMAIAAEAKVPVVEIKAQQL 749
+I E+V L++P+ F+ +G + P+G+L+ G G+GKT +A A+A E I ++
Sbjct: 218 QIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCINGPEI 277
Query: 750 EAGMWVGQSASNVRELFQTARDLAPVILFVEDFDLFAGVRGKFIHTENQDHEAFINQLLV 809
+ + G+S SN+R+ F+ A AP I+F+++ D A R K T + ++QLL
Sbjct: 278 MSKL-AGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREK---TNGEVERRIVSQLLT 333
Query: 810 ELDGFEKQDGVVLMATTRNLKQIDEALQRPGRMDRIFHLQRPTQAERENILYSAAKETMD 869
+DG + + V++M T ID AL+R GR DR + P + R +L T +
Sbjct: 334 LMDGLKSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIGRLEVL---RIHTKN 390
Query: 870 DQLVEYVDWKKVAEKT--------------ALLRPIELKLVPIALEGSAFRSKVLDTDEI 915
+L E VD +++++ T A L+ I K+ I LE + +++L++ +
Sbjct: 391 MKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMDVIDLEDDSIDAEILNSMAV 450
Query: 916 MSYCSFFAGMENTHYLCLDFQFFDAPMAEANQ--NCQEVKQNVGES 959
+ F + N++ L + P N + VK+ + E+
Sbjct: 451 TNE-HFHTALGNSNPSALRETVVEVPNVSWNDIGGLENVKRELQET 495
>At5g20000 26S proteasome AAA-ATPase subunit RPT6a - like protein
Length = 419
Score = 117 bits (292), Expect = 5e-26
Identities = 79/241 (32%), Positives = 125/241 (51%), Gaps = 11/241 (4%)
Query: 659 VDPIKTAFEHMKRVKKPPIPLNNFSSIDSMKEEISEVVAF-LQNPRAFQEMGARAPRGVL 717
VDP+ MK K P + +D +EI EV+ +++P F+ +G P+GVL
Sbjct: 143 VDPLVNL---MKVEKVPDSTYDMIGGLDQQIKEIKEVIELPIKHPELFESLGIAQPKGVL 199
Query: 718 IVGERGTGKTSLAMAIAAEAKVPVVEIKAQQLEAGMWVGQSASNVRELFQTARDLAPVIL 777
+ G GTGKT LA A+A + + +L ++G+ + VRELF AR+ AP I+
Sbjct: 200 LYGPPGTGKTLLARAVAHHTDCTFIRVSGSEL-VQKYIGEGSRMVRELFVMAREHAPSII 258
Query: 778 FVEDFDLFAGVRGKFIHTENQDHEA--FINQLLVELDGFEKQDGVVLMATTRNLKQIDEA 835
F+++ D R + + N D E + +LL +LDGFE + + ++ T + +D+A
Sbjct: 259 FMDEIDSIGSARMES-GSGNGDSEVQRTMLELLNQLDGFEASNKIKVLMATNRIDILDQA 317
Query: 836 LQRPGRMDRIFHLQRPTQAERENILYSAAKETMDDQLVEYVDWKKVAEKTALLRPIELKL 895
L RPGR+DR P + R +IL +++ L+ +D KK+AEK ELK
Sbjct: 318 LLRPGRIDRKIEFPNPNEESRFDILKIHSRKM---NLMRGIDLKKIAEKMNGASGAELKA 374
Query: 896 V 896
V
Sbjct: 375 V 375
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.332 0.143 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,293,352
Number of Sequences: 26719
Number of extensions: 1097309
Number of successful extensions: 5027
Number of sequences better than 10.0: 187
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 104
Number of HSP's that attempted gapping in prelim test: 4680
Number of HSP's gapped (non-prelim): 339
length of query: 1184
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1074
effective length of database: 8,379,506
effective search space: 8999589444
effective search space used: 8999589444
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC135461.15


