

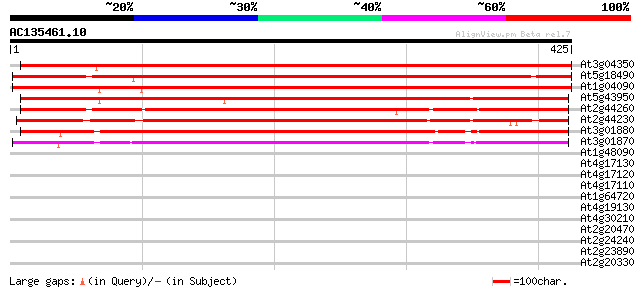
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.10 + phase: 0
(425 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g04350 unknown protein 651 0.0
At5g18490 Unknown protein 612 e-176
At1g04090 unknown protein 555 e-158
At5g43950 unknown protein 530 e-151
At2g44260 unknown protein 422 e-118
At2g44230 putative protein 406 e-113
At3g01880 hypothetical protein 370 e-102
At3g01870 hypothetical protein 364 e-101
At1g48090 unknown protein 40 0.002
At4g17130 hypothetical protein 35 0.060
At4g17120 hypothetical protein 33 0.30
At4g17110 hypothetical protein 33 0.39
At1g64720 membrane related protein CP5, putative 30 3.3
At4g19130 replication A protein-like 29 4.3
At4g30210 NADPH-ferrihemoprotein reductase (ATR2) 29 5.6
At2g20470 putative protein kinase 29 5.6
At2g24240 unknown protein 28 7.4
At2g23890 hypothetical protein 28 7.4
At2g20330 putative WD-40 repeat protein 28 7.4
>At3g04350 unknown protein
Length = 567
Score = 651 bits (1680), Expect = 0.0
Identities = 296/425 (69%), Positives = 354/425 (82%), Gaps = 8/425 (1%)
Query: 9 YFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKKNS---- 64
YFWLPNPP GY+A+G++VT P EP+ EEVRCVR DLTE CETS+++L + S K S
Sbjct: 143 YFWLPNPPVGYRAMGVIVTHEPGEPETEEVRCVREDLTESCETSEMILEVGSSKKSNGSS 202
Query: 65 --FQVWNTQPCDRGMLARGVSVGTFFCGTY-FDSEQVV-DVVCLKNLDSLLHAMPNLNQI 120
F VW+T+PC+RGML++GV+VG+FFC TY SE+ V D+ CLKNLD LHAMPNL+Q+
Sbjct: 203 SPFSVWSTRPCERGMLSQGVAVGSFFCCTYDLSSERTVPDIGCLKNLDPTLHAMPNLDQV 262
Query: 121 HALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGYNDG 180
HA+IEH+GPTVYFHP+E YMPSSV WFFKNGA+LY +G ++G+ I+ G+NLP GG ND
Sbjct: 263 HAVIEHFGPTVYFHPEEAYMPSSVQWFFKNGALLYRSGKSEGQPINSTGSNLPAGGCNDM 322
Query: 181 AFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLM 240
FWIDLP DE+A+SNLKKGN+ES+ELYVHVKPALGG FTDI MW+FCPFNGPATLK+ L
Sbjct: 323 DFWIDLPEDEEAKSNLKKGNLESSELYVHVKPALGGTFTDIVMWIFCPFNGPATLKIGLF 382
Query: 241 NIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIKENKPIVYSS 300
+ M +IGEHVGDWEHFT R+ NF+GELW +FFS+HSGG WV+A D+EF+K+NKP VYSS
Sbjct: 383 TLPMTRIGEHVGDWEHFTFRICNFSGELWQMFFSQHSGGGWVDASDIEFVKDNKPAVYSS 442
Query: 301 RHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWL 360
+HGHAS+PH G YLQGSSKLGIGVRND AKS +I+DSS RY IVAAEYLG G + EPCWL
Sbjct: 443 KHGHASFPHPGMYLQGSSKLGIGVRNDVAKSKYIVDSSQRYVIVAAEYLGKGAVIEPCWL 502
Query: 361 QYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWL 420
QYMREWGPTI YDS SEI KI+++LP+ VRFS+EN+ +LFP L GEEGPTGPKEKDNW
Sbjct: 503 QYMREWGPTIAYDSGSEINKIMNLLPLVVRFSIENIVDLFPIALYGEEGPTGPKEKDNWE 562
Query: 421 GDEYC 425
GDE C
Sbjct: 563 GDEMC 567
>At5g18490 Unknown protein
Length = 553
Score = 612 bits (1579), Expect = e-176
Identities = 281/427 (65%), Positives = 343/427 (79%), Gaps = 11/427 (2%)
Query: 3 SHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKK 62
S D YFWLPNPP GY+AVG++VT +EP+ +EVRCVR DLTE CET + +L +
Sbjct: 134 SSDSDCYFWLPNPPVGYRAVGVIVTDGSEEPEVDEVRCVREDLTESCETGEKVLGV---- 189
Query: 63 NSFQVWNTQPCDRGMLARGVSVGTFFCGTY---FDSEQVVDVVCLKNLDSLLHAMPNLNQ 119
SF VW+T+PC+RG+ +RGV VG+F C T D++ +++ CLKNLD L MPNL+Q
Sbjct: 190 GSFNVWSTKPCERGIWSRGVEVGSFVCSTNDLSSDNKAAMNIACLKNLDPSLQGMPNLDQ 249
Query: 120 IHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGYND 179
+HALI HYGP VYFHP+E YMPSSV WFFKNGA+L+ G ++G+ I+ G+NLP GG ND
Sbjct: 250 VHALIHHYGPMVYFHPEETYMPSSVPWFFKNGALLHRFGKSQGEPINSAGSNLPAGGEND 309
Query: 180 GAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSL 239
G+FWIDLP DE+ RSNLKKGNIES+ELYVHVKPALGG FTD+ MW+FCPFNGPATLK+ L
Sbjct: 310 GSFWIDLPEDEEVRSNLKKGNIESSELYVHVKPALGGIFTDVVMWIFCPFNGPATLKIGL 369
Query: 240 MNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIK-ENKPIVY 298
+ + MN++GEHVGDWEHFT R+SNF G+L +FFS+HSGG WV+ DLEF+K NKP+VY
Sbjct: 370 LTVPMNRLGEHVGDWEHFTFRISNFNGDLTQMFFSQHSGGGWVDVSDLEFVKGSNKPVVY 429
Query: 299 SSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPC 358
SS+HGHAS+PH G YLQG SKLGIGVRND AKS +++DSS RY+IVAAEYLG+G ++EP
Sbjct: 430 SSKHGHASFPHPGMYLQGPSKLGIGVRNDVAKSKYMVDSSQRYRIVAAEYLGEGAVSEPY 489
Query: 359 WLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDN 418
WLQ+MREWGPTIVYDS +EI KIID+LP+ +R S E+ LFP EL GEEGPTGPKEKDN
Sbjct: 490 WLQFMREWGPTIVYDSAAEINKIIDLLPLILRNSFES---LFPIELYGEEGPTGPKEKDN 546
Query: 419 WLGDEYC 425
W GDE C
Sbjct: 547 WEGDEIC 553
>At1g04090 unknown protein
Length = 572
Score = 555 bits (1431), Expect = e-158
Identities = 254/430 (59%), Positives = 327/430 (75%), Gaps = 7/430 (1%)
Query: 3 SHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKK 62
S EC YFWLP PP+GY+++G VVT +P+ EVRCVR DLT++CE ++++T S+
Sbjct: 143 SKSECGYFWLPQPPEGYRSIGFVVTKTSVKPELNEVRCVRADLTDICEPHNVIVTAVSES 202
Query: 63 NSFQ--VWNTQPCDRGMLARGVSVGTFFCGTYFDSEQV---VDVVCLKNLDSLLHAMPNL 117
+W T+P DRGM +GVS GTFFC T + + + + CLKNLD LHAMPN+
Sbjct: 203 LGVPLFIWRTRPSDRGMWGKGVSAGTFFCRTRLVAAREDLGIGIACLKNLDLSLHAMPNV 262
Query: 118 NQIHALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGY 177
+QI ALI+HYGPT+ FHP E Y+PSSVSWFFKNGA+L GN + ID +G+NLP GG
Sbjct: 263 DQIQALIQHYGPTLVFHPGETYLPSSVSWFFKNGAVLCEKGNPIEEPIDENGSNLPQGGS 322
Query: 178 NDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKV 237
ND FWIDLP D+ R +K+GN+ES++LY+H+KPALGG FTD+ W+FCPFNGPATLK+
Sbjct: 323 NDKQFWIDLPCDDQQRDFVKRGNLESSKLYIHIKPALGGTFTDLVFWIFCPFNGPATLKL 382
Query: 238 SLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIK-ENKPI 296
L++I + IG+HV DWEHFTLR+SNF+GEL+S++ S+HSGG+W+ A+DLE I NK +
Sbjct: 383 GLVDISLISIGQHVCDWEHFTLRISNFSGELYSIYLSQHSGGEWIEAYDLEIIPGSNKAV 442
Query: 297 VYSSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYL-GDGVIT 355
VYSS+HGHAS+P AGTYLQGS+ LGIG+RND A+S ++DSS RY+I+AAEYL G+ V+
Sbjct: 443 VYSSKHGHASFPRAGTYLQGSTMLGIGIRNDTARSELLVDSSSRYEIIAAEYLSGNSVLA 502
Query: 356 EPCWLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKE 415
EP WLQYMREWGP +VYDSR EIE++++ P VR S+ + P ELSGEEGPTGPKE
Sbjct: 503 EPPWLQYMREWGPKVVYDSREEIERLVNRFPRTVRVSLATVLRKLPVELSGEEGPTGPKE 562
Query: 416 KDNWLGDEYC 425
K+NW GDE C
Sbjct: 563 KNNWYGDERC 572
>At5g43950 unknown protein
Length = 566
Score = 530 bits (1366), Expect = e-151
Identities = 249/423 (58%), Positives = 323/423 (75%), Gaps = 9/423 (2%)
Query: 9 YFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKKNSFQ-- 66
YFWLP PP+GYK +G +VTT+P +P+ ++VRCVR DLT+ CE +++T S S
Sbjct: 143 YFWLPQPPQGYKPIGYLVTTSPAKPELDQVRCVRADLTDKCEAHKVIITAISDSLSIPMF 202
Query: 67 VWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVV-CLKNLDSLLHAMPNLNQIHALIE 125
+W T+P DRGM +GVS GTFFC T E + + CLKNLDS LHAMPN+ QIHA+I+
Sbjct: 203 IWKTRPSDRGMRGKGVSTGTFFCTTQSPEEDHLSTIACLKNLDSSLHAMPNIEQIHAMIQ 262
Query: 126 HYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAK---GKAIDYHGTNLPGGGYNDGAF 182
HYGP VYFHP+E Y+PSSVSWFFKNGA+L + N+ + ID G+NLP GG ND +
Sbjct: 263 HYGPRVYFHPNEVYLPSSVSWFFKNGALLCSNSNSSVINNEPIDETGSNLPHGGTNDKRY 322
Query: 183 WIDLP-TDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLMN 241
WIDLP D+ R +K+G++ES++LYVHVKPA GG FTD+A W+FCPFNGPATLK+ LM+
Sbjct: 323 WIDLPINDQQRREFIKRGDLESSKLYVHVKPAFGGTFTDLAFWIFCPFNGPATLKLGLMD 382
Query: 242 IEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIK-ENKPIVYSS 300
+ + K G+HV DWEHFT+R+SNF+GEL+S++FS+HSGG+W+ +LEF++ NK +VYSS
Sbjct: 383 LSLAKTGQHVCDWEHFTVRISNFSGELYSIYFSQHSGGEWIKPENLEFVEGSNKAVVYSS 442
Query: 301 RHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWL 360
++GHAS+ +G YLQGS+ LGIG+RND+AKS+ +DSS +Y+IVAAEYL G + EP WL
Sbjct: 443 KNGHASFSKSGMYLQGSALLGIGIRNDSAKSDLFVDSSLKYEIVAAEYL-RGAVVEPPWL 501
Query: 361 QYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWL 420
YMREWGP IVY+SRSEIEK+ + LP +R V+ + P ELSGEEGPTGPKEK+NW
Sbjct: 502 GYMREWGPKIVYNSRSEIEKLNERLPWRLRSWVDAVLRKIPVELSGEEGPTGPKEKNNWF 561
Query: 421 GDE 423
GDE
Sbjct: 562 GDE 564
>At2g44260 unknown protein
Length = 553
Score = 422 bits (1084), Expect = e-118
Identities = 200/418 (47%), Positives = 276/418 (65%), Gaps = 12/418 (2%)
Query: 9 YFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKKNSFQVW 68
YFW P PP GY+AVG++VT +P +++RC+R+DLTE CE + N +
Sbjct: 144 YFWQPVPPDGYQAVGLIVTNYSQKPPLDKLRCIRSDLTEQCEADTWIWGT----NGVNIS 199
Query: 69 NTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQIHALIEHYG 128
N +P RG A GV VGTF T S + CLKN MPN +QI L + +
Sbjct: 200 NLKPTTRGTQATGVYVGTFTWQTQNSSPPSLS--CLKNTKLDFSTMPNGSQIEELFQTFS 257
Query: 129 PTVYFHPDEKYMPSSVSWFFKNGAILYTAGN-AKGKAIDYHGTNLPGGGYNDGAFWIDLP 187
P +YFHPDE+Y+PSSV+W+F NGA+LY G +K I+ +G+NLP GG NDG++W+DLP
Sbjct: 258 PCIYFHPDEEYLPSSVTWYFNNGALLYKKGEESKPIPIESNGSNLPQGGSNDGSYWLDLP 317
Query: 188 TDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLMNIEMNKI 247
D++ + +KKG+++S ++Y+H+KP LG FTDI++W+F PFNGPA KV +N+ + +I
Sbjct: 318 IDKNGKERVKKGDLQSTKVYLHIKPMLGATFTDISIWIFYPFNGPAKAKVKFVNLPLGRI 377
Query: 248 GEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIK--ENKPIVYSSRHGHA 305
GEH+GDWEH TLR+SNFTGELW VF S+HSGG W++A DLEF NK + Y+S HGHA
Sbjct: 378 GEHIGDWEHTTLRISNFTGELWRVFLSQHSGGIWIDACDLEFQDGGNNKFVAYASLHGHA 437
Query: 306 SYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMRE 365
YP G LQG G+G+RND K +LD+ Y+++AAEY G GV+ EP W++Y R+
Sbjct: 438 MYPKPGLVLQGDD--GVGIRNDTGKGKKVLDTGLGYEVIAAEYDGGGVV-EPPWVKYFRK 494
Query: 366 WGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 423
WGP I Y+ E++ + +LP ++ + + P E+ GE+GPTGPK K NW GDE
Sbjct: 495 WGPKIDYNVDDEVKSVERILPGLLKKAFVKFVKKIPDEVYGEDGPTGPKLKSNWAGDE 552
>At2g44230 putative protein
Length = 542
Score = 406 bits (1044), Expect = e-113
Identities = 203/425 (47%), Positives = 282/425 (65%), Gaps = 23/425 (5%)
Query: 6 ECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKKNSF 65
E YFW P PP GY AVG++VTT+ ++P +++RCVR+DLT+ E L+ + N F
Sbjct: 133 ETGYFWQPVPPDGYNAVGLIVTTSDEKPPLDKIRCVRSDLTDQSEPDALIW----ETNGF 188
Query: 66 QVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQIHALIE 125
V +++P +RG A GVSVGTFF + + + CLKN + MP+ QI AL +
Sbjct: 189 SVSSSKPVNRGTQASGVSVGTFFSNSPNPA-----LPCLKNNNFDFSCMPSKPQIDALFQ 243
Query: 126 HYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA-IDYHGTNLPGGGYNDGAFWI 184
Y P +YFH DEKY+PSSV+WFF NGA+LY G+ ++ +G NLP G +NDG +W+
Sbjct: 244 TYAPWIYFHKDEKYLPSSVNWFFSNGALLYKKGDESNPVPVEPNGLNLPQGEFNDGLYWL 303
Query: 185 DLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLKVSLMNIEM 244
DLP DAR ++ G+++S E+Y+H+KP GG FTDIA+W+F PFNGP+ K+ +I +
Sbjct: 304 DLPVASDARKRVQCGDLQSMEVYLHIKPVFGGTFTDIAVWMFYPFNGPSRAKLKAASIPL 363
Query: 245 NKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFI-KENKPIVYSSRHG 303
+IGEH+GDWEHFTLR+SNF+G+L ++ S+HSGG W +A ++EF NKP+ Y+S +G
Sbjct: 364 GRIGEHIGDWEHFTLRISNFSGKLHRMYLSQHSGGSWADASEIEFQGGGNKPVAYASLNG 423
Query: 304 HASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYM 363
HA Y G LQG K +G+RND KS ++D++ R+++VAAEY+ G + EP WL YM
Sbjct: 424 HAMYSKPGLVLQG--KDNVGIRNDTGKSEKVIDTAVRFRVVAAEYM-RGELEEPAWLNYM 480
Query: 364 REWGPTIVYDSRSEI---EKII--DMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDN 418
R WGP I Y +EI EKI+ + L R +++ L P E+ GEEGPTGPK K N
Sbjct: 481 RHWGPKIDYGHENEIRGVEKIMVGESLKTTFRSAIKGL----PNEVFGEEGPTGPKLKRN 536
Query: 419 WLGDE 423
WLGDE
Sbjct: 537 WLGDE 541
>At3g01880 hypothetical protein
Length = 592
Score = 370 bits (949), Expect = e-102
Identities = 193/420 (45%), Positives = 257/420 (60%), Gaps = 16/420 (3%)
Query: 9 YFWLPNPPKGYKAVGIVVTTNPDEPKAEE--VRCVRTDLTEVCETSDLLLTIKSKKNSFQ 66
YFW P P GY AVG+ VTT+P +P + + CVR+DLTE E + IK S
Sbjct: 181 YFWQPLCPNGYHAVGLYVTTSPMKPSLGQNSISCVRSDLTEQSEADTWVWRIKDMTIS-- 238
Query: 67 VWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQIHALIEH 126
+ +P RG+ A GV GTF C + CLKN L +MP+ NQ L +
Sbjct: 239 --SLRPATRGVEATGVFTGTFSCKQLNFLPHPPPLFCLKNTKFDLSSMPSENQTRVLFKT 296
Query: 127 YGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA-IDYHGTNLPGGGYNDGAFWID 185
Y P +Y HP E ++PSSV+W F NGA+L+ GN I +G+NLP GG ND FW+D
Sbjct: 297 YSPWIYLHPKEDFLPSSVNWVFANGALLHKKGNESIPVPIHPNGSNLPQGGCNDDLFWLD 356
Query: 186 LPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLK-VSLMNIEM 244
D+ AR +K+G++ES ++Y+H+KP G FTDI +W+F P+NG A LK + + ++ +
Sbjct: 357 YLVDKKAREKVKRGDLESTKVYLHIKPMFGATFTDIVVWLFFPYNGNAHLKFLFIKSLSL 416
Query: 245 NKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIK-ENKPIVYSSRHG 303
IGEHVGDWEH TLR+SNF GELW V+FSEHSGG V+A DLEF++ NKP+VYSS HG
Sbjct: 417 GNIGEHVGDWEHVTLRISNFNGELWRVYFSEHSGGTLVDACDLEFMQGGNKPVVYSSLHG 476
Query: 304 HASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYM 363
HA + G LQG K GI RND A+S+ D+ Y+++A G GV+ EP WL Y
Sbjct: 477 HAMFSKPGVVLQGGGKSGI--RNDMARSDKCFDAGIGYEVIA----GPGVV-EPPWLNYF 529
Query: 364 REWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 423
R+WGP + Y + + +LPIF+R + L P E+ G++GPTGPK K W GDE
Sbjct: 530 RKWGPRVHYRIDIFLNSVAKILPIFLRKGLRKLINKIPLEMRGQDGPTGPKVKVTWTGDE 589
>At3g01870 hypothetical protein
Length = 583
Score = 364 bits (935), Expect = e-101
Identities = 189/425 (44%), Positives = 254/425 (59%), Gaps = 16/425 (3%)
Query: 3 SHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKA--EEVRCVRTDLTEVCETSDLLLTIKS 60
+ + +FW P P GY+AVG+ VTT+P +P E + CVR+DLTE ET + +
Sbjct: 171 NQEGAAFFWQPLCPNGYQAVGLYVTTSPIKPSLSQESISCVRSDLTEQSETDTWVWGTEE 230
Query: 61 KKNSFQVWNTQPCDRGMLARGVSVGTFFCGTYFDSEQVVDVVCLKNLDSLLHAMPNLNQI 120
S + +P +RG A GV GTF C + + CLKN L +MP+ NQ
Sbjct: 231 MTLS----SLRPANRGTEATGVHTGTFSCQP-LNIPPPPPLFCLKNTKFDLSSMPSHNQT 285
Query: 121 HALIEHYGPTVYFHPDEKYMPSSVSWFFKNGAILYTAGNAKGKA-IDYHGTNLPGGGYND 179
L + Y P +Y HPDE ++ SSV WFF NGA+L+ GN + G+NLP GG +D
Sbjct: 286 TVLFQSYSPWIYLHPDEDFISSSVDWFFSNGALLFQKGNESNPVPVQPDGSNLPQGGSDD 345
Query: 180 GAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFCPFNGPATLK-VS 238
G FW+D P D++A+ +K+G++ ++Y+H+KP GG FTDI +W+F PFNG A LK +
Sbjct: 346 GLFWLDYPADKNAKEWVKRGDLGHTKVYLHIKPMFGGTFTDIVVWIFYPFNGNARLKFLF 405
Query: 239 LMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDLEFIKENKPIVY 298
++ + IGEH+GDWEH TLR+SNF GELW +FSEHSGG V A DLEF NK + Y
Sbjct: 406 FKSLSLGDIGEHIGDWEHVTLRISNFNGELWRAYFSEHSGGTLVEACDLEFQGGNKLVSY 465
Query: 299 SSRHGHASYPHAGTYLQGSSKLGIGVRNDAAKSNFILDSSFRYKIVAAEYLGDGVITEPC 358
SS HGHA + G LQG G G+RND A+SN D+ Y++VA G G I EP
Sbjct: 466 SSLHGHAMFSKPGLVLQGDD--GNGIRNDMARSNKFFDAGVAYELVA----GPG-IQEPP 518
Query: 359 WLQYMREWGPTIVYDSRSEIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDN 418
WL Y R+WGP + +D + +E I LP +R NL P E+ E+GPTGPK K +
Sbjct: 519 WLNYFRKWGPLVPHDIQKNLEGIAKSLPGLLRKKFRNLINKIPREVLEEDGPTGPKVKRS 578
Query: 419 WLGDE 423
W GD+
Sbjct: 579 WTGDD 583
>At1g48090 unknown protein
Length = 4099
Score = 40.0 bits (92), Expect = 0.002
Identities = 19/67 (28%), Positives = 30/67 (44%)
Query: 8 VYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSDLLLTIKSKKNSFQV 67
V+ W P P GY ++G V++ + P + C R DL + +T S S Q+
Sbjct: 2342 VFCWFPVAPPGYVSLGCVLSKFDEAPHVDSFCCPRIDLVNQANIYEASVTRSSSSKSSQL 2401
Query: 68 WNTQPCD 74
W+ D
Sbjct: 2402 WSIWKVD 2408
Score = 28.9 bits (63), Expect = 5.6
Identities = 13/37 (35%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query: 5 DECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCV 41
+EC W+P P GY A+G V ++P V C+
Sbjct: 2145 NECS-LWMPVAPVGYTAMGCVANIGSEQPPDHIVYCL 2180
>At4g17130 hypothetical protein
Length = 747
Score = 35.4 bits (80), Expect = 0.060
Identities = 15/52 (28%), Positives = 19/52 (35%)
Query: 2 DSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCETSD 53
D D W P PKGY A+ VV++ P C+ C D
Sbjct: 608 DERDSSCSIWFPEAPKGYVALSCVVSSGSTPPSLASTFCILASSVSPCSLRD 659
Score = 32.3 bits (72), Expect = 0.51
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query: 5 DECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEV 48
D+ FW P+PP G+ ++G +T D+P + V V T+L V
Sbjct: 539 DQIYAFWRPHPPPGFASLGDYLTP-LDKPPTKGVLVVNTNLMRV 581
>At4g17120 hypothetical protein
Length = 1661
Score = 33.1 bits (74), Expect = 0.30
Identities = 18/73 (24%), Positives = 34/73 (45%), Gaps = 13/73 (17%)
Query: 6 ECVYFWLPNPPKGYKAVGIVVTTNPDEP-KAEEVRCVRTDLT--------EVCETSDLLL 56
E + FW+P P G+ ++G V +P ++RC R+D+ + +TSD
Sbjct: 191 ESISFWMPQAPPGFVSLGCVACKGSPKPYDFTKLRCARSDMVAGDHFADESLWDTSD--- 247
Query: 57 TIKSKKNSFQVWN 69
+ + F +W+
Sbjct: 248 -VWQRVEPFSIWS 259
>At4g17110 hypothetical protein
Length = 335
Score = 32.7 bits (73), Expect = 0.39
Identities = 16/50 (32%), Positives = 23/50 (46%)
Query: 1 MDSHDECVYFWLPNPPKGYKAVGIVVTTNPDEPKAEEVRCVRTDLTEVCE 50
+D + V W P P+G+ + G V EP+ V C+ T L E E
Sbjct: 222 LDDYISPVSIWHPRAPEGFVSPGCVAVAGFIEPELNTVYCMPTSLAEQTE 271
>At1g64720 membrane related protein CP5, putative
Length = 385
Score = 29.6 bits (65), Expect = 3.3
Identities = 15/45 (33%), Positives = 20/45 (44%), Gaps = 4/45 (8%)
Query: 330 KSNFILDSSFRYKIVAAEYLGDGVITEPCWLQYMREWGPTIVYDS 374
K+ F+ D FR+ E G PCW+Q M PT Y +
Sbjct: 85 KTGFVTDDDFRHLWKLVEVKDGG----PCWIQMMDRSTPTFSYQA 125
>At4g19130 replication A protein-like
Length = 717
Score = 29.3 bits (64), Expect = 4.3
Identities = 24/80 (30%), Positives = 36/80 (45%), Gaps = 4/80 (5%)
Query: 166 DYHGTNLPGGGYND--GA-FWIDLPTDEDARSNLKK-GNIESAELYVHVKPALGGAFTDI 221
D G +PGG +ND GA I T E S +++ NIE + P +G I
Sbjct: 111 DIIGHPVPGGKHNDQRGADSGIKFNTTEQQGSGIRQVNNIEPGRSNAAISPQVGAPPKII 170
Query: 222 AMWVFCPFNGPATLKVSLMN 241
+ P++G T+K + N
Sbjct: 171 PVNALSPYSGRWTIKARVTN 190
>At4g30210 NADPH-ferrihemoprotein reductase (ATR2)
Length = 711
Score = 28.9 bits (63), Expect = 5.6
Identities = 17/53 (32%), Positives = 23/53 (43%), Gaps = 1/53 (1%)
Query: 117 LNQIHALIEHYGPTVYFHPDEKYMPSSVSW-FFKNGAILYTAGNAKGKAIDYH 168
L ++ GPT + + +S W GA LY G+AKG A D H
Sbjct: 623 LAELSVAFSREGPTKEYVQHKMMDKASDIWNMISQGAYLYVCGDAKGMARDVH 675
>At2g20470 putative protein kinase
Length = 596
Score = 28.9 bits (63), Expect = 5.6
Identities = 14/35 (40%), Positives = 21/35 (60%)
Query: 174 GGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYV 208
GGG N A ++ TDE+A SN K + +A+ Y+
Sbjct: 28 GGGSNGSADEHNVETDEEAVSNTTKQKVAAAKQYI 62
>At2g24240 unknown protein
Length = 441
Score = 28.5 bits (62), Expect = 7.4
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 3/29 (10%)
Query: 48 VCETS---DLLLTIKSKKNSFQVWNTQPC 73
+C+ S D L + S++N F VW T PC
Sbjct: 410 ICDFSIGGDRLFALHSEENVFDVWETPPC 438
>At2g23890 hypothetical protein
Length = 546
Score = 28.5 bits (62), Expect = 7.4
Identities = 12/30 (40%), Positives = 14/30 (46%)
Query: 278 GGKWVNAFDLEFIKENKPIVYSSRHGHASY 307
G W FD+ K NKP Y+S H Y
Sbjct: 309 GDSWRELFDVVIAKANKPEFYTSEHPFRCY 338
>At2g20330 putative WD-40 repeat protein
Length = 648
Score = 28.5 bits (62), Expect = 7.4
Identities = 17/50 (34%), Positives = 23/50 (46%), Gaps = 2/50 (4%)
Query: 135 PDEKYMPSSVSWFFKNGAILYTAGNAKGKAIDYHGTNLPGGGYNDGAFWI 184
P E + SVSW +G L G+A+ K D G L G + G +I
Sbjct: 221 PSEGHQVRSVSWSPTSGQFLCVTGSAQAKIFDRDGLTL--GEFMKGDMYI 268
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,848,376
Number of Sequences: 26719
Number of extensions: 489756
Number of successful extensions: 968
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 900
Number of HSP's gapped (non-prelim): 25
length of query: 425
length of database: 11,318,596
effective HSP length: 102
effective length of query: 323
effective length of database: 8,593,258
effective search space: 2775622334
effective search space used: 2775622334
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135461.10


