

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135415.3 + phase: 0 /pseudo
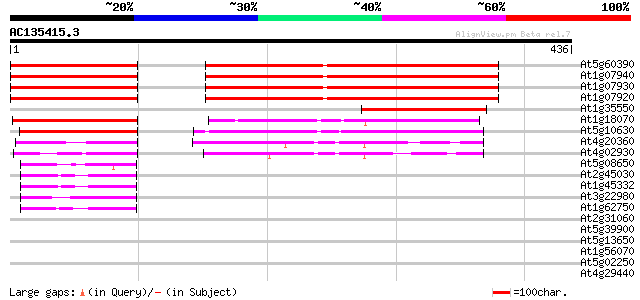
(436 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g60390 translation elongation factor eEF-1 alpha chain (gene A4) 301 5e-82
At1g07940 elongation factor 1-alpha 301 5e-82
At1g07930 elongation factor 1-alpha 301 5e-82
At1g07920 elongation factor 1-alpha 301 5e-82
At1g35550 elongation factor, putative 131 6e-31
At1g18070 unknown protein 94 2e-19
At5g10630 putative protein 88 8e-18
At4g20360 translation elongation factor EF-Tu precursor, chlorop... 60 2e-09
At4g02930 mitochondrial elongation factor Tu 59 5e-09
At5g08650 GTP-binding protein LepA homolog 54 1e-07
At2g45030 putative mitochondrial translation elongation factor G 46 4e-05
At1g45332 mitochondrial elongation factor, putative 46 4e-05
At3g22980 eukaryotic translation elongation factor 2, putative 45 6e-05
At1g62750 elongation factor G, putative 43 3e-04
At2g31060 putative GTP-binding protein 40 0.003
At5g39900 GTP-binding protein-like 40 0.003
At5g13650 GTP-binding protein typA (tyrosine phosphorylated prot... 38 0.013
At1g56070 elongation factor like protein 35 0.062
At5g02250 ribonuclease II-like protein 33 0.24
At4g29440 putative protein 33 0.31
>At5g60390 translation elongation factor eEF-1 alpha chain (gene A4)
Length = 449
Score = 301 bits (771), Expect = 5e-82
Identities = 152/228 (66%), Positives = 179/228 (77%), Gaps = 2/228 (0%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG++KPGMV+TFAPT L +
Sbjct: 216 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RGYVASNS+D+PA AA FTS+VI
Sbjct: 276 EVKSVEMHHESLLEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKGAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKF+++ TK DR +G E+EK+PK LKNGDAG+VKM
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI SV K TK
Sbjct: 394 PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKSVDKKDPTGAKVTK 441
Score = 155 bits (393), Expect = 3e-38
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>At1g07940 elongation factor 1-alpha
Length = 449
Score = 301 bits (771), Expect = 5e-82
Identities = 152/228 (66%), Positives = 179/228 (77%), Gaps = 2/228 (0%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG++KPGMV+TFAPT L +
Sbjct: 216 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RGYVASNS+D+PA AA FTS+VI
Sbjct: 276 EVKSVEMHHESLLEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKGAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKF+++ TK DR +G E+EK+PK LKNGDAG+VKM
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI SV K TK
Sbjct: 394 PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKSVDKKDPTGAKVTK 441
Score = 155 bits (393), Expect = 3e-38
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>At1g07930 elongation factor 1-alpha
Length = 449
Score = 301 bits (771), Expect = 5e-82
Identities = 152/228 (66%), Positives = 179/228 (77%), Gaps = 2/228 (0%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG++KPGMV+TFAPT L +
Sbjct: 216 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RGYVASNS+D+PA AA FTS+VI
Sbjct: 276 EVKSVEMHHESLLEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKGAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKF+++ TK DR +G E+EK+PK LKNGDAG+VKM
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI SV K TK
Sbjct: 394 PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKSVDKKDPTGAKVTK 441
Score = 155 bits (393), Expect = 3e-38
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>At1g07920 elongation factor 1-alpha
Length = 449
Score = 301 bits (771), Expect = 5e-82
Identities = 152/228 (66%), Positives = 179/228 (77%), Gaps = 2/228 (0%)
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG++KPGMV+TFAPT L +
Sbjct: 216 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTT 275
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RGYVASNS+D+PA AA FTS+VI
Sbjct: 276 EVKSVEMHHESLLEALPGDNVGFNVKNVAVKD--LKRGYVASNSKDDPAKGAANFTSQVI 333
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKF+++ TK DR +G E+EK+PK LKNGDAG+VKM
Sbjct: 334 IMNHPGQIGNGYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATK 380
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI SV K TK
Sbjct: 394 PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKSVDKKDPTGAKVTK 441
Score = 155 bits (393), Expect = 3e-38
Identities = 77/99 (77%), Positives = 83/99 (83%)
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 1 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 60
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 61 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 99
>At1g35550 elongation factor, putative
Length = 104
Score = 131 bits (330), Expect = 6e-31
Identities = 62/97 (63%), Positives = 74/97 (75%)
Query: 274 INHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIP 333
+NH I GYTP+LDCHTSH+AVKF+++ TK D TG E+EK+PK LKN +A I+ M P
Sbjct: 1 MNHLGQIKNGYTPVLDCHTSHIAVKFSEILTKIDWRTGHEIEKEPKFLKNSEAAIINMTP 60
Query: 334 MKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKK 370
KPMVVE + YP LGRFA+RDMRQTV VGVI SV K
Sbjct: 61 TKPMVVEAYSAYPPLGRFAIRDMRQTVGVGVIKSVVK 97
>At1g18070 unknown protein
Length = 532
Score = 93.6 bits (231), Expect = 2e-19
Identities = 59/213 (27%), Positives = 112/213 (51%), Gaps = 7/213 (3%)
Query: 155 QAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQSGV 214
+ +D I P R P R+P+ + K +GTV +G+VE+G ++ G L P K Q V
Sbjct: 315 EVLDSIEIPPRDPNGPFRMPI--IDKFKDMGTVVMGKVESGSIREGDSLVVMPNKEQVKV 372
Query: 215 KSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVIII 274
+I ++ + A P + + R+T + D++++ G+V S+ + P +F +++ I+
Sbjct: 373 VAIYCDEDKVKRAGPGENLRVRITG--IEDEDILSGFVLSSIVN-PVPAVTEFVAQLQIL 429
Query: 275 N--HPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
++ T GY IL H + +L ++ D +T ++KK +KNG A + ++
Sbjct: 430 ELLDNAIFTAGYKAILHIHAVVEECEIIELKSQIDLKTRKPMKKKVLFVKNGAAVVCRIQ 489
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVI 365
+ +E +++P LGRF +R +T+AVG +
Sbjct: 490 VTNSICIEKFSDFPQLGRFTLRTEGKTIAVGKV 522
Score = 80.9 bits (198), Expect = 1e-15
Identities = 36/97 (37%), Positives = 60/97 (61%)
Query: 3 NSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDK 62
N K ++N+VFIGHV++GKST G +L+ G + I+ KEA + S+ A+++D
Sbjct: 97 NKKRHLNVVFIGHVDAGKSTIGGQILFLSGQVDDRQIQKYEKEAKDKSRESWYMAYIMDT 156
Query: 63 LKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
+ ER +G T+++ FET T++DAPGH+ ++
Sbjct: 157 NEEERLKGKTVEVGRAHFETESTRFTILDAPGHKSYV 193
>At5g10630 putative protein
Length = 804
Score = 88.2 bits (217), Expect = 8e-18
Identities = 40/92 (43%), Positives = 60/92 (64%)
Query: 8 INIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAER 67
+N+ +GHV+SGKST +G LL+ LG I + + KEA SF YAW LD+ ER
Sbjct: 377 LNLAIVGHVDSGKSTLSGRLLHLLGRISQKQMHKYEKEAKLQGKGSFAYAWALDESAEER 436
Query: 68 ERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
ERGIT+ +++ F + ++ L+D+PGH+DF+
Sbjct: 437 ERGITMTVAVAYFNSKRHHVVLLDSPGHKDFV 468
Score = 74.7 bits (182), Expect = 9e-14
Identities = 57/226 (25%), Positives = 99/226 (43%), Gaps = 6/226 (2%)
Query: 144 SWCEANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPV-GRVETGVLKPGMV 202
SW + LL A+D + P R KPL +P+ + G V G++E G ++PG
Sbjct: 580 SWYQGPCLL--DAVDSVKSPDRDVSKPLLMPICDAVRSTSQGQVSACGKLEAGAVRPGSK 637
Query: 203 LTFAPTKLQSGVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAM 262
+ P+ Q ++S+++ + A D V L + +M G V + D P
Sbjct: 638 VMVMPSGDQGTIRSLERDSQACTIARAGDNVALALQG--IDANQVMAGDVLCHP-DFPVS 694
Query: 263 EAAKFTSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLK 322
A V+++ + I G H + A KL D +TG +K P+ L
Sbjct: 695 VATHLELMVLVLEGATPILLGSQLEFHVHHAKEAATVVKLVAMLDPKTGQPTKKSPRCLT 754
Query: 323 NGDAGIVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSV 368
+ ++++ P+ VE +E +LGR +R +TVA+G + +
Sbjct: 755 AKQSAMLEVSLQNPVCVETFSESRALGRVFLRSSGRTVAMGKVTRI 800
>At4g20360 translation elongation factor EF-Tu precursor,
chloroplast
Length = 476
Score = 60.5 bits (145), Expect = 2e-09
Identities = 61/234 (26%), Positives = 100/234 (42%), Gaps = 27/234 (11%)
Query: 143 HSWCEANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMV 202
+ W + L+ D I P+R P L ++ V+ I G GTV GRVE G +K G
Sbjct: 260 NKWVDKIYELMDAVDDYIPIPQRQTELPFLLAVEDVFSITGRGTVATGRVERGTVKVGET 319
Query: 203 LTFAPTKLQSG--VKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEP 260
+ + V ++ FQ+ ++EAL D VG L + + ++ RG V ++
Sbjct: 320 VDLVGLRETRSYTVTGVEMFQKILDEALAGDNVGLLL--RGIQKADIQRGMVL--AKPGS 375
Query: 261 AMEAAKFTSRVIII-----NHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELE 315
KF + + ++ S GY P T+ V K K+ D E+
Sbjct: 376 ITPHTKFEAIIYVLKKEEGGRHSPFFAGYRPQFYMRTTDVTGKVTKIMNDKDEES----- 430
Query: 316 KKPKSLKNGDAGIVKMIPMKPMVVE-GINEYPSLGRFAVRDMRQTVAVGVIMSV 368
K + GD + + + P+ E G+ RFA+R+ +TV GVI ++
Sbjct: 431 ---KMVMPGDRVKIVVELIVPVACEQGM-------RFAIREGGKTVGAGVIGTI 474
Score = 50.8 bits (120), Expect = 1e-06
Identities = 30/95 (31%), Positives = 44/95 (45%), Gaps = 15/95 (15%)
Query: 5 KPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLK 64
KP++NI IGHV+ GK+T L L I S + + +D
Sbjct: 77 KPHVNIGTIGHVDHGKTTLTAALTMALASIGSSVAKKYDE---------------IDAAP 121
Query: 65 AERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
ER RGITI+ + ++ET +D PGH D++
Sbjct: 122 EERARGITINTATVEYETENRHYAHVDCPGHADYV 156
>At4g02930 mitochondrial elongation factor Tu
Length = 454
Score = 58.9 bits (141), Expect = 5e-09
Identities = 59/229 (25%), Positives = 99/229 (42%), Gaps = 34/229 (14%)
Query: 151 LLLQQAIDQ-ISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPG-----MVLT 204
L L A+D+ I +P R KP +P++ V+ I G GTV GR+E GV+K G + L
Sbjct: 247 LKLMDAVDEYIPDPVRVLDKPFLMPIEDVFSIQGRGTVATGRIEQGVIKVGEEVEILGLR 306
Query: 205 FAPTKLQSGVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEA 264
L+S V ++ F++ ++ D VG L + L +++ RG V ++
Sbjct: 307 EGGVPLKSTVTGVEMFKKILDNGQAGDNVGLLL--RGLKREDIQRGMVI--AKPGSCKTY 362
Query: 265 AKFTSRVIII-----NHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPK 319
KF + + ++ + Y P T+ + K VEL + K
Sbjct: 363 KKFEAEIYVLTKDEGGRHTAFFSNYRPQFYLRTADITGK-------------VELPENVK 409
Query: 320 SLKNGDAGIVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSV 368
+ GD + P+ +E + RFA+R+ +TV GV+ V
Sbjct: 410 MVMPGDNVTAVFELIMPVPLE------TGQRFALREGGRTVGAGVVSKV 452
Score = 51.2 bits (121), Expect = 1e-06
Identities = 31/96 (32%), Positives = 50/96 (51%), Gaps = 15/96 (15%)
Query: 4 SKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKL 63
+KP++N+ IGHV+ GK+T +AI + E + K +F +DK
Sbjct: 64 NKPHVNVGTIGHVDHGKTTLT------------AAITKVLAEEGKAKAIAFDE---IDKA 108
Query: 64 KAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
E++RGITI + ++ET K +D PGH D++
Sbjct: 109 PEEKKRGITIATAHVEYETAKRHYAHVDCPGHADYV 144
>At5g08650 GTP-binding protein LepA homolog
Length = 675
Score = 54.3 bits (129), Expect = 1e-07
Identities = 35/94 (37%), Positives = 45/94 (47%), Gaps = 20/94 (21%)
Query: 9 NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERE 68
N I H++ GKST A LL G +Q +MK + LD + ERE
Sbjct: 82 NFSIIAHIDHGKSTLADKLLQVTGTVQNR----------DMKEQ------FLDNMDLERE 125
Query: 69 RGITIDISMCK----FETTKYCCTLIDAPGHRDF 98
RGITI + + +E T +C LID PGH DF
Sbjct: 126 RGITIKLQAARMRYVYEDTPFCLNLIDTPGHVDF 159
>At2g45030 putative mitochondrial translation elongation factor G
Length = 754
Score = 46.2 bits (108), Expect = 4e-05
Identities = 30/90 (33%), Positives = 42/90 (46%), Gaps = 10/90 (11%)
Query: 9 NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERE 68
NI H++SGK+T +L+ G I + E G++ KM D + ERE
Sbjct: 67 NIGISAHIDSGKTTLTERVLFYTGRIHEIH-EVRGRDGVGAKM---------DSMDLERE 116
Query: 69 RGITIDISMCKFETTKYCCTLIDAPGHRDF 98
+GITI + Y +ID PGH DF
Sbjct: 117 KGITIQSAATYCTWKDYKVNIIDTPGHVDF 146
>At1g45332 mitochondrial elongation factor, putative
Length = 754
Score = 46.2 bits (108), Expect = 4e-05
Identities = 30/90 (33%), Positives = 42/90 (46%), Gaps = 10/90 (11%)
Query: 9 NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERE 68
NI H++SGK+T +L+ G I + E G++ KM D + ERE
Sbjct: 67 NIGISAHIDSGKTTLTERVLFYTGRIHEIH-EVRGRDGVGAKM---------DSMDLERE 116
Query: 69 RGITIDISMCKFETTKYCCTLIDAPGHRDF 98
+GITI + Y +ID PGH DF
Sbjct: 117 KGITIQSAATYCTWKDYKVNIIDTPGHVDF 146
>At3g22980 eukaryotic translation elongation factor 2, putative
Length = 1015
Score = 45.4 bits (106), Expect = 6e-05
Identities = 29/90 (32%), Positives = 39/90 (43%), Gaps = 13/90 (14%)
Query: 9 NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERE 68
NI + HV+ GK+T A HL+ GG + R +D L E+
Sbjct: 11 NICILAHVDHGKTTLADHLIASSGG-------------GVLHPRLAGKLRFMDYLDEEQR 57
Query: 69 RGITIDISMCKFETTKYCCTLIDAPGHRDF 98
R IT+ S + Y LID+PGH DF
Sbjct: 58 RAITMKSSSISLKYKDYSLNLIDSPGHMDF 87
>At1g62750 elongation factor G, putative
Length = 783
Score = 43.1 bits (100), Expect = 3e-04
Identities = 30/90 (33%), Positives = 44/90 (48%), Gaps = 13/90 (14%)
Query: 9 NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERE 68
NI + H+++GK+TT +LY G K I + + A M D ++ E+E
Sbjct: 98 NIGIMAHIDAGKTTTTERILYYTGRNYK--IGEVHEGTATM-----------DWMEQEQE 144
Query: 69 RGITIDISMCKFETTKYCCTLIDAPGHRDF 98
RGITI + K+ +ID PGH DF
Sbjct: 145 RGITITSAATTTFWDKHRINIIDTPGHVDF 174
>At2g31060 putative GTP-binding protein
Length = 664
Score = 40.0 bits (92), Expect = 0.003
Identities = 25/90 (27%), Positives = 35/90 (38%), Gaps = 19/90 (21%)
Query: 9 NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERE 68
N+ I HV+ GK+T LL + G + +D + ERE
Sbjct: 61 NVAVIAHVDHGKTTLMDRLLRQCGA-------------------DIPHERAMDSINLERE 101
Query: 69 RGITIDISMCKFETTKYCCTLIDAPGHRDF 98
RGITI + ++D PGH DF
Sbjct: 102 RGITISSKVTSIFWKDNELNMVDTPGHADF 131
>At5g39900 GTP-binding protein-like
Length = 661
Score = 39.7 bits (91), Expect = 0.003
Identities = 32/99 (32%), Positives = 39/99 (39%), Gaps = 27/99 (27%)
Query: 9 NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERE 68
N I H++ GKST A L+ G I+K LDKL +RE
Sbjct: 68 NFSIIAHIDHGKSTLADRLMELTGTIKK----------------GHGQPQYLDKL--QRE 109
Query: 69 RGITIDISMCKF---------ETTKYCCTLIDAPGHRDF 98
RGIT+ E + Y LID PGH DF
Sbjct: 110 RGITVKAQTATMFYENKVEDQEASGYLLNLIDTPGHVDF 148
>At5g13650 GTP-binding protein typA (tyrosine phosphorylated
protein A)
Length = 609
Score = 37.7 bits (86), Expect = 0.013
Identities = 25/90 (27%), Positives = 35/90 (38%), Gaps = 15/90 (16%)
Query: 9 NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERE 68
NI + HV+ GK+T +L ++A + ++D ERE
Sbjct: 18 NIAIVAHVDHGKTTLVDSML---------------RQAKVFRDNQVMQERIMDSNDLERE 62
Query: 69 RGITIDISMCKFETTKYCCTLIDAPGHRDF 98
RGITI +ID PGH DF
Sbjct: 63 RGITILSKNTSITYKNTKVNIIDTPGHSDF 92
>At1g56070 elongation factor like protein
Length = 843
Score = 35.4 bits (80), Expect = 0.062
Identities = 36/115 (31%), Positives = 50/115 (43%), Gaps = 32/115 (27%)
Query: 1 MVNSKPNI-NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWV 59
+++ K NI N+ I HV+ GKST L+ G I + A +++M
Sbjct: 12 IMDYKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVRMT------- 57
Query: 60 LDKLKAERERGITI------------DISMCKF----ETTKYCCTLIDAPGHRDF 98
D E ERGITI D S+ F + +Y LID+PGH DF
Sbjct: 58 -DTRADEAERGITIKSTGISLYYEMTDESLKSFTGARDGNEYLINLIDSPGHVDF 111
>At5g02250 ribonuclease II-like protein
Length = 803
Score = 33.5 bits (75), Expect = 0.24
Identities = 20/75 (26%), Positives = 36/75 (47%), Gaps = 7/75 (9%)
Query: 131 GWSDLGTCSTCVHSWCEANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVG 190
G SD +CSTC+HS L++ +++ R +G +R+ K+ G V
Sbjct: 60 GNSDAPSCSTCIHS-------LVESVSEELESISRRKGSRMRVRASVKVKLTSYGEVLED 112
Query: 191 RVETGVLKPGMVLTF 205
++ L+ G++L F
Sbjct: 113 KLVNQELEAGLLLEF 127
>At4g29440 putative protein
Length = 1071
Score = 33.1 bits (74), Expect = 0.31
Identities = 53/239 (22%), Positives = 87/239 (36%), Gaps = 28/239 (11%)
Query: 207 PTKLQSGVKSIQKFQENINEALPADIVGF---RLTNKTLSDKNLMRGYVASNSEDEPAME 263
PT L SGV +++ E +P R T+KT + +S+DE E
Sbjct: 767 PTHLHSGVS-----HKDLEEEIPTRASTRSQDRRTHKTTPASASASYFHTMSSDDEDEKE 821
Query: 264 AAKFTSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVEL--EKKPKSL 321
+ T+ I P + T + S V K K++ FD E+ +L E KP +
Sbjct: 822 VHRDTAH--IQTRPYISISRRTKGQERRPSLVTAKIDKVS--FDEESPPKLSPEAKPLTK 877
Query: 322 KNGDAGIVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGY-------- 373
+ G A + +P V +P LG A ++Q + + + K
Sbjct: 878 QQGSASSLSYLPKTEKVSHDQESHPKLGLGAKPLIKQQGSASSLSFLPKTNKASPDQDSP 937
Query: 374 -----KNKAATKMTGSTTDQKDDAET-SSKKRKEGPFETVSAETPSKNVSGLVIGDDGL 426
K K A K GS + +T + ++ P + + E P+ G L
Sbjct: 938 PKLVPKEKPAAKQRGSASSLSFLPKTDKASPDQDSPPKLLPKEKPAAKQQGSATSSSSL 996
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,320,996
Number of Sequences: 26719
Number of extensions: 383079
Number of successful extensions: 1090
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1036
Number of HSP's gapped (non-prelim): 43
length of query: 436
length of database: 11,318,596
effective HSP length: 102
effective length of query: 334
effective length of database: 8,593,258
effective search space: 2870148172
effective search space used: 2870148172
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135415.3


