

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135320.16 + phase: 0
(153 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
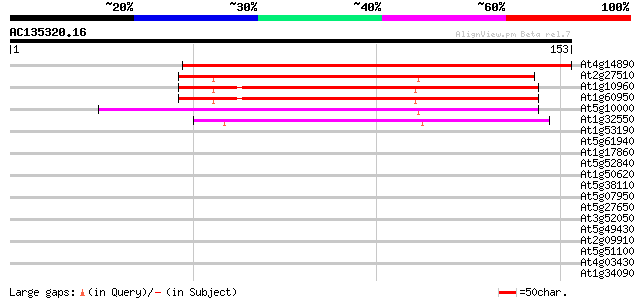
Score E
Sequences producing significant alignments: (bits) Value
At4g14890 ferredoxin 189 5e-49
At2g27510 putative ferredoxin 91 3e-19
At1g10960 ferredoxin precusor isolog 75 1e-14
At1g60950 ferrodoxin precursor 75 1e-14
At5g10000 putative protein 74 3e-14
At1g32550 ferredoxin, putative 62 1e-10
At1g53190 RING finger like protein 30 0.67
At5g61940 putative protein 28 1.5
At1g17860 Unknown protein (F2H15.9) 28 1.5
At5g52840 unknown protein 28 1.9
At1g50620 hypothetical protein 27 3.3
At5g38110 anti-silencing factor 1-like protein 27 4.3
At5g07950 putative protein 27 4.3
At5g27650 putative protein 27 5.7
At3g52050 putative protein 27 5.7
At5g49430 WD-40 repeat protein-like 26 7.4
At2g09910 pseudogene 26 7.4
At5g51100 unknown protein 26 9.7
At4g03430 putative pre-mRNA splicing factor 26 9.7
At1g34090 putative protein 26 9.7
>At4g14890 ferredoxin
Length = 154
Score = 189 bits (480), Expect = 5e-49
Identities = 87/106 (82%), Positives = 100/106 (94%)
Query: 48 VRSSYKVVIEHEGKTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTVDQ 107
+ +YKVV+EH+GKTT+LEVEPDETILSKALDSGLDVP+DC LGVCMTCPA+L++GTVDQ
Sbjct: 49 IARAYKVVVEHDGKTTELEVEPDETILSKALDSGLDVPYDCNLGVCMTCPAKLVTGTVDQ 108
Query: 108 SDGMLSDDVVERGYALLCASYPRSDCHIRVIPEDELLSMQLATAND 153
S GMLSDDVVERGY LLCASYP SDCHI++IPE+ELLS+QLATAND
Sbjct: 109 SGGMLSDDVVERGYTLLCASYPTSDCHIKMIPEEELLSLQLATAND 154
>At2g27510 putative ferredoxin
Length = 155
Score = 90.5 bits (223), Expect = 3e-19
Identities = 43/99 (43%), Positives = 65/99 (65%), Gaps = 2/99 (2%)
Query: 47 TVRSSYKV-VIEHEGKTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTV 105
T+ + YKV ++ +G+ + EV+ D+ IL A ++G+D+P+ C+ G C TC +++SG V
Sbjct: 56 TMSAVYKVKLLGPDGQEDEFEVQDDQYILDAAEEAGVDLPYSCRAGACSTCAGQIVSGNV 115
Query: 106 DQSDG-MLSDDVVERGYALLCASYPRSDCHIRVIPEDEL 143
DQSDG L D +E+GY L C +YP+SDC I E EL
Sbjct: 116 DQSDGSFLEDSHLEKGYVLTCVAYPQSDCVIHTHKETEL 154
>At1g10960 ferredoxin precusor isolog
Length = 148
Score = 75.5 bits (184), Expect = 1e-14
Identities = 38/100 (38%), Positives = 61/100 (61%), Gaps = 3/100 (3%)
Query: 47 TVRSSYKV-VIEHEGKTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTV 105
T ++YKV I EG+ ++E E D +L A ++GLD+P+ C+ G C +C +++SG++
Sbjct: 50 TAMATYKVKFITPEGEQ-EVECEEDVYVLDAAEEAGLDLPYSCRAGSCSSCAGKVVSGSI 108
Query: 106 DQSD-GMLSDDVVERGYALLCASYPRSDCHIRVIPEDELL 144
DQSD L D+ + GY L C +YP SD I E+ ++
Sbjct: 109 DQSDQSFLDDEQMSEGYVLTCVAYPTSDVVIETHKEEAIM 148
>At1g60950 ferrodoxin precursor
Length = 148
Score = 75.1 bits (183), Expect = 1e-14
Identities = 37/100 (37%), Positives = 63/100 (63%), Gaps = 3/100 (3%)
Query: 47 TVRSSYKV-VIEHEGKTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTV 105
T ++YKV I EG+ ++E + D +L A ++G+D+P+ C+ G C +C +++SG+V
Sbjct: 50 TAMATYKVKFITPEGEL-EVECDDDVYVLDAAEEAGIDLPYSCRAGSCSSCAGKVVSGSV 108
Query: 106 DQSD-GMLSDDVVERGYALLCASYPRSDCHIRVIPEDELL 144
DQSD L D+ + G+ L CA+YP SD I E++++
Sbjct: 109 DQSDQSFLDDEQIGEGFVLTCAAYPTSDVTIETHKEEDIV 148
>At5g10000 putative protein
Length = 148
Score = 73.9 bits (180), Expect = 3e-14
Identities = 39/121 (32%), Positives = 64/121 (52%), Gaps = 1/121 (0%)
Query: 25 SQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEGKTTQLEVEPDETILSKALDSGLDV 84
++LN G S + F S +I EG+ ++E D IL A ++GL++
Sbjct: 28 TRLNNTTYFGLSSSRGNFGKVFAKESRKVKLISPEGEEQEIEGNEDCCILESAENAGLEL 87
Query: 85 PHDCKLGVCMTCPARLISGTVDQSDG-MLSDDVVERGYALLCASYPRSDCHIRVIPEDEL 143
P+ C+ G C TC +L+SG VDQS G L ++ +++GY L C + P DC + + +L
Sbjct: 88 PYSCRSGTCGTCCGKLVSGKVDQSLGSFLEEEQIQKGYILTCIALPLEDCVVYTHKQSDL 147
Query: 144 L 144
+
Sbjct: 148 I 148
>At1g32550 ferredoxin, putative
Length = 181
Score = 62.0 bits (149), Expect = 1e-10
Identities = 33/100 (33%), Positives = 52/100 (52%), Gaps = 3/100 (3%)
Query: 51 SYKVVIE--HEGKTTQLEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTVDQS 108
S+KV + G + EV D+ IL A + +P C+ G C +C R+ SG + Q
Sbjct: 58 SHKVTVHDRQRGVVHEFEVPEDQYILHSAESQNISLPFACRHGCCTSCAVRVKSGELRQP 117
Query: 109 DGM-LSDDVVERGYALLCASYPRSDCHIRVIPEDELLSMQ 147
+ +S ++ +GYALLC +P SD + EDE+ +Q
Sbjct: 118 QALGISAELKSQGYALLCVGFPTSDLEVETQDEDEVYWLQ 157
>At1g53190 RING finger like protein
Length = 494
Score = 29.6 bits (65), Expect = 0.67
Identities = 17/63 (26%), Positives = 23/63 (35%)
Query: 19 TKLPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEGKTTQLEVEPDETILSKAL 78
T YPSQLN R S H +P +R + H + V P +
Sbjct: 241 TSFMYPSQLNPRDHYYSHHHHPAPPPVQGMRGQNATLYPHTASSASYRVPPGSFTPQNTM 300
Query: 79 DSG 81
+SG
Sbjct: 301 NSG 303
>At5g61940 putative protein
Length = 1094
Score = 28.5 bits (62), Expect = 1.5
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 5/88 (5%)
Query: 23 YPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEGKTTQLEVEPDETILSKALDSGL 82
Y S+ N R G G S ++ + +V+E E T+ E+ ET +KA D +
Sbjct: 960 YGSRYNQRTEEGCGKTNSVQHISSRCPPIFTIVLEWENSATENEI--SET--TKAFDWEI 1015
Query: 83 DVPHDCKLGVCMTCPARLISGTVDQSDG 110
D+ + G+ RL+S V S+G
Sbjct: 1016 DISRLYEEGLAPNTKYRLVS-MVGYSEG 1042
>At1g17860 Unknown protein (F2H15.9)
Length = 196
Score = 28.5 bits (62), Expect = 1.5
Identities = 23/69 (33%), Positives = 31/69 (44%), Gaps = 11/69 (15%)
Query: 54 VVIEHEGKTTQLEVEPDETILSKALDSG-----LDVPHDCKLGVCM------TCPARLIS 102
V I H G TT+ VEP + I K+L +G L V G+ M TCP +I
Sbjct: 13 VFISHRGVTTEAAVEPVKDINGKSLLTGVNYYILPVIRGRGGGLTMSNLKTETCPTSVIQ 72
Query: 103 GTVDQSDGM 111
+ S G+
Sbjct: 73 DQFEVSQGL 81
>At5g52840 unknown protein
Length = 169
Score = 28.1 bits (61), Expect = 1.9
Identities = 15/44 (34%), Positives = 24/44 (54%)
Query: 99 RLISGTVDQSDGMLSDDVVERGYALLCASYPRSDCHIRVIPEDE 142
R + V Q+ G++ DVV A+L Y ++ I+ +PEDE
Sbjct: 8 RPLLAKVKQTTGIVGLDVVPNARAVLIDLYSKTLKEIQAVPEDE 51
>At1g50620 hypothetical protein
Length = 629
Score = 27.3 bits (59), Expect = 3.3
Identities = 15/47 (31%), Positives = 25/47 (52%), Gaps = 8/47 (17%)
Query: 94 MTCPARLISGTVDQSDGMLSDDVVERGYALLC------ASYPRSDCH 134
MTC ++ GT+++ + +L D E+GY L C P+S+ H
Sbjct: 322 MTC--QICQGTINEIETVLICDACEKGYHLKCLHAHNIKGVPKSEWH 366
>At5g38110 anti-silencing factor 1-like protein
Length = 218
Score = 26.9 bits (58), Expect = 4.3
Identities = 19/77 (24%), Positives = 29/77 (36%), Gaps = 1/77 (1%)
Query: 11 LFRQKHPTTKLPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEGKTTQLEVEPD 70
+ K TK P LG G P+ P V V +E K ++E D
Sbjct: 139 ILTDKPRVTKFPINFHPENEQTLGDGPAPTEPFADSVVNGEAPVFLEQPQKLQEIEQFDD 198
Query: 71 ETILSKALDSGLDVPHD 87
+ +A+ + LD P +
Sbjct: 199 SDVNGEAI-ALLDQPQN 214
>At5g07950 putative protein
Length = 303
Score = 26.9 bits (58), Expect = 4.3
Identities = 17/56 (30%), Positives = 26/56 (46%), Gaps = 7/56 (12%)
Query: 94 MTCPARLISGTVDQSDGMLSDDVVERGYALLCASYPRSDCHIRVIPE---DELLSM 146
+ C +L +G++ Q G D A C P CH+ +IPE +EL+ M
Sbjct: 138 LRCYTKLSTGSLIQFSGTKEDS----NDAGDCGGIPFGTCHVEMIPEKMAEELVEM 189
>At5g27650 putative protein
Length = 1063
Score = 26.6 bits (57), Expect = 5.7
Identities = 13/68 (19%), Positives = 32/68 (46%), Gaps = 6/68 (8%)
Query: 54 VVIEHEGKTTQLEVEPDETILS------KALDSGLDVPHDCKLGVCMTCPARLISGTVDQ 107
V+++ +V PD+T++ +A+D ++ +L +T AR++ +
Sbjct: 6 VIVQQTDSIQDPKVTPDDTVVDSSGDVHEAIDDDVEASSPMELDSAVTNDARVLESERSE 65
Query: 108 SDGMLSDD 115
DG++ +
Sbjct: 66 KDGVVGSE 73
>At3g52050 putative protein
Length = 384
Score = 26.6 bits (57), Expect = 5.7
Identities = 19/66 (28%), Positives = 30/66 (44%), Gaps = 14/66 (21%)
Query: 59 EGKTTQ-LEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTVDQSDGMLSDDVV 117
EGK + L D+ ILSK LD+G + + L+ D ++G +D V+
Sbjct: 324 EGKIKESLIASADQAILSKKLDNGEKL-------------SSLLIAIADYAEGFSADPVI 370
Query: 118 ERGYAL 123
R + L
Sbjct: 371 RRAFRL 376
>At5g49430 WD-40 repeat protein-like
Length = 1576
Score = 26.2 bits (56), Expect = 7.4
Identities = 17/51 (33%), Positives = 22/51 (42%)
Query: 65 LEVEPDETILSKALDSGLDVPHDCKLGVCMTCPARLISGTVDQSDGMLSDD 115
+E EP+ ILS DS +VP + G C SG S G +D
Sbjct: 673 MEWEPEVDILSDENDSEYNVPEEYSSGKEQECLNSSTSGESGSSSGESYED 723
>At2g09910 pseudogene
Length = 985
Score = 26.2 bits (56), Expect = 7.4
Identities = 12/46 (26%), Positives = 24/46 (52%)
Query: 14 QKHPTTKLPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHE 59
Q + ++ P+ L+ R GSG HP++P + + + +I+ E
Sbjct: 36 QSSRSNRIEIPTLLDEPDREGSGGHPAAPEDSVDFLAQFDPLIDLE 81
>At5g51100 unknown protein
Length = 305
Score = 25.8 bits (55), Expect = 9.7
Identities = 12/28 (42%), Positives = 16/28 (56%)
Query: 57 EHEGKTTQLEVEPDETILSKALDSGLDV 84
E EG T+ E PD+ + LDS +DV
Sbjct: 274 EQEGTETEDEENPDDEVPEVYLDSDIDV 301
>At4g03430 putative pre-mRNA splicing factor
Length = 1029
Score = 25.8 bits (55), Expect = 9.7
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 4/64 (6%)
Query: 60 GKTTQLEVEPDETILS---KALDSGLDVPHD-CKLGVCMTCPARLISGTVDQSDGMLSDD 115
GKT ++V P+ T +S + DVP + + M P+R+ + D +LSD
Sbjct: 9 GKTLSIDVNPNSTTISAFEQLAHQRSDVPQSFLRYSLRMRNPSRVFVDSKDSDSILLSDL 68
Query: 116 VVER 119
V R
Sbjct: 69 GVSR 72
>At1g34090 putative protein
Length = 761
Score = 25.8 bits (55), Expect = 9.7
Identities = 18/70 (25%), Positives = 30/70 (42%), Gaps = 4/70 (5%)
Query: 7 TPLTLFRQKHPTTKLPYPSQLNTRPRLGSGSHPSSPSLTFTVRSSYKVVIEHEGKTTQL- 65
TP T R +T+ P S + T+P + S P+ + T + +S + + TT +
Sbjct: 106 TPTTPQRSSTESTETPMSSPVITQPSPSASSIPTMHAATSSASTSQQSSVASNKSTTDVV 165
Query: 66 ---EVEPDET 72
E P T
Sbjct: 166 QIQEASPKST 175
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,458,232
Number of Sequences: 26719
Number of extensions: 142986
Number of successful extensions: 383
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 366
Number of HSP's gapped (non-prelim): 21
length of query: 153
length of database: 11,318,596
effective HSP length: 90
effective length of query: 63
effective length of database: 8,913,886
effective search space: 561574818
effective search space used: 561574818
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC135320.16


