

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.8 - phase: 0 /pseudo
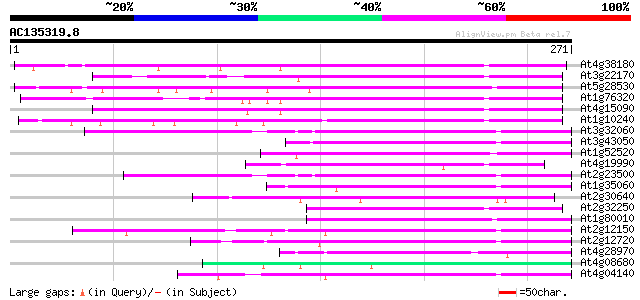
(271 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 75 5e-14
At3g22170 far-red impaired response protein, putative 74 8e-14
At5g28530 far-red impaired response protein (FAR1) - like 70 1e-12
At1g76320 putative phytochrome A signaling protein 62 4e-10
At4g15090 unknown protein 60 9e-10
At1g10240 unknown protein 60 9e-10
At3g32060 hypothetical protein 59 4e-09
At3g43050 putative protein 54 1e-07
At1g52520 F6D8.26 54 1e-07
At4g19990 putative protein 52 3e-07
At2g23500 Mutator-like transposase 52 3e-07
At1g35060 hypothetical protein 51 8e-07
At2g30640 Mutator-like transposase 50 1e-06
At2g32250 Mutator-like transposase 50 2e-06
At1g80010 hypothetical protein 48 5e-06
At2g12150 pseudogene 48 6e-06
At2g12720 putative protein 47 1e-05
At4g28970 putative protein 46 2e-05
At4g08680 putative MuDR-A-like transposon protein 46 2e-05
At4g04140 putative transposon protein 45 3e-05
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 74.7 bits (182), Expect = 5e-14
Identities = 63/279 (22%), Positives = 120/279 (42%), Gaps = 16/279 (5%)
Query: 3 ELVCERSG-----EHKLPKTKVKHEATESRKCGCLFKVRGYVVRENNAWKLTILNGVHNH 57
+ VC + G E + ++K T +R GC + ++++ W ++ HNH
Sbjct: 118 QFVCAKEGFRNMNEKRTKDREIKRPRTITR-VGCKASL-SVKMQDSGKWLVSGFVKDHNH 175
Query: 58 EMVRYVAGHLLAG--RLMEDDKKIVHDLTDSLVKPKNILTNLKKK----RKESITNIKQV 111
E+V H L ++ K ++ L + + P+ I++ L K+ K T +
Sbjct: 176 ELVPPDQVHCLRSHRQISGPAKTLIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDCR 235
Query: 112 YNERHKFKKTKRGDLTEM-QYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNT 170
R+ +K+ G++ + YL +N Y + E Q+V ++FW P ++ F
Sbjct: 236 NYMRNNRQKSIEGEIQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDFTH 295
Query: 171 FPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLE 230
F + D+TY++N YR+P GV +F+ E E +F W L +
Sbjct: 296 FGDTVTFDTTYRSNRYRLPFAPFTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAAM- 354
Query: 231 PNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHVGK 269
++ P + T+ D + A+ + P + C +H+ K
Sbjct: 355 -SAHPPVSITTDHDAVIRAAIMHVFPGARHRFCKWHILK 392
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 73.9 bits (180), Expect = 8e-14
Identities = 55/228 (24%), Positives = 99/228 (43%), Gaps = 19/228 (8%)
Query: 41 RENNAWKLTILNGVHNHEMVRYVAGHLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKK 100
R + W + HNHE+ L A + E +KI + + K +++ LK
Sbjct: 166 RPDGKWVIHSFVREHNHEL-------LPAQAVSEQTRKIYAAMAKQFAEYKTVIS-LKSD 217
Query: 101 RKESITNIKQVYNERHKFKKTKRGDLTEMQYLISKLEE-NGYVHYVREKPESQTVQDIFW 159
K S E+ + + GD + +S+++ N Y + + Q V+++FW
Sbjct: 218 SKSSF--------EKGRTLSVETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFW 269
Query: 160 THPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFT 219
S + +F V+ +D+TY N Y+MPL VGV Y + + ++ E ++
Sbjct: 270 VDAKSRHNYGSFCDVVSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYS 329
Query: 220 WALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHV 267
W ++ L+ + PKV++T D M V P++ L +HV
Sbjct: 330 WLMETWLRAI--GGQAPKVLITELDVVMNSIVPEIFPNTRHCLFLWHV 375
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 70.1 bits (170), Expect = 1e-12
Identities = 66/290 (22%), Positives = 130/290 (44%), Gaps = 30/290 (10%)
Query: 3 ELVCERSGEHKLPKTKVKHEATESRK---CGCLFKVRGYVVREN----NAWKLTILNGVH 55
+ VC RSG ++ P+ K E RK CGC K+ Y+ +E + W ++ + VH
Sbjct: 114 DFVCYRSGFNQ-PRKKANVEHPRERKSVRCGCDGKL--YLTKEVVDGVSHWYVSQFSNVH 170
Query: 56 NHEMVRYVAGHLLAG--RLMEDDKKI----------VHDLTDSLVKPKNILTN----LKK 99
NHE++ LL ++ + D++ V+ + L K +++ ++K
Sbjct: 171 NHELLEDDQVRLLPAYRKIQQSDQERILLLSKAGFPVNRIVKLLELEKGVVSGQLPFIEK 230
Query: 100 KRKESITNIKQVYNERHKFKKTKR-GDLTEMQYLISKLEENGYVH-YVREKPESQTVQDI 157
+ + K+ E F KR D E+ L E Y E+Q V++I
Sbjct: 231 DVRNFVRACKKSVQENDAFMTEKRESDTLELLECCKGLAERDMDFVYDCTSDENQKVENI 290
Query: 158 FWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDN 217
W + SV+ ++ F V++ D++Y++ Y + L G+ + + + E +
Sbjct: 291 AWAYGDSVRGYSLFGDVVVFDTSYRSVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRS 350
Query: 218 FTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHV 267
FTWALQ ++ + P+ ++T+ D + A+ +P+++ ++ H+
Sbjct: 351 FTWALQTFVRFMRGRH--PQTILTDIDTGLKDAIGREMPNTNHVVFMSHI 398
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 61.6 bits (148), Expect = 4e-10
Identities = 56/278 (20%), Positives = 118/278 (42%), Gaps = 35/278 (12%)
Query: 6 CERSGEHKLPKTKVKHEATESRKCGCLFKVRGYVVRENNAWKLTILNGVHNHEMVRYVAG 65
C R G + + A+ C V+ R + W + HNH+++ A
Sbjct: 45 CIRYGSKQQSDDAINPRASPKIGCKASMHVKR---RPDGKWYVYSFVKEHNHDLLPEQAH 101
Query: 66 HLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKKRKESITNIKQV--YNE--------R 115
+ + R E LVK + + L++K+ +T+ K + Y++ R
Sbjct: 102 YFRSHRNTE------------LVKSND--SRLRRKKNTPLTDCKHLSAYHDLDFIDGYMR 147
Query: 116 HKFKKTKR-----GDLTEM-QYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFN 169
++ K +R GD + ++L+ EEN + + E ++++FW ++ +
Sbjct: 148 NQHDKGRRLVLDTGDAEILLEFLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYK 207
Query: 170 TFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLL 229
+F V+ +++Y + Y++PL VGV + + + + W +Q L +
Sbjct: 208 SFSDVVSFETSYFVSKYKVPLVLFVGVNHHVQPVLLGCGLLADDTVYTYVWLMQSWLVAM 267
Query: 230 EPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHV 267
PKV++T+++ ++ A+ LP++ C +HV
Sbjct: 268 --GGQKPKVMLTDQNNAIKAAIAAVLPETRHCYCLWHV 303
>At4g15090 unknown protein
Length = 768
Score = 60.5 bits (145), Expect = 9e-10
Identities = 49/235 (20%), Positives = 102/235 (42%), Gaps = 10/235 (4%)
Query: 41 RENNAWKLTILNGVHNHEMVRYVAGHLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKK 100
R + W + HNHE++ +A H R ++ +K D+ ++ + + +
Sbjct: 75 RPDGKWIIHEFVKDHNHELLPALAYHFRIQRNVKLAEKNNIDILHAVSERTKKMYVEMSR 134
Query: 101 RKESITNIKQVYN-------ERHKFKKTKRGDLTEM-QYLISKLEENGYVHYVREKPESQ 152
+ NI + ++ ++ + GD + +Y +EN Y + E Q
Sbjct: 135 QSGGYKNIGSLLQTDVSSQVDKGRYLALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQ 194
Query: 153 TVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTA 212
++++FW S + +F V+ D+TY ++PL +GV + + +
Sbjct: 195 RLRNLFWADAKSRDDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVAD 254
Query: 213 EKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHV 267
E + F W ++ L+ + PKV++T++D +M AV+ LP++ +HV
Sbjct: 255 ESMETFVWLIKTWLRAM--GGRAPKVILTDQDKFLMSAVSELLPNTRHCFALWHV 307
>At1g10240 unknown protein
Length = 680
Score = 60.5 bits (145), Expect = 9e-10
Identities = 63/278 (22%), Positives = 121/278 (42%), Gaps = 20/278 (7%)
Query: 5 VCERSGEHKLPKTKVKHEATESRK---CGCLFKVRGYVVRE--NNAWKLTILNGVHNHEM 59
VC R+G + KT + + +R+ CGC +R + E + W++T HNHE+
Sbjct: 96 VCHRAGNTPI-KTLSEGKPQRNRRSSRCGCQAYLRISKLTELGSTEWRVTGFANHHNHEL 154
Query: 60 VRYVAGHLL-AGRLMEDDKK---IVHDLTDSLVKPKNILTNLKKKRKESITNI--KQVYN 113
+ L A R + D K ++ T V+ L L+K + K V N
Sbjct: 155 LEPNQVRFLPAYRSISDADKSRILMFSKTGISVQQMMRLLELEKCVEPGFLPFTEKDVRN 214
Query: 114 ERHKFKKT----KRGDLTEMQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFN 169
FKK + D M I + + N + + + +++I W++ +S++ +
Sbjct: 215 LLQSFKKLDPEDENIDFLRMCQSIKEKDPNFKFEFTLDANDK--LENIAWSYASSIQSYE 272
Query: 170 TFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLL 229
F ++ D+T++ + MPL VGV + + + E +++WALQ +
Sbjct: 273 LFGDAVVFDTTHRLSAVEMPLGIWVGVNNYGVPCFFGCVLLRDENLRSWSWALQAFTGFM 332
Query: 230 EPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHV 267
N P+ ++T+ + + +A+ +P + LC + V
Sbjct: 333 --NGKAPQTILTDHNMCLKEAIAGEMPATKHALCIWMV 368
>At3g32060 hypothetical protein
Length = 487
Score = 58.5 bits (140), Expect = 4e-09
Identities = 50/235 (21%), Positives = 108/235 (45%), Gaps = 11/235 (4%)
Query: 37 GYVVRENNAWKLTILNGVHNHEMVRYVAGHLLAGRLMEDDKKIVHDLTDSLVKPKNILTN 96
GYV+++ + + + N ++V V G++ +I+ +L + + K +
Sbjct: 138 GYVIKKLDKERYVLKKRKGNPQLVAAVLHDHFLGQVETPVPRIIMELVQTKLGVKVSYST 197
Query: 97 LKKKRKESITNIKQVYNERHKFKKTKRGDLTEMQYLISKLEENGYVHYVREKPESQTVQD 156
+ + + +I ++K E +K D+ Y++ K+ + G V Y++ E+ Q
Sbjct: 198 VLRGKYHAIYDLKGSREESYK-------DINCYLYMLKKVND-GTVTYLK-LDENDKFQY 248
Query: 157 IFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKED 216
+F S++ F VLI+D+T+ N Y L Y + F+ + E +
Sbjct: 249 VFVALGASIEGFRVMRKVLIVDATHLKNGYGGVLVFASAQDPNRHHYIIAFAVLDGENDA 308
Query: 217 NFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHVGKNV 271
++ W + L ++ S++ V +T+R+ S++KA+ N + C +H+ +NV
Sbjct: 309 SWEWFFEKLKTVVPDTSEL--VFMTDRNASLIKAIRNVYTAAHHGYCIWHLSQNV 361
>At3g43050 putative protein
Length = 608
Score = 53.5 bits (127), Expect = 1e-07
Identities = 34/138 (24%), Positives = 69/138 (49%), Gaps = 3/138 (2%)
Query: 134 SKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEI 193
+K+ G + +V E E + Q +F+ ++ FN V+I+D+T+ L
Sbjct: 295 TKITNEGTISFV-ELDEKKNFQYLFFALGACIEGFNAMRKVIIVDATHLKIAQGGVLVVA 353
Query: 194 VGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTN 253
+ + Y +DF + EK+ +++W + L ++ +S++ V V++R+ S+MK+
Sbjct: 354 LAQDLEHHHYPIDFGVLDGEKDVSWSWFFEKLKSVIPDSSEL--VFVSDRNQSLMKSQRE 411
Query: 254 ALPDSSAILCYFHVGKNV 271
P S C +H+ +NV
Sbjct: 412 LYPLSQHGCCIYHLCQNV 429
>At1g52520 F6D8.26
Length = 703
Score = 53.5 bits (127), Expect = 1e-07
Identities = 32/151 (21%), Positives = 70/151 (46%), Gaps = 4/151 (2%)
Query: 122 KRGDLTEMQYLISKLE-ENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDST 180
KRGD + +++ N Y+ + + ++++FW S + F V+ +DS+
Sbjct: 244 KRGDSAAIYNYFCRMQLTNPNFFYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVIFIDSS 303
Query: 181 YKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVV 240
Y + + +PL GV T + F+ E +++ W L++ L +++ + P+ +V
Sbjct: 304 YISGKFEIPLVTFTGVNHHGKTTLLSCGFLAGETMESYHWLLKVWLSVMKRS---PQTIV 360
Query: 241 TNRDPSMMKAVTNALPDSSAILCYFHVGKNV 271
T+R + A++ P S H+ + +
Sbjct: 361 TDRCKPLEAAISQVFPRSHQRFSLTHIMRKI 391
>At4g19990 putative protein
Length = 672
Score = 52.4 bits (124), Expect = 3e-07
Identities = 36/145 (24%), Positives = 65/145 (44%), Gaps = 5/145 (3%)
Query: 115 RHKFKKTKRGDLTEMQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTV 174
R K +K + E++ KLE+ + + Q++++IFW + F V
Sbjct: 149 RRKLEKLNGAIVKEVKS--RKLEDGDVERLLNFFTDMQSLRNIFWVDAKGRFDYTCFSDV 206
Query: 175 LIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFS-FMTAEKEDNFTWALQMLLKLLEPNS 233
+ +D+T+ N Y++PL GV + F +T E + F W + LK + +
Sbjct: 207 VSIDTTFIKNEYKLPLVAFTGVNHHGQFLLLGFGLLLTDESKSGFVWLFRAWLKAM--HG 264
Query: 234 DMPKVVVTNRDPSMMKAVTNALPDS 258
P+V++T D + +AV P S
Sbjct: 265 CRPRVILTKHDQMLKEAVLEVFPSS 289
>At2g23500 Mutator-like transposase
Length = 784
Score = 52.0 bits (123), Expect = 3e-07
Identities = 47/216 (21%), Positives = 97/216 (44%), Gaps = 11/216 (5%)
Query: 56 NHEMVRYVAGHLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKKRKESITNIKQVYNER 115
N ++V + G+L I+ +L + + K + + + +I ++K E
Sbjct: 287 NPQLVAALLHDHFPGQLETPVPSIIMELVQTKLGVKVSYSTALRGKYHAIYDLKGSPEES 346
Query: 116 HKFKKTKRGDLTEMQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVL 175
+K D+ Y++ K+ + G V Y++ E+ Q +F S++ F VL
Sbjct: 347 YK-------DINCYLYMLKKVND-GTVTYLK-LDENDKFQYVFVALGASIEGFRVMRKVL 397
Query: 176 IMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDM 235
I+D+T+ N Y L Y + F+ + E + ++ W + L ++ S++
Sbjct: 398 IVDATHLKNGYGGVLVFASAQDPNRHHYIIAFAVLDGENDASWEWFFEKLKTVVPDTSEL 457
Query: 236 PKVVVTNRDPSMMKAVTNALPDSSAILCYFHVGKNV 271
V +T+R+ S++KA+ N + C +H+ +NV
Sbjct: 458 --VFMTDRNASLIKAIRNVYTAAHHGYCIWHLSQNV 491
>At1g35060 hypothetical protein
Length = 873
Score = 50.8 bits (120), Expect = 8e-07
Identities = 32/152 (21%), Positives = 71/152 (46%), Gaps = 8/152 (5%)
Query: 125 DLTEMQYLISKLEENGYVHYVREKPESQTVQD-----IFWTHPTSVKLFNTFPTVLIMDS 179
+L E +L+ KL G + ++ +P+ + + +F S++ F VL++D
Sbjct: 472 NLAEYLHLL-KLTNPGTITHIETEPDIEDERKERFLYMFLAFGASIQGFKHLRRVLVVDG 530
Query: 180 TYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVV 239
T+ Y+ L G + + Y + F+ + +E +D +TW L +++ N+ + +
Sbjct: 531 THLKGKYKGVLLTASGQDANFQVYPLAFAVVDSENDDAWTWFFTKLERIIADNNTL--TI 588
Query: 240 VTNRDPSMMKAVTNALPDSSAILCYFHVGKNV 271
+++R S+ V P + C H+ +N+
Sbjct: 589 LSDRHESIKVGVKKVFPQAHHGACIIHLCRNI 620
>At2g30640 Mutator-like transposase
Length = 754
Score = 50.1 bits (118), Expect = 1e-06
Identities = 46/184 (25%), Positives = 81/184 (44%), Gaps = 10/184 (5%)
Query: 89 KPKNILTNLKKKRKESITNIKQVYNERHKFKKTKRGDLTEMQYLISKLEEN------GYV 142
KPK IL + + ++T KQ + + + RG E L+ + + G V
Sbjct: 283 KPKEILEQIHQVHGITLT-YKQAWRGKERIMAAVRGSYEEDYRLLPRYCDQIRRTNPGSV 341
Query: 143 HYVREKPESQTVQDIFWTHPTSVKLF-NTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYL 201
V P + Q F + S+ F N ++ +D T + Y L G
Sbjct: 342 AVVHGSPVDGSFQQFFISFQASICGFLNACRPLIGLDRTVLKSKYVGTLLLATGFDGEGA 401
Query: 202 TYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSD-MPKV-VVTNRDPSMMKAVTNALPDSS 259
+ + F+ ++ E + ++ W L L +LLE NS+ MPK+ ++++RD S++ V P +
Sbjct: 402 VFPLAFAIISEENDSSWQWFLSELRQLLEVNSENMPKLTILSSRDQSIVDGVDTNFPTAF 461
Query: 260 AILC 263
LC
Sbjct: 462 HGLC 465
>At2g32250 Mutator-like transposase
Length = 684
Score = 49.7 bits (117), Expect = 2e-06
Identities = 28/124 (22%), Positives = 55/124 (43%), Gaps = 2/124 (1%)
Query: 144 YVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTY 203
Y + + V+++FW + + +F V++ D+ Y N YR+P +GV+
Sbjct: 189 YAVDFDSDKRVRNVFWLDAKAKHDYCSFSDVVLFDTFYVRNGYRIPFAPFIGVSHHRQYV 248
Query: 204 SVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILC 263
+ + + E ++W + LK + P V++T++D + V PD I C
Sbjct: 249 LLGCALIGEVSESTYSWLFRTWLKAV--GGQAPGVMITDQDKLLSDIVVEVFPDVRHIFC 306
Query: 264 YFHV 267
+ V
Sbjct: 307 LWSV 310
>At1g80010 hypothetical protein
Length = 696
Score = 48.1 bits (113), Expect = 5e-06
Identities = 27/128 (21%), Positives = 58/128 (45%), Gaps = 2/128 (1%)
Query: 144 YVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTY 203
Y+ + + +++++FW + ++ F VL+ D+T +N Y +PL VG+ T
Sbjct: 262 YLMDLADDGSLRNVFWIDARARAAYSHFGDVLLFDTTCLSNAYELPLVAFVGINHHGDTI 321
Query: 204 SVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILC 263
+ + + + + W + L + P++ +T + +M AV+ P + L
Sbjct: 322 LLGCGLLADQSFETYVWLFRAWLTCMLGRP--PQIFITEQCKAMRTAVSEVFPRAHHRLS 379
Query: 264 YFHVGKNV 271
HV N+
Sbjct: 380 LTHVLHNI 387
>At2g12150 pseudogene
Length = 942
Score = 47.8 bits (112), Expect = 6e-06
Identities = 48/256 (18%), Positives = 107/256 (41%), Gaps = 23/256 (8%)
Query: 31 CLFKVRGYVVRENNAWKLTILNGVH-----NHEMVRYVAGHLLAGRLMEDDKKIVHDLTD 85
C + + +V+ + +K+T + +H + R A + L G +M++
Sbjct: 422 CSWHLTAHVIPNSKCFKITGYDSIHVCKIDTRKDYRKHATYKLLGEVMKNRYSSSQGGPR 481
Query: 86 SLVKPKNILTNLKKKRKESITNIKQVYNERHKFKKTKRGD-------LTEMQYLISKLEE 138
++ P+ +L +L + S + R + RGD L YL+ +L
Sbjct: 482 AVDLPRLVLNDLNVRISYS-----SAWRAREVAVNSIRGDDMASYRFLPTYLYLL-QLAN 535
Query: 139 NGYVHYVREKPES---QTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVG 195
G + ++ PE Q + +F + S+K + V+++D T Y+ L
Sbjct: 536 PGTICHLHSTPEDKGRQRFKYVFVSLSASIKGWKYMRKVIVVDGTQLVGRYKGCLLIACA 595
Query: 196 VTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNAL 255
+ + + F + E + ++ W + L +++ D+ ++V++R S+ K V+
Sbjct: 596 QDGNFQIFPLAFGVVDGETDASWIWFFEKLSEIVPDTDDL--MIVSDRHSSIYKGVSVVY 653
Query: 256 PDSSAILCYFHVGKNV 271
P ++ C H+ +N+
Sbjct: 654 PKANHGACIVHLERNI 669
>At2g12720 putative protein
Length = 819
Score = 46.6 bits (109), Expect = 1e-05
Identities = 41/194 (21%), Positives = 85/194 (43%), Gaps = 18/194 (9%)
Query: 88 VKPKNILTNLKKKRKESITNIKQVYNERHKFKKTKRGDLTEMQYLISKLEENGYVHYVRE 147
VKPK+I + K + +K Y++ +K + R +LT S E Y++ +R
Sbjct: 359 VKPKSIGIIITKHFR-----VKMSYSKSYKTLRFAR-ELTLGTPDSSFEELPSYLYMIRR 412
Query: 148 K----------PESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVT 197
ES +F S+ F+ V+++D T+ Y+ L +
Sbjct: 413 ANPGTVARLQIDESGRFNYMFIVFGASIAGFHYMRRVVVVDGTFLHGSYKGTLLTALAQD 472
Query: 198 STYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALPD 257
+ + + F + E +D++ W L ++ +D+ ++++R S+ KA+ P
Sbjct: 473 GNFQIFPLAFGVVDTENDDSWRWLFTQLKVVIPDATDL--AIISDRHKSIGKAIGEVYPL 530
Query: 258 SSAILCYFHVGKNV 271
++ +C +H+ KN+
Sbjct: 531 AARGICTYHLYKNI 544
>At4g28970 putative protein
Length = 914
Score = 45.8 bits (107), Expect = 2e-05
Identities = 35/142 (24%), Positives = 70/142 (48%), Gaps = 6/142 (4%)
Query: 131 YLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPL 190
Y++ K+ G V YV + E + + +F ++ F V+++D+T+ +Y L
Sbjct: 369 YMLEKVNP-GTVTYVELEGEKK-FKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGML 426
Query: 191 FEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVV-VTNRDPSMMK 249
+ Y + F + +EK+ ++ W L+ KL SD+P +V +++R S+ K
Sbjct: 427 VIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLE---KLKTVYSDVPGLVFISDRHQSIKK 483
Query: 250 AVTNALPDSSAILCYFHVGKNV 271
AV P++ C +H+ +N+
Sbjct: 484 AVKTVYPNALHAACIWHLCQNM 505
>At4g08680 putative MuDR-A-like transposon protein
Length = 761
Score = 45.8 bits (107), Expect = 2e-05
Identities = 41/202 (20%), Positives = 81/202 (39%), Gaps = 24/202 (11%)
Query: 94 LTNLKKKRKESITNIKQVYNERHKFKKT----KRGDLTEMQYLISKLEEN-----GYVHY 144
L L+ K +I++ ER K T +RG L ++ L + EE GYV
Sbjct: 201 LDKLRLNEKFMPMDIQEYIKERWKMVSTIPQCQRGRLLALKMLKKEYEEQFAHIRGYVEE 260
Query: 145 VREKPESQTVQDIFWTHPTSVKLFNTFPT---------------VLIMDSTYKTNLYRMP 189
+ + + + +FN F ++ +D T+ + +
Sbjct: 261 IHSQNPGSVAFIDTYRNEKGEDVFNRFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVKGV 320
Query: 190 LFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMK 249
L VG Y + ++ + +E +N+ W +Q + K L V++++R ++
Sbjct: 321 LLTAVGHDPNNQIYPIAWAVVQSENAENWLWFVQQIKKDLNLEDGSRFVILSDRSKGLLS 380
Query: 250 AVTNALPDSSAILCYFHVGKNV 271
AV LP++ +C H+ +N+
Sbjct: 381 AVKQELPNAEHRMCVKHIVENL 402
>At4g04140 putative transposon protein
Length = 946
Score = 45.4 bits (106), Expect = 3e-05
Identities = 41/195 (21%), Positives = 84/195 (43%), Gaps = 14/195 (7%)
Query: 82 DLTDSLVKPKNILTNLKK--KRKESITNIKQVYNERHKFKKTKRGDLTEMQYLISKLEEN 139
DL DSL++ N+ K K KE Q +E L + + +L
Sbjct: 430 DLPDSLLREHNVRMTYWKCWKAKELAVETAQGTDE-------SSFSLLPVYLHVLQLANP 482
Query: 140 GYVHYVREKPES---QTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGV 196
G V+++ + + + +F + SVK V+++D T+ Y L
Sbjct: 483 GTVYHLETELDDIGDDRFKYVFLSLGASVKRLKYIRRVVVVDGTHLFGKYLGCLLTASCQ 542
Query: 197 TSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAVTNALP 256
+ + + + F+ + +E + ++TW + L ++++ D+ V++R+ S+ K+V P
Sbjct: 543 DANFQIFPIAFAVVDSETDHSWTWFMNKLSEIIKDGPDL--TFVSDRNQSIFKSVGLVFP 600
Query: 257 DSSAILCYFHVGKNV 271
+ C H+ +NV
Sbjct: 601 QAHHGACLVHIRRNV 615
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,097,589
Number of Sequences: 26719
Number of extensions: 249591
Number of successful extensions: 869
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 809
Number of HSP's gapped (non-prelim): 88
length of query: 271
length of database: 11,318,596
effective HSP length: 98
effective length of query: 173
effective length of database: 8,700,134
effective search space: 1505123182
effective search space used: 1505123182
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135319.8


