

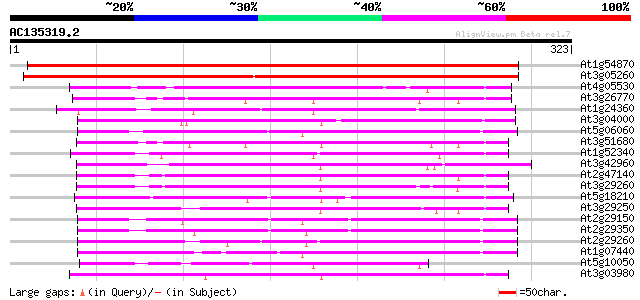
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.2 - phase: 0
(323 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g54870 putative protein (At1g54870) 461 e-130
At3g05260 putative glucose and ribitol dehydrogenase homolog 426 e-120
At4g05530 unknown protein 104 6e-23
At3g26770 short chain alcohol dehydrogenase like protein 96 2e-20
At1g24360 putative 3-oxoacyl [acyl-carrier protein] reductase 96 3e-20
At3g04000 short-chain type dehydrogenase/reductase like protein 94 8e-20
At5g06060 short chain alcohol dehydrogenase-like 93 2e-19
At3g51680 short-chain alcohol dehydrogenase-like protein 93 2e-19
At1g52340 short chain alcohol dehydrogenase, putative 93 2e-19
At3g42960 alcohol dehydrogenase (ATA1) 92 4e-19
At2g47140 alcohol dehydrogenase like protein 89 3e-18
At3g29260 short-chain alcohol dehydrogenase, putative 89 4e-18
At5g18210 Brn1-like protein 88 5e-18
At3g29250 short-chain alcohol dehydrogenase, putative 87 1e-17
At2g29150 pseudogene 87 1e-17
At2g29350 putative tropinone reductase 87 2e-17
At2g29260 tropinone reductase like protein 87 2e-17
At1g07440 tropinone reductase-I, putative 87 2e-17
At5g10050 unknown protein 86 2e-17
At3g03980 putative short-chain type dehydrogenase/reductase 85 5e-17
>At1g54870 putative protein (At1g54870)
Length = 288
Score = 461 bits (1187), Expect = e-130
Identities = 217/283 (76%), Positives = 253/283 (88%)
Query: 11 QKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVI 70
QKQ QPGKEH M +PQF+ DY+P+NKL+GK+A++TGGDSGIGRAV FA EGATV
Sbjct: 6 QKQHAQPGKEHVMESSPQFSSSDYQPSNKLRGKVALITGGDSGIGRAVGYCFASEGATVA 65
Query: 71 FTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDIL 130
FTYVKG E+KDA++TL MLK KT+++K+P+AIP DLGFDENCKRV+DE++NA+GRID+L
Sbjct: 66 FTYVKGQEEKDAQETLQMLKEVKTSDSKEPIAIPTDLGFDENCKRVVDEVVNAFGRIDVL 125
Query: 131 VNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINTTSVNAYKG 190
+NNAAEQYE ++EEIDEPRLERVFRTNIFSYFF+TRHALKHMKEGS+IINTTSVNAYKG
Sbjct: 126 INNAAEQYESSTIEEIDEPRLERVFRTNIFSYFFLTRHALKHMKEGSSIINTTSVNAYKG 185
Query: 191 NSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSD 250
N++L+DYT+TKGAIVAFTR L+LQL KGIRVNGVAPGPIWTPLIPASFNEEK FGS+
Sbjct: 186 NASLLDYTATKGAIVAFTRGLALQLAEKGIRVNGVAPGPIWTPLIPASFNEEKIKNFGSE 245
Query: 251 VPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLINA 293
VPMKRAGQP+EVAPS+VFLA N CSSY TGQVLHPNG ++NA
Sbjct: 246 VPMKRAGQPIEVAPSYVFLACNHCSSYFTGQVLHPNGGAVVNA 288
>At3g05260 putative glucose and ribitol dehydrogenase homolog
Length = 289
Score = 426 bits (1096), Expect = e-120
Identities = 201/285 (70%), Positives = 246/285 (85%), Gaps = 1/285 (0%)
Query: 9 PPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGAT 68
PPQKQ+TQPG +H M PTP+F+ +YKP+NKL GK+A+VTGGDSGIG+AVC+ +ALEGA+
Sbjct: 6 PPQKQETQPGIQHVMEPTPEFSSSNYKPSNKLHGKVALVTGGDSGIGKAVCHCYALEGAS 65
Query: 69 VIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRID 128
V FTYVKG EDKDA +TL +L KT AK+P+ I DLGF+ENCKRV++E++N++GRID
Sbjct: 66 VAFTYVKGREDKDAEETLRLLHEVKTREAKEPIMIATDLGFEENCKRVVEEVVNSFGRID 125
Query: 129 ILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINTTSVNAY 188
+LVN AAEQ+E S+E+IDE RLERVFRTNIFS FF+ ++ALKHMKEGS+IINTTSV AY
Sbjct: 126 VLVNCAAEQHEV-SIEDIDEARLERVFRTNIFSQFFLVKYALKHMKEGSSIINTTSVVAY 184
Query: 189 KGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFG 248
GNS+L++YT+TKGAIV+FTR L+LQL KGIRVNGVAPGP+WTPLIPASF+EE QFG
Sbjct: 185 AGNSSLLEYTATKGAIVSFTRGLALQLAPKGIRVNGVAPGPVWTPLIPASFSEEAIKQFG 244
Query: 249 SDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLINA 293
S+ PMKRA QPVEVAPS+VFLA N CSSY TGQ+LHPNG +++NA
Sbjct: 245 SETPMKRAAQPVEVAPSYVFLACNHCSSYYTGQILHPNGGLIVNA 289
>At4g05530 unknown protein
Length = 254
Score = 104 bits (260), Expect = 6e-23
Identities = 75/258 (29%), Positives = 129/258 (49%), Gaps = 14/258 (5%)
Query: 35 KPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKT 94
K +L+GK+A+VT GIG + F LEGA+V+ V + + + + LK
Sbjct: 4 KLPRRLEGKVAIVTASTQGIGFGITERFGLEGASVV---VSSRKQANVDEAVAKLK---- 56
Query: 95 ANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERV 154
+ D I + ++ + ++++ + YG+IDI+V NAA + E L+++
Sbjct: 57 SKGIDAYGIVCHVSNAQHRRNLVEKTVQKYGKIDIVVCNAAANPSTDPILSSKEAVLDKL 116
Query: 155 FRTNIFSYFFMTRHALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
+ N+ S + + H+++GS++I TS+ + + Y TK A++ T+AL+ +
Sbjct: 117 WEINVKSSILLLQDMAPHLEKGSSVIFITSIAGFSPQGAMAMYGVTKTALLGLTKALAAE 176
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASF---NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLAS 271
+ + RVN VAPG + P ASF + E + R G ++A + FLAS
Sbjct: 177 M-APDTRVNAVAPG--FVPTHFASFITGSSEVREGIEEKTLLNRLGTTGDMAAAAAFLAS 233
Query: 272 NQCSSYITGQVLHPNGDM 289
+ SSYITG+ L G M
Sbjct: 234 DD-SSYITGETLVVAGGM 250
>At3g26770 short chain alcohol dehydrogenase like protein
Length = 306
Score = 96.3 bits (238), Expect = 2e-20
Identities = 73/266 (27%), Positives = 124/266 (46%), Gaps = 24/266 (9%)
Query: 37 ANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTAN 96
+ KL+GK+A++TGG SG+G+A + F GA V+ D DA K AK
Sbjct: 38 SKKLEGKVALITGGASGLGKATASEFLRHGARVVIA------DLDAETG---TKTAKELG 88
Query: 97 AKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNA--AEQYECGSVEEIDEPRLERV 154
++ + D+ + + ++ + YG++D++ NNA S+ ++D ERV
Sbjct: 89 SEAEF-VRCDVTVEADIAGAVEMTVERYGKLDVMYNNAGIVGPMTPASISQLDMTEFERV 147
Query: 155 FRTNIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALS 212
R N+F +HA K M I+ T+SV G YT +K ++ +
Sbjct: 148 MRINVFGVVSGIKHAAKFMIPARSGCILCTSSVAGVTGGLAPHSYTISKFTTPGIVKSAA 207
Query: 213 LQLVSKGIRVNGVAPGPIWTPL-------IPASFNEEKTAQFGSDVPMKRAG--QPVEVA 263
+L G+R+N ++PG + TPL + +EEK + + + + +VA
Sbjct: 208 SELCEHGVRINCISPGTVATPLTLSYLQKVFPKVSEEKLRETVKGMGELKGAECEEADVA 267
Query: 264 PSFVFLASNQCSSYITGQVLHPNGDM 289
+ ++LASN Y+TG L +G M
Sbjct: 268 KAALYLASND-GKYVTGHNLVVDGGM 292
>At1g24360 putative 3-oxoacyl [acyl-carrier protein] reductase
Length = 319
Score = 95.5 bits (236), Expect = 3e-20
Identities = 74/271 (27%), Positives = 126/271 (46%), Gaps = 17/271 (6%)
Query: 28 QFTCPDYKPAN---KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARD 84
Q T + P K++ + V+TG GIG+A+ G V+ Y AR
Sbjct: 59 QATATEQSPGEVVQKVESPVVVITGASRGIGKAIALALGKAGCKVLVNY--------ARS 110
Query: 85 TLDMLKMAKTANAKDPMAIP--ADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGS 142
+ ++AK AI D+ + ++ ++ +G ID++VNNA +
Sbjct: 111 AKEAEEVAKQIEEYGGQAITFGGDVSKATDVDAMMKTALDKWGTIDVVVNNAGITRDTLL 170
Query: 143 VEEIDEPRLERVFRTNIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTST 200
+ + + + + V N+ F T+ A+K M K+ IIN +SV GN +Y +
Sbjct: 171 IR-MKQSQWDEVIALNLTGVFLCTQAAVKIMMKKKRGRIINISSVVGLIGNIGQANYAAA 229
Query: 201 KGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPV 260
KG +++F++ + + S+ I VN V PG I + + A E+ + +P+ R G+
Sbjct: 230 KGGVISFSKTAAREGASRNINVNVVCPGFIASDM-TAELGEDMEKKILGTIPLGRYGKAE 288
Query: 261 EVAPSFVFLASNQCSSYITGQVLHPNGDMLI 291
EVA FLA + +SYITGQ +G + I
Sbjct: 289 EVAGLVEFLALSPAASYITGQAFTIDGGIAI 319
>At3g04000 short-chain type dehydrogenase/reductase like protein
Length = 272
Score = 94.4 bits (233), Expect = 8e-20
Identities = 74/262 (28%), Positives = 119/262 (45%), Gaps = 13/262 (4%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANA-- 97
L G++A+VTG GIGRA+ A GA V+ Y + + T +K A
Sbjct: 14 LAGRVAIVTGSSRGIGRAIAIHLAELGARVVVNYSTSPVEAEKVATAITTNCSKDAEVAG 73
Query: 98 KDP--MAIPADLGFDENCKRVIDEIINAY-GRIDILVNNAA-EQYECGSVEEIDEPRLER 153
K P + + AD+ K + DE + + ILVN+AA ++ ++ +R
Sbjct: 74 KSPRVIVVKADISEPSQVKSLFDEAERVFESPVHILVNSAAIADPNHSTISDMSVELFDR 133
Query: 154 VFRTNIFSYFFMTRHALKHMKEGSN----IINTTSVNAYKGNSTLIDYTSTKGAIVAFTR 209
+ N F R A +K G +++T+ V N+ YT++K A+ A +
Sbjct: 134 IISVNTRGAFICAREAANRLKRGGGGRIILLSTSLVQTL--NTNYGSYTASKAAVEAMAK 191
Query: 210 ALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFL 269
L+ +L I VN V+PGP+ T + + E + S R G+ ++AP FL
Sbjct: 192 ILAKELKGTEITVNCVSPGPVATEMFYTGLSNEIVEKVKSQNLFGRIGETKDIAPVVGFL 251
Query: 270 ASNQCSSYITGQVLHPNGDMLI 291
AS+ +I GQV+ NG L+
Sbjct: 252 ASD-AGEWINGQVIMANGGCLL 272
>At5g06060 short chain alcohol dehydrogenase-like
Length = 264
Score = 93.2 bits (230), Expect = 2e-19
Identities = 81/257 (31%), Positives = 121/257 (46%), Gaps = 13/257 (5%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L GK A+VTGG GIGRAV A GA V H ++ L+ AN
Sbjct: 9 LAGKTALVTGGTRGIGRAVVEELAKFGAKV-------HTCSRNQEELNACLNDWKANGLV 61
Query: 100 PMAIPADLGFDENCKRVIDEIINAY-GRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
D + +++I E +A+ G+++IL+NN +VE E ++ TN
Sbjct: 62 VSGSVCDASVRDQREKLIQEASSAFSGKLNILINNVGTNVRKPTVEYSSE-EYAKIMSTN 120
Query: 159 IFSYFFMTR--HALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
+ S F +++ H L +I+ +SV S+ Y +TKGA+ TR L+ +
Sbjct: 121 LESAFHLSQIAHPLLKASGVGSIVFISSVAGLVHLSSGSIYGATKGALNQLTRNLACEWA 180
Query: 217 SKGIRVNGVAPGPIWTPLIPASFNEEKTAQ-FGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
S IR N VAP I T L+ +++ + S P+ R G+P EV+ FL S
Sbjct: 181 SDNIRTNCVAPWYIKTSLVETLLEKKEFVEAVVSRTPLGRVGEPEEVSSLVAFLCL-PAS 239
Query: 276 SYITGQVLHPNGDMLIN 292
SYITGQV+ +G +N
Sbjct: 240 SYITGQVISVDGGFTVN 256
>At3g51680 short-chain alcohol dehydrogenase-like protein
Length = 303
Score = 92.8 bits (229), Expect = 2e-19
Identities = 77/270 (28%), Positives = 134/270 (49%), Gaps = 29/270 (10%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L+GK+A++TGG GIG+A LFA GATV+ V D A + L + +++
Sbjct: 31 RLEGKVAIITGGAHGIGKATVMLFARHGATVVIADV----DNVAGSS---LAKSLSSHKT 83
Query: 99 DPMA--IPADLGFDENCKRVIDEIINAYGRIDILVNNA---AEQYECGSVEEIDEPRLER 153
PM I D+ + + + +++ + YGR+DIL NNA +Q + S+ + D +
Sbjct: 84 SPMVAFISCDVSVEADVENLVNVTVARYGRLDILFNNAGVLGDQKKHKSILDFDADEFDH 143
Query: 154 VFRTNIFSYFFMTRHALKHM-KEGSN--IINTTSVNAYKGNSTLIDYTSTKGAIVAFTRA 210
V R N+ +H + M K G II+T SV G YT++K AIV T+
Sbjct: 144 VMRVNVRGVGLGMKHGARAMIKRGFKGCIISTASVAGVMGGMGPHAYTASKHAIVGLTKN 203
Query: 211 LSLQLVSKGIRVNGVAPGPIWTPLIPASFNE-----------EKTAQFGSDVPMKRAG-- 257
+ +L GIRVN ++P + T ++ ++ + E+ +F + +
Sbjct: 204 AACELGKYGIRVNCISPFGVATSMLVNAWRKTSGGDVEDDDVEEMEEFVRSLANLKGETL 263
Query: 258 QPVEVAPSFVFLASNQCSSYITGQVLHPNG 287
+ ++A + ++LAS++ S Y+ G L +G
Sbjct: 264 RANDIAEAALYLASDE-SKYVNGHNLVVDG 292
>At1g52340 short chain alcohol dehydrogenase, putative
Length = 285
Score = 92.8 bits (229), Expect = 2e-19
Identities = 78/269 (28%), Positives = 127/269 (46%), Gaps = 27/269 (10%)
Query: 36 PANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTL--DMLKMAK 93
P+ +L GK+A++TGG +GIG ++ LF GA V D +D L ++ K
Sbjct: 14 PSQRLLGKVALITGGATGIGESIVRLFHKHGAKVCIV--------DLQDDLGGEVCKSLL 65
Query: 94 TANAKDP-MAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAE-QYECGSVEEIDEPRL 151
+K+ I D+ +++ +D + +G +DIL+NNA C +
Sbjct: 66 RGESKETAFFIHGDVRVEDDISNAVDFAVKNFGTLDILINNAGLCGAPCPDIRNYSLSEF 125
Query: 152 ERVFRTNIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFT 208
E F N+ F +HA + M K+GS I++ SV G Y +K A++ T
Sbjct: 126 EMTFDVNVKGAFLSMKHAARVMIPEKKGS-IVSLCSVGGVVGGVGPHSYVGSKHAVLGLT 184
Query: 209 RALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQ---------FGSDVPMKRAGQP 259
R+++ +L GIRVN V+P + T L A EE+ + ++ +K
Sbjct: 185 RSVAAELGQHGIRVNCVSPYAVATKLALAHLPEEERTEDAFVGFRNFAAANANLKGVELT 244
Query: 260 V-EVAPSFVFLASNQCSSYITGQVLHPNG 287
V +VA + +FLAS+ S YI+G L +G
Sbjct: 245 VDDVANAVLFLASDD-SRYISGDNLMIDG 272
>At3g42960 alcohol dehydrogenase (ATA1)
Length = 272
Score = 92.0 bits (227), Expect = 4e-19
Identities = 76/278 (27%), Positives = 128/278 (45%), Gaps = 29/278 (10%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L K+A++TGG GIG A LF GA VI + +E A +
Sbjct: 7 RLFEKVAIITGGARGIGAATARLFTENGAYVIVADILENEG------------ILVAESI 54
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
+ D+ + + + ++ + GR+D++ NNA GS+ +D + ++ N
Sbjct: 55 GGCYVHCDVSKEADVEAAVELAMRRKGRLDVMFNNAGMSLNEGSIMGMDVDMVNKLVSVN 114
Query: 159 IFSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQL 215
+ +HA K M +G +II T+S + G YT +KGAI R + +L
Sbjct: 115 VNGVLHGIKHAAKAMIKGGRGGSIICTSSSSGLMGGLGGHAYTLSKGAINGVVRTTACEL 174
Query: 216 VSKGIRVNGVAPGPIWTPLIPASF----NEEK---------TAQFGSDVPMKRAGQPVEV 262
S GIRVN ++P + T ++ ++ N +K A+ GS + RAG +V
Sbjct: 175 GSHGIRVNSISPHGVPTDILVNAYRKFLNHDKLNVAEVTDIIAEKGS-LLTGRAGTVEDV 233
Query: 263 APSFVFLASNQCSSYITGQVLHPNGDMLINASNVIYLF 300
A + +FLAS + S +ITG L +G S + +++
Sbjct: 234 AQAALFLASQESSGFITGHNLVVDGGYTSATSTMRFIY 271
>At2g47140 alcohol dehydrogenase like protein
Length = 257
Score = 89.0 bits (219), Expect = 3e-18
Identities = 73/256 (28%), Positives = 119/256 (45%), Gaps = 17/256 (6%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L GKI ++TGG SGIG LF GA V+ D +D L +A +
Sbjct: 5 RLDGKIVIITGGASGIGAESVRLFTEHGARVVIV--------DVQDELGQ-NVAVSIGED 55
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
D+ + + + + YG++D+L +NA S+ +++ L+R N
Sbjct: 56 KASYYHCDVTNETEVENAVKFTVEKYGKLDVLFSNAGVIEPFVSILDLNLNELDRTIAIN 115
Query: 159 IFSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQL 215
+ +HA + M E +I+ TTSV A + YT++K ++ ++ S L
Sbjct: 116 LRGTAAFIKHAARAMVEKGIRGSIVCTTSVAAEIAGTAPHGYTTSKHGLLGLIKSASGGL 175
Query: 216 VSKGIRVNGVAPGPIWTPLIPASFN-EEKTAQFGSDVPMKRAG---QPVEVAPSFVFLAS 271
GIRVNGVAP + TPL+ F E + + G + VA + +FLAS
Sbjct: 176 GKYGIRVNGVAPFGVATPLVCNGFKMEPNVVEQNTSASANLKGIVLKARHVAEAALFLAS 235
Query: 272 NQCSSYITGQVLHPNG 287
++ S+Y++GQ L +G
Sbjct: 236 DE-SAYVSGQNLAVDG 250
>At3g29260 short-chain alcohol dehydrogenase, putative
Length = 259
Score = 88.6 bits (218), Expect = 4e-18
Identities = 77/258 (29%), Positives = 118/258 (44%), Gaps = 22/258 (8%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L GKI ++TGG SGIG LF GA V+ D ++ L +A +
Sbjct: 5 RLDGKIVIITGGASGIGAEAARLFTDHGAKVVIV--------DLQEELGQ-NVAVSIGLD 55
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
D+ + + + + +G++D+L +NA GS+ ++D +R N
Sbjct: 56 KASFYRCDITDETEVENAVKFTVEKHGKLDVLFSNAGVMEPHGSILDLDLEAFDRTMAVN 115
Query: 159 IFSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQL 215
+ +HA + M +I+ TTSV A G YT++K A++ R+ L
Sbjct: 116 VRGAAAFIKHAARSMVASGTRGSIVCTTSVTAEIGGPGPHSYTASKHALLGLVRSACGGL 175
Query: 216 VSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRA------GQPVEVAPSFVFL 269
GIRVNGVAP + T L S+NEE T + D A + VA + +FL
Sbjct: 176 GKYGIRVNGVAPYGVATGL--TSYNEE-TVKMVEDYCSATAILKGVVLKARHVADAALFL 232
Query: 270 ASNQCSSYITGQVLHPNG 287
AS+ S YI+GQ L +G
Sbjct: 233 ASDD-SVYISGQNLGVDG 249
>At5g18210 Brn1-like protein
Length = 261
Score = 88.2 bits (217), Expect = 5e-18
Identities = 75/262 (28%), Positives = 118/262 (44%), Gaps = 16/262 (6%)
Query: 38 NKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANA 97
+ L G++A+VTG GIGRA+ A GA ++ Y + D + ++ A T
Sbjct: 6 SSLAGRVAIVTGSSRGIGRAIAIHLAELGAKIVINYTTRSTEAD-QVAAEINSSAGTVPQ 64
Query: 98 KDPMAIPADLGFDENCKRVIDEIINAYGR-IDILVNNAA---EQYECGSVEEIDEPRLER 153
+ AD+ K + D A+ + ILVN+A Y + I+E +R
Sbjct: 65 PIAVVFLADISEPSQIKSLFDAAEKAFNSPVHILVNSAGILNPNYPTIANTPIEE--FDR 122
Query: 154 VFRTNIFSYFFMTRHALKHMKEGSN---IINTTSVNA--YKGNSTLIDYTSTKGAIVAFT 208
+F+ N F + A K +K G I+ T+S+ G YT++K A+ A
Sbjct: 123 IFKVNTRGSFLCCKEAAKRLKRGGGGRIILLTSSLTEALIPGQGA---YTASKAAVEAMV 179
Query: 209 RALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVF 268
+ L+ +L GI N V+PGP+ T + +EE P R G+ ++A F
Sbjct: 180 KILAKELKGLGITANCVSPGPVATEMFFDGKSEETVMNIIERSPFGRLGETKDIASVVGF 239
Query: 269 LASNQCSSYITGQVLHPNGDML 290
LAS+ +I GQV+ NG L
Sbjct: 240 LASDG-GEWINGQVIVANGAFL 260
>At3g29250 short-chain alcohol dehydrogenase, putative
Length = 260
Score = 87.0 bits (214), Expect = 1e-17
Identities = 73/258 (28%), Positives = 123/258 (47%), Gaps = 21/258 (8%)
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMA-KTANA 97
+L GKIA++TGG SGIG LF GA V+ ++ ++ ++ + K + N
Sbjct: 5 RLDGKIAIITGGASGIGAEAVRLFTDHGAKVVIVDIQEELGQNLAVSIGLDKASFYRCNV 64
Query: 98 KDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
D + + + + + +G++D+L +NA GSV ++D +R
Sbjct: 65 TD----------ETDVENAVKFTVEKHGKLDVLFSNAGVLEAFGSVLDLDLEAFDRTMAV 114
Query: 158 NIFSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
N+ +HA + M +I+ TTS+ A G YT++K A++ R+
Sbjct: 115 NVRGAAAFIKHAARSMVASGTRGSIVCTTSIAAEIGGPGPHSYTASKHALLGLIRSACAG 174
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASFNEEKT---AQFGSDVPMKRAG--QPVEVAPSFVFL 269
L GIRVNGVAP + T + A +NEE ++G + + + +A + +FL
Sbjct: 175 LGQYGIRVNGVAPYGVATGMTSA-YNEEAVKMLEEYGEALGNLKGVVLKARHIAEAALFL 233
Query: 270 ASNQCSSYITGQVLHPNG 287
AS+ S YI+GQ L +G
Sbjct: 234 ASDD-SVYISGQNLVVDG 250
>At2g29150 pseudogene
Length = 268
Score = 87.0 bits (214), Expect = 1e-17
Identities = 79/259 (30%), Positives = 122/259 (46%), Gaps = 18/259 (6%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK- 98
L+G A+VTGG G+G AV A+ GA V ARD + + + AK
Sbjct: 16 LEGMTALVTGGSKGLGEAVVEELAMLGARV---------HTCARDETQLQERLREWQAKG 66
Query: 99 -DPMAIPADLGFDENCKRVIDEIINAY-GRIDILVNNAAEQYECGSVEEIDEPRLERVFR 156
+ D+ E +++++ + + + G+++ILVNNA S E E +
Sbjct: 67 FEVTTSVCDVSSREQREKLMETVSSVFQGKLNILVNNAGTGIIKPSTEYTAED-YSFLMA 125
Query: 157 TNIFSYFFMTR--HALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
TN+ S F +++ H L +I+ +SV I Y ++KGA+ R+L+ +
Sbjct: 126 TNLESAFHLSQIAHPLLKASGSGSIVFMSSVAGLVHTGASI-YGASKGAMNQLGRSLACE 184
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSD-VPMKRAGQPVEVAPSFVFLASNQ 273
S IRVN V P I TPL F++EK + D PM R G+ EV+ FL
Sbjct: 185 WASDNIRVNSVCPWVITTPLTSFIFSDEKLRKAVEDKTPMGRVGEANEVSSLVAFLCF-P 243
Query: 274 CSSYITGQVLHPNGDMLIN 292
+SYITGQ + +G +N
Sbjct: 244 AASYITGQTICVDGGASVN 262
>At2g29350 putative tropinone reductase
Length = 269
Score = 86.7 bits (213), Expect = 2e-17
Identities = 80/259 (30%), Positives = 119/259 (45%), Gaps = 18/259 (6%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L G A+VTGG GIG AV A+ GA V ARD + + + AK
Sbjct: 15 LGGMTALVTGGSKGIGEAVVEELAMLGAKV---------HTCARDETQLQERLREWQAKG 65
Query: 100 PMAIPA--DLGFDENCKRVIDEIINAY-GRIDILVNNAAEQYECGSVEEIDEPRLERVFR 156
+ D+ + ++++ + + Y G+++ILVNN + E E V
Sbjct: 66 FQVTTSVCDVSSRDQRVKLMETVSSLYQGKLNILVNNVGTSIFKPTTEYTAED-FSFVMA 124
Query: 157 TNIFSYFFMTRHA---LKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSL 213
TN+ S F +++ A LK GS ++ +++ N I Y +TKGA+ R L+
Sbjct: 125 TNLESAFHLSQLAHPLLKASGSGSIVLISSAAGVVHVNVGSI-YGATKGAMNQLARNLAC 183
Query: 214 QLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQ 273
+ S IR N V P I TPL F+EE + PM R G+ EV+P FL
Sbjct: 184 EWASDNIRTNSVCPWYITTPLSNDFFDEEFKKEAVRTTPMGRVGEANEVSPLVAFLCL-P 242
Query: 274 CSSYITGQVLHPNGDMLIN 292
+SYITGQ + +G +N
Sbjct: 243 SASYITGQTICVDGGATVN 261
>At2g29260 tropinone reductase like protein
Length = 322
Score = 86.7 bits (213), Expect = 2e-17
Identities = 79/259 (30%), Positives = 119/259 (45%), Gaps = 17/259 (6%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L G A+VTGG GIGRA+ A GA V +E ++ + + D
Sbjct: 68 LNGMSALVTGGTRGIGRAIVEELAGLGAEVHTCARNEYELENCLSDWNRSGFRVAGSVCD 127
Query: 100 PMAIPADLGFDENCKRVIDEIINAY--GRIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
D + + + E +++ G++ ILVNN VE + T
Sbjct: 128 VS--------DRSQREALMETVSSVFEGKLHILVNNVGTNIRKPMVE-FTAGEFSTLMST 178
Query: 158 NIFSYFFMTRHA---LKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
N S F + + A L+ K GS ++ +SV+ + + +STKGAI TR+L+ +
Sbjct: 179 NFESVFHLCQLAYPLLRESKAGS-VVFISSVSGFVSLKNMSVQSSTKGAINQLTRSLACE 237
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASF-NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQ 273
IR+N VAP I T ++ N+E + S P+ R G+P EV+ + FL
Sbjct: 238 WAKDNIRINAVAPWYIKTSMVEQVLSNKEYLEEVYSVTPLGRLGEPREVSSAVAFLCL-P 296
Query: 274 CSSYITGQVLHPNGDMLIN 292
SSYITGQ+L +G M IN
Sbjct: 297 ASSYITGQILCVDGGMSIN 315
>At1g07440 tropinone reductase-I, putative
Length = 266
Score = 86.7 bits (213), Expect = 2e-17
Identities = 74/255 (29%), Positives = 114/255 (44%), Gaps = 10/255 (3%)
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+ K +VTGG GIG A+ FA GA + +E + T + D
Sbjct: 12 LKAKTVLVTGGTKGIGHAIVEEFAGFGAVIHTCARNEYELNECLSKWQKKGFQVTGSVCD 71
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
P E + + + G++DIL+NN +++ E + TN+
Sbjct: 72 ASLRPER----EKLMQTVSSMFG--GKLDILINNLGAIRSKPTLDYTAEDFSFHI-STNL 124
Query: 160 FSYFFMTR--HALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVS 217
S + +++ H L NII +S+ S Y++TKGA+ R L+ + S
Sbjct: 125 ESAYHLSQLAHPLLKASGCGNIIFMSSIAGVVSASVGSIYSATKGALNQLARNLACEWAS 184
Query: 218 KGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSY 277
GIR N VAP I TPL A +++E S P+ R G+P EV+ FL +SY
Sbjct: 185 DGIRANAVAPAVIATPLAEAVYDDEFKKVVISRKPLGRFGEPEEVSSLVAFLCM-PAASY 243
Query: 278 ITGQVLHPNGDMLIN 292
ITGQ + +G + +N
Sbjct: 244 ITGQTICVDGGLTVN 258
>At5g10050 unknown protein
Length = 279
Score = 86.3 bits (212), Expect = 2e-17
Identities = 66/207 (31%), Positives = 99/207 (46%), Gaps = 19/207 (9%)
Query: 41 QGKIAVVTG-GDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
+ + ++TG GIG A+ F +G V+ T +R T+ L+ K+
Sbjct: 6 ESPVVLITGCSQGGIGHALAREFTEKGCRVVAT-------SRSRSTMTDLEQDSRLFVKE 58
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNI 159
D+ D+N +V+ E+I+ +G+ID+LVNNA Q G + E +E F TN+
Sbjct: 59 -----LDVQSDQNVSKVLSEVIDKFGKIDVLVNNAGVQC-VGPLAETPISAMENTFNTNV 112
Query: 160 FSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVS 217
F MT+ + HM K+ I+N S+ YT+TK AI A T L L+L
Sbjct: 113 FGSMRMTQAVVPHMVSKKKGKIVNVGSITVMAPGPWAGVYTATKAAIHALTDTLRLELRP 172
Query: 218 KGIRVNGVAPGPIWTPL---IPASFNE 241
GI V V PG I T + A+FN+
Sbjct: 173 FGIDVINVVPGGIRTNIANSAVATFNK 199
>At3g03980 putative short-chain type dehydrogenase/reductase
Length = 270
Score = 85.1 bits (209), Expect = 5e-17
Identities = 68/260 (26%), Positives = 111/260 (42%), Gaps = 8/260 (3%)
Query: 35 KPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKD-ARDTLDMLKMAK 93
+P L G++A+VTG GIGRA+ A GA ++ Y D + ++ + +
Sbjct: 9 QPPLPLAGRVAIVTGSSRGIGRAIAIHLAELGARIVINYTSKAADAERVASEINDFPVRE 68
Query: 94 TANAKDPMAIPADLGFDE--NCKRVIDEIINAY-GRIDILVNNAAE-QYECGSVEEIDEP 149
K P AI E K + D +A+ + ILVN+A + ++ +
Sbjct: 69 EITGKGPRAIVVQANVSEPSQVKSMFDAAESAFEAPVHILVNSAGILDPKYPTIADTSVE 128
Query: 150 RLERVFRTNIFSYFFMTRHALKHMKEGSN--IINTTSVNAYKGNSTLIDYTSTKGAIVAF 207
+ F N F ++ A +K+G II TS Y ++K A+
Sbjct: 129 DFDHTFSVNTKGAFLCSKEAANRLKQGGGGRIILLTSSQTRSLKPGFGAYAASKAAVETM 188
Query: 208 TRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFV 267
+ L+ +L GI N VAPGPI T + E + ++ P R G+ +V P
Sbjct: 189 VKILAKELKGTGITANCVAPGPIATEMFFDGKTPELVEKIAAESPFGRVGEAKDVVPLVG 248
Query: 268 FLASNQCSSYITGQVLHPNG 287
FLA + ++ GQ++ NG
Sbjct: 249 FLAGDG-GEWVNGQIIPVNG 267
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,324,307
Number of Sequences: 26719
Number of extensions: 310033
Number of successful extensions: 898
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 71
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 714
Number of HSP's gapped (non-prelim): 106
length of query: 323
length of database: 11,318,596
effective HSP length: 99
effective length of query: 224
effective length of database: 8,673,415
effective search space: 1942844960
effective search space used: 1942844960
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135319.2


