

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.10 + phase: 1 /pseudo/partial
(430 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
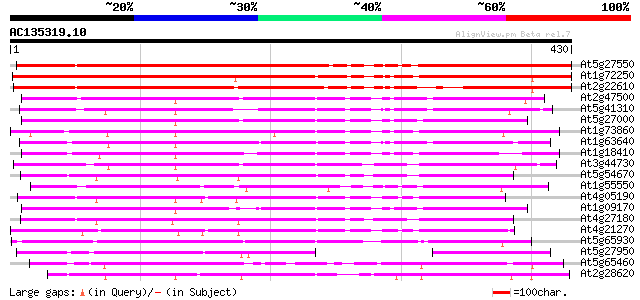
Score E
Sequences producing significant alignments: (bits) Value
At5g27550 kinesin-like protein 495 e-140
At1g72250 kinesin, putative 362 e-100
At2g22610 putative kinesin heavy chain 320 7e-88
At2g47500 kinesin heavy chain like protein 229 2e-60
At5g41310 kinesin-like protein 224 9e-59
At5g27000 kinesin-like heavy chain (KATD) 224 9e-59
At1g73860 kinesin-related protein 222 3e-58
At1g63640 kinesin-like protein 216 3e-56
At1g18410 kinesin-related protein, putative 216 3e-56
At3g44730 kinesin-like protein heavy chain (KP1) 210 1e-54
At5g54670 heavy chain polypeptide of kinesin-like protein (katC) 209 2e-54
At1g55550 207 1e-53
At4g05190 kinesin - like protein 202 3e-52
At1g09170 putative kinesin 201 6e-52
At4g27180 heavy chain polypeptide of kinesin-like protein (katB) 199 2e-51
At4g21270 kinesin-related protein katA 198 4e-51
At5g65930 kinesin-like calmodulin-binding protein 181 5e-46
At5g27950 unknown protein 157 1e-38
At5g65460 putative protein 156 2e-38
At2g28620 putative kinesin-like spindle protein 152 4e-37
>At5g27550 kinesin-like protein
Length = 425
Score = 495 bits (1274), Expect = e-140
Identities = 272/426 (63%), Positives = 326/426 (75%), Gaps = 20/426 (4%)
Query: 6 LKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSE-ELQ 64
L+++YLE S ER+RL NEVIELKGNIRVFCRCRPLN+ EIANG A SV F++ E ELQ
Sbjct: 15 LEKQYLEESSERKRLYNEVIELKGNIRVFCRCRPLNQAEIANGCA-SVAEFDTTQENELQ 73
Query: 65 VVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTME 124
++ SDSSKK FKFDHVFKP+D QE VFAQTKPIV SVLDG+NVCIFAYGQTGTGKTFTME
Sbjct: 74 ILSSDSSKKHFKFDHVFKPDDGQETVFAQTKPIVTSVLDGYNVCIFAYGQTGTGKTFTME 133
Query: 125 GTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKLEV 184
GTPE+RGVNYRTLEELFR SE + +K+EL VSMLEVYNEKI+DLL NS++ KKLEV
Sbjct: 134 GTPENRGVNYRTLEELFRCSESKSHLMKFELSVSMLEVYNEKIRDLLVDNSNQPPKKLEV 193
Query: 185 KQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVDD 244
KQ+A+GTQEVPGLVE VY DGVW++LK G VRSVGST+ANE SSRSH + + +
Sbjct: 194 KQSAEGTQEVPGLVEAQVYNTDGVWDLLKKGYAVRSVGSTAANEQSSRSHCLLRVTVKGE 253
Query: 245 KKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*ISLY 304
+++ LW + +A ++V V V G+ +LKE N S +S
Sbjct: 254 N--LINGQRTRSHLW--LVDLAGSERVG-------KVEVEGE-RLKESQFINKS--LSAL 299
Query: 305 HHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLN 364
++ L ++ +++ RNSKLTH+LQ+SLGGDCKTLMFVQISPSS DL ETLCSLN
Sbjct: 300 GDVI----SALASKTSHIPYRNSKLTHMLQNSLGGDCKTLMFVQISPSSADLGETLCSLN 355
Query: 365 FATRVRGIESGPARKQVDLTELLKYKQMAEKSKHDEKEARKLQDNLQSVQMRLATREFMC 424
FA+RVRGIESGPARKQ D++ELLK KQMAEK KH+EKE +KLQDN+QS+Q+RL RE +C
Sbjct: 356 FASRVRGIESGPARKQADVSELLKSKQMAEKLKHEEKETKKLQDNVQSLQLRLTAREHIC 415
Query: 425 RNLQDK 430
R LQDK
Sbjct: 416 RGLQDK 421
>At1g72250 kinesin, putative
Length = 1195
Score = 362 bits (930), Expect = e-100
Identities = 205/434 (47%), Positives = 284/434 (65%), Gaps = 24/434 (5%)
Query: 3 YEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEE 62
+E LK K++ ER+ L N+++ELKGNIRVFCRCRPLN E G ++ + + + E
Sbjct: 464 HENLKVKFVAGEKERKELYNKILELKGNIRVFCRCRPLNFEETEAGVSMGIDVESTKNGE 523
Query: 63 LQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFT 122
+ V+ + KK FKFD VF P +Q VF T P SV+DG+NVCIFAYGQTGTGKTFT
Sbjct: 524 VIVMSNGFPKKSFKFDSVFGPNASQADVFEDTAPFATSVIDGYNVCIFAYGQTGTGKTFT 583
Query: 123 MEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLL--AGNSSEATK 180
MEGT RGVNYRTLE LFR+ + R+ YE+ VS+LEVYNE+I+DLL A S+ A K
Sbjct: 584 MEGTQHDRGVNYRTLENLFRIIKAREHRYNYEISVSVLEVYNEQIRDLLVPASQSASAPK 643
Query: 181 KLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYI 240
+ E++Q ++G VPGLVE V + VW++LK+G+ R+VG T+ANE SSRSH +
Sbjct: 644 RFEIRQLSEGNHHVPGLVEAPVKSIEEVWDVLKTGSNARAVGKTTANEHSSRSH--CIHC 701
Query: 241 IVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS* 300
++ + +L+ LW + +A ++V V G+ +LKE +N++
Sbjct: 702 VMVKGENLLNGECTKSKLW--LVDLAGSERVA-------KTEVQGE-RLKET--QNINKS 749
Query: 301 ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETL 360
+S ++ L N+ +++ RNSKLTH+LQ SLGGD KTLMFVQISP+ D +ETL
Sbjct: 750 LSALGDVIF----ALANKSSHIPFRNSKLTHLLQDSLGGDSKTLMFVQISPNENDQSETL 805
Query: 361 CSLNFATRVRGIESGPARKQVDLTELLKYKQMAEKSKHD----EKEARKLQDNLQSVQMR 416
CSLNFA+RVRGIE GPA+KQ+D TELLKYKQM EK K D +++ RK+++ + ++ +
Sbjct: 806 CSLNFASRVRGIELGPAKKQLDNTELLKYKQMVEKWKQDMKGKDEQIRKMEETMYGLEAK 865
Query: 417 LATREFMCRNLQDK 430
+ R+ + LQDK
Sbjct: 866 IKERDTKNKTLQDK 879
>At2g22610 putative kinesin heavy chain
Length = 1068
Score = 320 bits (821), Expect = 7e-88
Identities = 191/432 (44%), Positives = 273/432 (62%), Gaps = 51/432 (11%)
Query: 4 EILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSE-E 62
E LK+KY E +R+ L N + E KGNIRVFCRCRPLN E + SA ++V+F+ + E
Sbjct: 401 EDLKQKYSEEQAKRKELYNHIQETKGNIRVFCRCRPLNTEETSTKSA-TIVDFDGAKDGE 459
Query: 63 LQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFT 122
L V+ ++SKK FKFD V+ P+D Q VFA P+V SVLDG+NVCIFAYGQTGTGKTFT
Sbjct: 460 LGVITGNNSKKSFKFDRVYTPKDGQVDVFADASPMVVSVLDGYNVCIFAYGQTGTGKTFT 519
Query: 123 MEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKL 182
MEGTP++RGVNYRT+E+LF V+ ER+ TI Y + VS+LEVYNE+I+DLLA +S +KKL
Sbjct: 520 MEGTPQNRGVNYRTVEQLFEVARERRETISYNISVSVLEVYNEQIRDLLA--TSPGSKKL 577
Query: 183 EVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIV 242
E+KQ++DG+ VPGLVE +V + VW +L++G+ RSVGS + NE SSRSH + ++
Sbjct: 578 EIKQSSDGSHHVPGLVEANVENINEVWNVLQAGSNARSVGSNNVNEHSSRSHCMLSIMV- 636
Query: 243 DDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*IS 302
K +++ LW + +A +++ V G+ +LKE +N++ +S
Sbjct: 637 -KAKNLMNGDCTKSKLW--LVDLAGSERLA-------KTDVQGE-RLKEA--QNINRSLS 683
Query: 303 LYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCS 362
++ L +K +HI SPS D++ETL S
Sbjct: 684 ALGDVIYALA--------------TKSSHI---------------PYSPSEHDVSETLSS 714
Query: 363 LNFATRVRGIESGPARKQVDLTELLKYKQMAEKSKHD----EKEARKLQDNLQSVQMRLA 418
LNFATRVRG+E GPARKQVD E+ K K M EK++ + ++ +K+++N+Q+++ +
Sbjct: 715 LNFATRVRGVELGPARKQVDTGEIQKLKAMVEKARQESRSKDESIKKMEENIQNLEGKNK 774
Query: 419 TREFMCRNLQDK 430
R+ R+LQ+K
Sbjct: 775 GRDNSYRSLQEK 786
>At2g47500 kinesin heavy chain like protein
Length = 983
Score = 229 bits (585), Expect = 2e-60
Identities = 152/408 (37%), Positives = 232/408 (56%), Gaps = 29/408 (7%)
Query: 10 YLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVCSD 69
Y V E R+L N+V +LKG+IRV+CR RP + + S + N E ++ +
Sbjct: 379 YHRVLEENRKLYNQVQDLKGSIRVYCRVRPFLPGQSSFSSTIG--NMEDDTIGINTASRH 436
Query: 70 S-SKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEG--- 125
S K F F+ VF P QE VF+ +P++ SVLDG+NVCIFAYGQTG+GKTFTM G
Sbjct: 437 GKSLKSFTFNKVFGPSATQEEVFSDMQPLIRSVLDGYNVCIFAYGQTGSGKTFTMSGPRD 496
Query: 126 -TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKLEV 184
T + +GVNYR L +LF ++E+R+ T +Y++ V M+E+YNE+++DLL + S K+LE+
Sbjct: 497 LTEKSQGVNYRALGDLFLLAEQRKDTFRYDIAVQMIEIYNEQVRDLLVTDGS--NKRLEI 554
Query: 185 KQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVDD 244
+ ++ VP V V +++K+G++ R+VGST+ N+ SSRSH + V
Sbjct: 555 RNSSQKGLSVPDASLVPVSSTFDVIDLMKTGHKNRAVGSTALNDRSSRSH-SCLTVHVQG 613
Query: 245 KKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*ISLY 304
+ + AV G + + +A ++V +T +LKE ++++ +S
Sbjct: 614 RDLTSGAVLRGCM---HLVDLAGSERVDKSEVT--------GDRLKEA--QHINRSLSAL 660
Query: 305 HHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLN 364
++ L P+ P RNSKLT +LQ SLGG KTLMFV ISP + + ET+ +L
Sbjct: 661 GDVIASLAHKNPHVPY----RNSKLTQLLQDSLGGQAKTLMFVHISPEADAVGETISTLK 716
Query: 365 FATRVRGIESGPARKQVDLTELLKYKQMAE--KSKHDEKEARKLQDNL 410
FA RV +E G AR D +++ + K+ K+ KEA Q+N+
Sbjct: 717 FAERVATVELGAARVNNDTSDVKELKEQIATLKAALARKEAESQQNNI 764
>At5g41310 kinesin-like protein
Length = 967
Score = 224 bits (570), Expect = 9e-59
Identities = 155/423 (36%), Positives = 233/423 (54%), Gaps = 57/423 (13%)
Query: 8 RKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVC 67
+ Y + E RRL NEV ELKGNIRV+CR RP + + +++ E E ++V
Sbjct: 406 KNYQIIIEENRRLYNEVQELKGNIRVYCRIRPFLQGQNKKQTSI-----EYTGENGELVV 460
Query: 68 SDSSK------KQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTF 121
++ K + FKF+ VF PE QE VF T+P++ S+LDG+NVCIFAYGQTG+GKT+
Sbjct: 461 ANPLKQGKDTYRLFKFNKVFGPESTQEEVFLDTRPMIRSILDGYNVCIFAYGQTGSGKTY 520
Query: 122 TMEG----TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSE 177
TM G + E RGVNYR L +LF +++ RQ ++ YE+ V M+E+YNE+++DLL
Sbjct: 521 TMSGPSITSEEDRGVNYRALNDLFHLTQSRQNSVMYEVGVQMVEIYNEQVRDLL------ 574
Query: 178 ATKKLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQI 237
+Q+VP V + V E++ G R+VG+T+ NE SSRSH +
Sbjct: 575 -------------SQDVPDASMHSVRSTEDVLELMNIGLMNRTVGATTLNEKSSRSH-SV 620
Query: 238 FYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNL 297
+ V + +V G L + +A ++V +T G+ +LKE +++
Sbjct: 621 LSVHVRGVDVKTESVLRGSL---HLVDLAGSERVGRSEVT-------GE-RLKEA--QHI 667
Query: 298 SS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLT 357
+ +S ++ L P+ P RNSKLT +LQ+SLGG KTLMFVQI+P
Sbjct: 668 NKSLSALGDVIFALAHKNPHVPY----RNSKLTQVLQNSLGGQAKTLMFVQINPDEDSYA 723
Query: 358 ETLCSLNFATRVRGIESGPARKQV---DLTELL-KYKQMAEKSKHDEKEARKLQDNLQSV 413
ET+ +L FA RV G+E G AR D+ +L+ + + + ++E +K Q N+ +
Sbjct: 724 ETVSTLKFAERVSGVELGAARSYKEGRDVRQLMEQVSNLKDMIAKKDEELQKFQ-NINGI 782
Query: 414 QMR 416
Q R
Sbjct: 783 QKR 785
>At5g27000 kinesin-like heavy chain (KATD)
Length = 987
Score = 224 bits (570), Expect = 9e-59
Identities = 150/393 (38%), Positives = 220/393 (55%), Gaps = 25/393 (6%)
Query: 10 YLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANG-SAVSVVNFESNSEELQVVCS 68
Y V E R+L N V +LKGNIRV+CR RP + + G SAV ++ + + +
Sbjct: 374 YKRVLEENRKLYNLVQDLKGNIRVYCRVRPFLPGQESGGLSAVEDIDEGTITIRVPSKYG 433
Query: 69 DSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEG--- 125
+ +K F F+ VF P QE VF+ +P+V SVLDG+NVCIFAYGQTG+GKTFTM G
Sbjct: 434 KAGQKPFMFNKVFGPSATQEEVFSDMQPLVRSVLDGYNVCIFAYGQTGSGKTFTMTGPKE 493
Query: 126 -TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKLEV 184
T E GVNYR L +LF +S +R+ T YE+ V MLE+YNE+++DLLA + TK+LE+
Sbjct: 494 LTEESLGVNYRALADLFLLSNQRKDTTSYEISVQMLEIYNEQVRDLLAQDGQ--TKRLEI 551
Query: 185 KQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVDD 244
+ + VP V D V +++ G+ R+V ST+ N+ SSRSH + V
Sbjct: 552 RNNSHNGINVPEASLVPVSSTDDVIQLMDLGHMNRAVSSTAMNDRSSRSH-SCVTVHVQG 610
Query: 245 KKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*ISLY 304
+ + ++ G + + +A ++V +T +LKE ++++ +S
Sbjct: 611 RDLTSGSILHGSM---HLVDLAGSERVDKSEVT--------GDRLKEA--QHINKSLSAL 657
Query: 305 HHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLN 364
++ L + +++ RNSKLT +LQ SLGG KTLMFV ISP L ET+ +L
Sbjct: 658 GDVISSLS----QKTSHVPYRNSKLTQLLQDSLGGSAKTLMFVHISPEPDTLGETISTLK 713
Query: 365 FATRVRGIESGPARKQVDLTELLKYKQMAEKSK 397
FA RV +E G AR D +E+ + K+ K
Sbjct: 714 FAERVGSVELGAARVNKDNSEVKELKEQIANLK 746
>At1g73860 kinesin-related protein
Length = 1050
Score = 222 bits (566), Expect = 3e-58
Identities = 159/448 (35%), Positives = 246/448 (54%), Gaps = 48/448 (10%)
Query: 1 DEYEILKRKYLEVS----------LERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSA 50
D++ L +K +E+S E R+L NE+ ELKGNIRVFCR RP A G+A
Sbjct: 490 DQFSQLGKKLIELSNAAENYHAVLTENRKLFNELQELKGNIRVFCRVRPFLP---AQGAA 546
Query: 51 VSVVNFESNSEELQVVCSDSSKK----QFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHN 106
+VV + EL V K QFKF+ V+ P +Q VF+ +P+V SVLDG+N
Sbjct: 547 NTVVEYVGEDGELVVTNPTRPGKDGLRQFKFNKVYSPTASQADVFSDIRPLVRSVLDGYN 606
Query: 107 VCIFAYGQTGTGKTFTMEG----TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEV 162
VCIFAYGQTG+GKT+TM G + E GVNYR L +LF++S+ R+G I YE+ V M+E+
Sbjct: 607 VCIFAYGQTGSGKTYTMTGPDGSSEEDWGVNYRALNDLFKISQSRKGNISYEVGVQMVEI 666
Query: 163 YNEKIKDLLAGNSSEATKKLEVKQAADGTQEVPGLVETH-----VYGADGVWEILKSGNR 217
YNE++ DLL+ ++S+ + T + GL V V ++ G +
Sbjct: 667 YNEQVLDLLSDDNSQKKYPFVLNPGILSTTQQNGLAVPDASMYPVTSTSDVITLMDIGLQ 726
Query: 218 VRSVGSTSANELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT 277
R+VGST+ NE SSRSH I + V K + +V G L + +A ++V +T
Sbjct: 727 NRAVGSTALNERSSRSH-SIVTVHVRGKDLKTGSVLYGNL---HLVDLAGSERVDRSEVT 782
Query: 278 *LAVSV*GKLKLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSL 337
+L+E ++++ +S ++ L ++ +++ RNSKLT +LQ+SL
Sbjct: 783 --------GDRLREA--QHINKSLSSLGDVIF----SLASKSSHVPYRNSKLTQLLQTSL 828
Query: 338 GGDCKTLMFVQISPSSVDLTETLCSLNFATRVRGIESGPA---RKQVDLTELL-KYKQMA 393
GG KTLMFVQ++P + +E++ +L FA RV G+E G A ++ D+ +L+ + +
Sbjct: 829 GGRAKTLMFVQLNPDATSYSESMSTLKFAERVSGVELGAAKTSKEGKDVRDLMEQLASLK 888
Query: 394 EKSKHDEKEARKLQDNLQSVQMRLATRE 421
+ ++E +LQ Q +Q + R+
Sbjct: 889 DTIARKDEEIERLQHQPQRLQKSMMRRK 916
>At1g63640 kinesin-like protein
Length = 1071
Score = 216 bits (549), Expect = 3e-56
Identities = 151/415 (36%), Positives = 225/415 (53%), Gaps = 33/415 (7%)
Query: 8 RKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVC 67
+ Y V E RRL NEV ELKGNIRV+CR RP + S + + + + EL V
Sbjct: 450 KNYHVVLEENRRLYNEVQELKGNIRVYCRIRPFLPGQ---NSRQTTIEYIGETGELVVAN 506
Query: 68 SDSSKKQ----FKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTM 123
K FKF+ VF QE VF T+P++ S+LDG+NVCIFAYGQTG+GKT+TM
Sbjct: 507 PFKQGKDTHRLFKFNKVFDQAATQEEVFLDTRPLIRSILDGYNVCIFAYGQTGSGKTYTM 566
Query: 124 EG----TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEAT 179
G + E GVNYR L +LF +++ RQ T+ YE+ V M+E+YNE+++D+L+ S
Sbjct: 567 SGPSITSKEDWGVNYRALNDLFLLTQSRQNTVMYEVGVQMVEIYNEQVRDILSDGGSSRR 626
Query: 180 KKLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFY 239
++ +G VP V + V E++ G R+VG+T+ NE SSRSH +
Sbjct: 627 YRIWNTALPNGL-AVPDASMHCVRSTEDVLELMNIGLMNRTVGATALNERSSRSHC-VLS 684
Query: 240 IIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS 299
+ V + ++ G L + +A ++V T G+ +LKE ++++
Sbjct: 685 VHVRGVDVETDSILRGSL---HLVDLAGSERVDRSEAT-------GE-RLKEA--QHINK 731
Query: 300 *ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTET 359
+S ++ L P+ P RNSKLT +LQSSLGG KTLMFVQ++P ET
Sbjct: 732 SLSALGDVIFALAHKNPHVPY----RNSKLTQVLQSSLGGQAKTLMFVQVNPDGDSYAET 787
Query: 360 LCSLNFATRVRGIESGPARKQVDLTELLKYKQMAEKSKHDEKEARKLQDNLQSVQ 414
+ +L FA RV G+E G A+ + ++ +Q+ E+ + + K + LQ+ Q
Sbjct: 788 VSTLKFAERVSGVELGAAKSSKEGRDV---RQLMEQVSNLKDVIAKKDEELQNFQ 839
>At1g18410 kinesin-related protein, putative
Length = 1081
Score = 216 bits (549), Expect = 3e-56
Identities = 151/420 (35%), Positives = 228/420 (53%), Gaps = 55/420 (13%)
Query: 10 YLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVC-- 67
Y EV E ++L NE+ ELKGNIRV+CR RP + G++ +VV + EL V+
Sbjct: 612 YHEVLTENQKLFNELQELKGNIRVYCRVRPFLRGQ---GASKTVVEHIGDHGELVVLNPT 668
Query: 68 --SDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEG 125
+ ++F+F+ V+ P Q VF+ KP++ SVLDG+NVCIFAYGQTG+GKT+TM G
Sbjct: 669 KPGKDAHRKFRFNKVYSPASTQAEVFSDIKPLIRSVLDGYNVCIFAYGQTGSGKTYTMTG 728
Query: 126 ----TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKK 181
+ E GVNYR L +LFR+S+ R+ I YE+ V M+E+YNE+++DLL+G S +
Sbjct: 729 PDGASEEEWGVNYRALNDLFRISQSRKSNIAYEVGVQMVEIYNEQVRDLLSGILSTTQQ- 787
Query: 182 LEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYII 241
VP V V E++ G + R V ST+ NE SSRSH I +
Sbjct: 788 --------NGLAVPDASMYPVTSTSDVLELMSIGLQNRVVSSTALNERSSRSH-SIVTVH 838
Query: 242 VDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*I 301
V K + + G L + +A ++V +T +LKE ++++ +
Sbjct: 839 VRGKDLKTGSALYGNL---HLVDLAGSERVDRSEVT--------GDRLKEA--QHINKSL 885
Query: 302 SLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLC 361
S ++ L ++ +++ RNSKLT +LQSSLGG KTLMFVQ++P +E++
Sbjct: 886 SALGDVIF----SLASKSSHVPYRNSKLTQLLQSSLGGRAKTLMFVQLNPDITSYSESMS 941
Query: 362 SLNFATRVRGIESGPARKQVDLTELLKYKQMAEKSKHDEKEARKLQDNLQSVQMRLATRE 421
+L FA RV G+E G A KS D ++ R+L + L S++ +A ++
Sbjct: 942 TLKFAERVSGVELG-----------------AAKSSKDGRDVRELMEQLGSLKDTIARKD 984
>At3g44730 kinesin-like protein heavy chain (KP1)
Length = 1087
Score = 210 bits (534), Expect = 1e-54
Identities = 149/429 (34%), Positives = 232/429 (53%), Gaps = 40/429 (9%)
Query: 4 EILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEEL 63
E+ Y +V E R L NEV +LKG IRV+CR RP + + S V + N
Sbjct: 351 EVTSSSYHKVLEENRLLYNEVQDLKGTIRVYCRVRPFFQEQKDMQSTVDYIGENGN---- 406
Query: 64 QVVCSDSSKKQ------FKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGT 117
++ ++ K++ F F+ VF +QE ++ T+P++ SVLDG NVCIFAYGQTG+
Sbjct: 407 -IIINNPFKQEKDARKIFSFNKVFGQTVSQEQIYIDTQPVIRSVLDGFNVCIFAYGQTGS 465
Query: 118 GKTFTMEG----TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAG 173
GKT+TM G T GVNYR L +LF++S R + YE+ V M+E+YNE+++DLL
Sbjct: 466 GKTYTMSGPDLMTETTWGVNYRALRDLFQLSNARTHVVTYEIGVQMIEIYNEQVRDLLVS 525
Query: 174 NSSEATKKLEVKQAAD-GTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSR 232
+ S +++L+++ + VP V V ++++ G + R+VG+T+ NE SSR
Sbjct: 526 DGS--SRRLDIRNNSQLNGLNVPDANLIPVSNTRDVLDLMRIGQKNRAVGATALNERSSR 583
Query: 233 SHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEK 292
SH + + V K++ ++ G L + R +K + +V +LK +
Sbjct: 584 SH-SVLTVHVQGKELASGSILRGCLHLVDLAGSERVEK---------SEAVGERLKEAQH 633
Query: 293 G*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPS 352
++LS+ + + L + +++ RNSKLT +LQ SLGG KTLMFV I+P
Sbjct: 634 INKSLSALGDVIY--------ALAQKSSHVPYRNSKLTQVLQDSLGGQAKTLMFVHINPE 685
Query: 353 SVDLTETLCSLNFATRVRGIESGPARKQVDLTEL--LKYKQMAEKSKHDEKEARKLQDNL 410
+ ET+ +L FA RV IE G AR + E+ LK + + KS ++KEA + L
Sbjct: 686 VNAVGETISTLKFAQRVASIELGAARSNKETGEIRDLKDEISSLKSAMEKKEAE--LEQL 743
Query: 411 QSVQMRLAT 419
+S +R T
Sbjct: 744 RSGSIRNTT 752
>At5g54670 heavy chain polypeptide of kinesin-like protein (katC)
Length = 754
Score = 209 bits (532), Expect = 2e-54
Identities = 150/392 (38%), Positives = 209/392 (53%), Gaps = 33/392 (8%)
Query: 9 KYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQV--- 65
K +E R++L+N ++ELKGNIRVFCR RPL E NG +++ ++ E L
Sbjct: 375 KLVEGEKLRKKLHNTILELKGNIRVFCRVRPLLPGE-NNGDEGKTISYPTSLEALGRGID 433
Query: 66 VCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEG 125
+ ++ K F FD VF P +QE VF + +V S LDG+ VCIFAYGQTG+GKT+TM G
Sbjct: 434 LMQNAQKHAFTFDKVFAPTASQEDVFTEISQLVQSALDGYKVCIFAYGQTGSGKTYTMMG 493
Query: 126 TP---EHRGVNYRTLEELFRVSEE-RQGTIKYELLVSMLEVYNEKIKDLLAGN------- 174
P E +G+ R LE++F + R KYEL VSMLE+YNE I+DLL+ N
Sbjct: 494 RPGNVEEKGLIPRCLEQIFETRQSLRSQGWKYELQVSMLEIYNETIRDLLSTNKEAVRTD 553
Query: 175 SSEATKKLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSH 234
S + +K +K A G V L V + V +L R RSVG T NE SSRSH
Sbjct: 554 SGVSPQKHAIKHDASGNTHVAELTILDVKSSREVSFLLDHAARNRSVGKTQMNEQSSRSH 613
Query: 235 WQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG* 294
+ +F + + G L + +A ++++ + S +LK +
Sbjct: 614 F-VFTLRISGVNESTEQQVQGVL---NLIDLAGSERLS------KSGSTGDRLKETQAIN 663
Query: 295 RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSV 354
++LSS + L + ++ RNSKLT++LQ LGGD KTLMFV I+P S
Sbjct: 664 KSLSSLGDVIF--------ALAKKEDHVPFRNSKLTYLLQPCLGGDAKTLMFVNIAPESS 715
Query: 355 DLTETLCSLNFATRVRGIESGPARKQVDLTEL 386
E+LCSL FA RV E G R+Q ++ L
Sbjct: 716 STGESLCSLRFAARVNACEIGTPRRQTNIKPL 747
>At1g55550
Length = 887
Score = 207 bits (526), Expect = 1e-53
Identities = 142/416 (34%), Positives = 220/416 (52%), Gaps = 52/416 (12%)
Query: 17 RRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVCSDSSKKQFK 76
RR++ NE ++LKGNIRVFCR +PL E S ++ + + S++ +K +
Sbjct: 78 RRQILNEFLDLKGNIRVFCRVKPLGATEKLRPPVAS------DTRNVIIKLSETKRKTYN 131
Query: 77 FDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEGTPEHRGVNYRT 136
FD VF+P+ +Q+ VF + +P++ SV+DG+N CIFAYGQTGTGKT+TMEG P G+ R
Sbjct: 132 FDRVFQPDSSQDDVFLEIEPVIKSVIDGYNACIFAYGQTGTGKTYTMEGLPNSPGIVPRA 191
Query: 137 LEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATK-------KLEVKQAAD 189
++ LF+ EE + + SMLE+Y +KDLL SEATK L + +
Sbjct: 192 IKGLFKQVEE--SNHMFTIHFSMLEIYMGNLKDLLL---SEATKPISPIPPSLSIHTDPN 246
Query: 190 GTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVD----DK 245
G ++ LV+ V + + + K G R R+ ST++N +SSRSH I + ++
Sbjct: 247 GEIDIENLVKLKVDDFNEILRLYKVGCRSRATASTNSNSVSSRSHCMIRVSVTSLGAPER 306
Query: 246 KMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*ISLYH 305
+ + +W + + ++V L G+ + E NLS +S
Sbjct: 307 RRETNKIW--------LVDLGGSERV-------LKTRATGR-RFDEGKAINLS--LSALG 348
Query: 306 HLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNF 365
++ LQ + +++ RNSKLT +L+ SLG D KTLM V ISP DL ET+CSLNF
Sbjct: 349 DVINSLQ----RKNSHIPYRNSKLTQVLKDSLGQDSKTLMLVHISPKEDDLCETICSLNF 404
Query: 366 ATRVRGIESG-------PARKQVDLTELLKYKQMAEKSKH-DEKEARKLQDNLQSV 413
ATR + I G A+K+ + L K + E+ + ++ R L + L+ +
Sbjct: 405 ATRAKNIHLGQDESTEEQAKKEAVMMNLQKMMEKIEQEREMSLRKMRNLNETLEKL 460
>At4g05190 kinesin - like protein
Length = 777
Score = 202 bits (514), Expect = 3e-52
Identities = 147/393 (37%), Positives = 211/393 (53%), Gaps = 39/393 (9%)
Query: 7 KRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQV- 65
+R+ E L R++L+N ++ELKGNIRVFCR RPL ++ A SV+ + +++E L
Sbjct: 392 ERQLFEGELLRKKLHNTILELKGNIRVFCRVRPLLPDDGGRQEA-SVIAYPTSTESLGRG 450
Query: 66 --VCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTM 123
V +K F FD VF +QE VF + +V S LDG+ VCIFAYGQTG+GKT+TM
Sbjct: 451 IDVVQSGNKHPFTFDKVFDHGASQEEVFFEISQLVQSALDGYKVCIFAYGQTGSGKTYTM 510
Query: 124 EG---TPEHRGVNYRTLEELFRVSE--ERQGTIKYELLVSMLEVYNEKIKDLLA------ 172
G TPE +G+ R+LE++F+ S+ QG KY++ VSMLE+YNE I+DLL+
Sbjct: 511 MGRPETPEQKGLIPRSLEQIFKTSQSLSTQGW-KYKMQVSMLEIYNESIRDLLSTSRTIA 569
Query: 173 -----GNSSEATKKLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSAN 227
+SS + ++ + +G V L V + +L+ + RSVG T N
Sbjct: 570 IESVRADSSTSGRQYTITHDVNGNTHVSDLTIVDVCSIGQISSLLQQAAQSRSVGKTHMN 629
Query: 228 ELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKL 287
E SSRSH+ +F + + G L + +A ++++ T
Sbjct: 630 EQSSRSHF-VFTLRISGVNESTEQQVQGVL---NLIDLAGSERLSRSGAT--------GD 677
Query: 288 KLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFV 347
+LKE N S +S ++ L + ++ RNSKLT++LQ LGGD KTLMFV
Sbjct: 678 RLKETQAINKS--LSALSDVIF----ALAKKEDHVPFRNSKLTYLLQPCLGGDSKTLMFV 731
Query: 348 QISPSSVDLTETLCSLNFATRVRGIESGPARKQ 380
ISP E+LCSL FA RV E G R+Q
Sbjct: 732 NISPDPSSTGESLCSLRFAARVNACEIGIPRRQ 764
>At1g09170 putative kinesin
Length = 1046
Score = 201 bits (511), Expect = 6e-52
Identities = 142/392 (36%), Positives = 206/392 (52%), Gaps = 39/392 (9%)
Query: 10 YLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVCSD 69
Y V E R+L N+V +LKG+IRV+CR RP + + + V + + S
Sbjct: 414 YQRVLEENRKLYNQVQDLKGSIRVYCRVRPFLPGQKSVLTTVDHLEDSTLSIATPSKYGK 473
Query: 70 SSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEG---- 125
+K F F+ VF P +QEAVFA T+P++ SVLDG+NVCIFAYGQTG+GKTFTM G
Sbjct: 474 EGQKTFTFNKVFGPSASQEAVFADTQPLIRSVLDGYNVCIFAYGQTGSGKTFTMMGPNEL 533
Query: 126 TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKLEVK 185
T E GVNYR L +LF +S R+ T Y + V MLE+YNE+I+ NS++
Sbjct: 534 TDETLGVNYRALSDLFHLSSVRKETFSYNISVQMLEIYNEQIR-----NSTQ-------- 580
Query: 186 QAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVDDK 245
DG VP V V ++ G + R+V +T+ N+ SSRSH + V K
Sbjct: 581 ---DGI-NVPEATLVPVSTTSDVIHLMNIGQKNRAVSATAMNDRSSRSH-SCLTVHVQGK 635
Query: 246 KMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*ISLYH 305
+ G + + +A +++ +T +LKE ++++ +S
Sbjct: 636 DLTSGVTLRGSM---HLVDLAGSERIDKSEVT--------GDRLKEA--QHINKSLSALG 682
Query: 306 HLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNF 365
++ L + ++ RNSKLT +LQ +LGG KTLMF+ ISP DL ETL +L F
Sbjct: 683 DVIASLS----QKNNHIPYRNSKLTQLLQDALGGQAKTLMFIHISPELEDLGETLSTLKF 738
Query: 366 ATRVRGIESGPARKQVDLTELLKYKQMAEKSK 397
A RV ++ G AR D +E+ + K+ K
Sbjct: 739 AERVATVDLGAARVNKDTSEVKELKEQIASLK 770
>At4g27180 heavy chain polypeptide of kinesin-like protein (katB)
Length = 745
Score = 199 bits (507), Expect = 2e-51
Identities = 145/392 (36%), Positives = 207/392 (51%), Gaps = 33/392 (8%)
Query: 9 KYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQV--- 65
K +E R++L+N + ELKGNIRVFCR RPL E ++ A +++ ++ E L
Sbjct: 366 KLIEGEKLRKKLHNTIQELKGNIRVFCRVRPLLSGENSSEEA-KTISYPTSLEALGRGID 424
Query: 66 VCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTM-- 123
+ + F FD VF P +QE VF + +V S LDG+ VCIFAYGQTG+GKT+TM
Sbjct: 425 LLQNGQSHCFTFDKVFVPSASQEDVFVEISQLVQSALDGYKVCIFAYGQTGSGKTYTMMG 484
Query: 124 -EGTPEHRGVNYRTLEELFRVSEE-RQGTIKYELLVSMLEVYNEKIKDLLAGN------- 174
G P+ +G+ R LE++F+ + R KYEL VSMLE+YNE I+DLL+ N
Sbjct: 485 RPGNPDEKGLIPRCLEQIFQTRQSLRSQGWKYELQVSMLEIYNETIRDLLSTNKEAVRAD 544
Query: 175 SSEATKKLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSH 234
+ + +K +K A G V L V + V +L R RSVG T+ NE SSRSH
Sbjct: 545 NGVSPQKYAIKHDASGNTHVVELTVVDVRSSKQVSFLLDHAARNRSVGKTAMNEQSSRSH 604
Query: 235 WQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG* 294
+ +F + + G L + +A ++++ + S +LK +
Sbjct: 605 F-VFTLKISGFNESTEQQVQGVL---NLIDLAGSERLS------KSGSTGDRLKETQAIN 654
Query: 295 RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSV 354
++LSS + L + ++ RNSKLT++LQ LGGD KTLMFV I+P
Sbjct: 655 KSLSSLGDVIF--------ALAKKEDHVPFRNSKLTYLLQPCLGGDSKTLMFVNITPEPS 706
Query: 355 DLTETLCSLNFATRVRGIESGPARKQVDLTEL 386
E+LCSL FA RV E G A + V+ L
Sbjct: 707 STGESLCSLRFAARVNACEIGTAHRHVNARPL 738
>At4g21270 kinesin-related protein katA
Length = 793
Score = 198 bits (504), Expect = 4e-51
Identities = 149/406 (36%), Positives = 213/406 (51%), Gaps = 40/406 (9%)
Query: 1 DEYEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVV---NFE 57
D ++ + E L R++L+N ++ELKGNIRVFCR RPL ++ A + + E
Sbjct: 402 DRLADMEHQLCEGELLRKKLHNTILELKGNIRVFCRVRPLLPDDGGRHEATVIAYPTSTE 461
Query: 58 SNSEELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGT 117
+ + +V S +K F FD VF E +QE VF + +V S LDG+ VCIFAYGQTG+
Sbjct: 462 AQGRGVDLVQS-GNKHPFTFDKVFNHEASQEEVFFEISQLVQSALDGYKVCIFAYGQTGS 520
Query: 118 GKTFTMEGTPE---HRGVNYRTLEELFRVSEE--RQGTIKYELLVSMLEVYNEKIKDLLA 172
GKT+TM G PE +G+ R+LE++F+ S+ QG KY++ VSMLE+YNE I+DLL+
Sbjct: 521 GKTYTMMGRPEAPDQKGLIPRSLEQIFQASQSLGAQGW-KYKMQVSMLEIYNETIRDLLS 579
Query: 173 GN-----------SSEATKKLEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSV 221
N S + K+ + +G V L V + +L+ + RSV
Sbjct: 580 TNRTTSMDLVRADSGTSGKQYTITHDVNGHTHVSDLTIFDVCSVGKISSLLQQAAQSRSV 639
Query: 222 GSTSANELSSRSHWQIFYIIVDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAV 281
G T NE SSRSH+ +F + + G L + +A ++++ T
Sbjct: 640 GKTQMNEQSSRSHF-VFTMRISGVNESTEQQVQGVL---NLIDLAGSERLSKSGAT---- 691
Query: 282 SV*GKLKLKEKG*RNLSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDC 341
+LKE N S +S ++ L + ++ RNSKLT++LQ LGGD
Sbjct: 692 ----GDRLKETQAINKS--LSALSDVIF----ALAKKEDHVPFRNSKLTYLLQPCLGGDS 741
Query: 342 KTLMFVQISPSSVDLTETLCSLNFATRVRGIESGPARKQVDLTELL 387
KTLMFV ISP E+LCSL FA RV E G R+Q T+LL
Sbjct: 742 KTLMFVNISPDPTSAGESLCSLRFAARVNACEIGIPRRQTS-TKLL 786
>At5g65930 kinesin-like calmodulin-binding protein
Length = 1260
Score = 181 bits (460), Expect = 5e-46
Identities = 133/405 (32%), Positives = 209/405 (50%), Gaps = 30/405 (7%)
Query: 2 EYEILKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSE 61
E EIL Y E + R+R N + ++KG IRV+CR RPLNE E + + + +
Sbjct: 863 ELEIL---YKEEQVLRKRYYNTIEDMKGKIRVYCRIRPLNEKESSEREKQMLTTVDEFTV 919
Query: 62 ELQVVCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTF 121
E D +KQ +D VF +Q+ +F TK +V S +DG+NVCIFAYGQTG+GKTF
Sbjct: 920 EHP--WKDDKRKQHIYDRVFDMRASQDDIFEDTKYLVQSAVDGYNVCIFAYGQTGSGKTF 977
Query: 122 TMEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKK 181
T+ G + G+ R +ELF + + + L M+E+Y + + DLL S+ K
Sbjct: 978 TIYGHESNPGLTPRATKELFNILKRDSKRFSFSLKAYMVELYQDTLVDLLLPKSARRL-K 1036
Query: 182 LEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYII 241
LE+K+ + G V + + + + IL+ G+ R V T+ NE SSRSH I ++
Sbjct: 1037 LEIKKDSKGMVFVENVTTIPISTLEELRMILERGSERRHVSGTNMNEESSRSH-LILSVV 1095
Query: 242 VDDKKMVLHAVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*I 301
++ + + G L + + R +K G + K ++++ +
Sbjct: 1096 IESIDLQTQSAARGKLSFVDLAGSERVKKS-------------GSAGCQLKEAQSINKSL 1142
Query: 302 SLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLC 361
S ++ L NQ ++ RN KLT ++ SLGG+ KTLMFV +SP+ +L ET
Sbjct: 1143 SALGDVIGALSS--GNQ--HIPYRNHKLTMLMSDSLGGNAKTLMFVNVSPAESNLDETYN 1198
Query: 362 SLNFATRVRGIESGP-----ARKQVDLTELLKY-KQMAEKSKHDE 400
SL +A+RVR I + P +++ V L +L+ Y K+ A K +E
Sbjct: 1199 SLLYASRVRTIVNDPSKHISSKEMVRLKKLVAYWKEQAGKKGEEE 1243
>At5g27950 unknown protein
Length = 625
Score = 157 bits (397), Expect = 1e-38
Identities = 99/235 (42%), Positives = 133/235 (56%), Gaps = 12/235 (5%)
Query: 6 LKRKYLEVSLERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQV 65
LK K + +R+++ N++I+ KG+IRVFCR RP E V+F ++ V
Sbjct: 55 LKLKLKSLDEKRKQVLNKIIDTKGSIRVFCRVRPFLLTE--RRPIREPVSFGPDNV---V 109
Query: 66 VCSDSSKKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEG 125
+ S S K+F+FD VF QE VF + KPI+ S LDGHNVC+ AYGQTGTGKTFTM+G
Sbjct: 110 IRSAGSSKEFEFDKVFHQSATQEEVFGEVKPILRSALDGHNVCVLAYGQTGTGKTFTMDG 169
Query: 126 TPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSS----EATKK 181
T E G+ R ++ELF + Q T +SMLE+Y +KDLL+ S EA+ K
Sbjct: 170 TSEQPGLAPRAIKELFNEASMDQ-THSVTFRMSMLEIYMGNLKDLLSARQSLKSYEASAK 228
Query: 182 --LEVKQAADGTQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSH 234
L ++ + G+ E+ GL E V G RVRS T+ NE SSRSH
Sbjct: 229 CNLNIQVDSKGSVEIEGLTEVEVMDFTKARWWYNKGRRVRSTSWTNVNETSSRSH 283
Score = 63.9 bits (154), Expect = 2e-10
Identities = 35/91 (38%), Positives = 52/91 (56%), Gaps = 1/91 (1%)
Query: 325 RNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATRVRGIESGPA-RKQVDL 383
RNSKLT IL+ SLG K LM V ISP D+ ET+CSL+F R R +ES ++
Sbjct: 357 RNSKLTQILKDSLGTRSKVLMLVHISPRDEDVGETICSLSFTKRARAVESNRGLTAELQK 416
Query: 384 TELLKYKQMAEKSKHDEKEARKLQDNLQSVQ 414
K ++ E+ + ++ +K++ LQ V+
Sbjct: 417 LREKKISELEEEMEETQEGCKKIKARLQEVE 447
>At5g65460 putative protein
Length = 1264
Score = 156 bits (394), Expect = 2e-38
Identities = 125/420 (29%), Positives = 210/420 (49%), Gaps = 50/420 (11%)
Query: 16 ERRRLNNEVIELKGNIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVCSDSS---- 71
E++RL N+++ KGN++VFCR RPL E+E S++ F N ++V SD +
Sbjct: 124 EKKRLFNDLLTTKGNVKVFCRARPLFEDE-----GPSIIEFPDNCT-IRVNTSDDTLSNP 177
Query: 72 KKQFKFDHVFKPEDNQEAVFAQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEGTPEHRG 131
KK+F+FD V+ P+ Q ++F+ +P V S LDG NV IFAYGQT GKT+TMEG+ + RG
Sbjct: 178 KKEFEFDRVYGPQVGQASLFSDVQPFVQSALDGSNVSIFAYGQTHAGKTYTMEGSNQDRG 237
Query: 132 VNYRTLEELFRV-SEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKLEVKQAADG 190
+ R EEL + + + ++ VS+ E+YNE+++DLL+G S K G
Sbjct: 238 LYARCFEELMDLANSDSTSASQFSFSVSVFELYNEQVRDLLSGCQSNLPK------INMG 291
Query: 191 TQEVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVDDKKMVLH 250
+E ++E D E + R + S N + +S + ++IV +H
Sbjct: 292 LRE--SVIELSQEKVDNPSEFM------RVLNSAFQNRGNDKSKSTVTHLIVS-----IH 338
Query: 251 AVWCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*RNLSS*ISLYHHLVML 310
+ ++ RE ++ L LA S L +++ +++ + + + + L
Sbjct: 339 ICYSN--------TITRENVISKLSLVDLAGS--EGLTVEDDNGDHVTDLLHVTNSISAL 388
Query: 311 LQPL--LPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATR 368
L L ++ + NS LT IL SLGG KTLM V I PS+ +L+E + LN+A R
Sbjct: 389 GDVLSSLTSKRDTIPYENSFLTRILADSLGGSSKTLMIVNICPSARNLSEIMSCLNYAAR 448
Query: 369 VRGIESGPARKQVDLTELLKYKQMAEKSKHD----EKEARKLQDNLQSVQMRLATREFMC 424
R + + K++ +A ++ + E+E ++L+ + ++ L C
Sbjct: 449 ARNTVPSLGNRDT----IKKWRDVANDARKEVLEKERENQRLKQEVTGLKQALKEANDQC 504
>At2g28620 putative kinesin-like spindle protein
Length = 1076
Score = 152 bits (383), Expect = 4e-37
Identities = 135/443 (30%), Positives = 206/443 (46%), Gaps = 64/443 (14%)
Query: 30 NIRVFCRCRPLNENEIANGSAVSVVNFESNSEELQVVCSDSSK---KQFKFDHVFKPEDN 86
NI+V RCRP N E +V+ +E+ V + + K K F FD VF P
Sbjct: 50 NIQVIVRCRPFNSEE-TRLQTPAVLTCNDRKKEVAVAQNIAGKQIDKTFLFDKVFGPTSQ 108
Query: 87 QEAVFAQT-KPIVASVLDGHNVCIFAYGQTGTGKTFTMEG--------TPEHRGVNYRTL 137
Q+ ++ Q PIV VLDG+N IFAYGQTGTGKT+TMEG P GV R +
Sbjct: 109 QKDLYHQAVSPIVFEVLDGYNCTIFAYGQTGTGKTYTMEGGARKKNGEIPSDAGVIPRAV 168
Query: 138 EELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSS-----EATKKLEVKQAADGTQ 192
+++F + E Q +Y L VS LE+YNE++ DLLA + ++ K L + + G
Sbjct: 169 KQIFDIL-EAQSAAEYSLKVSFLELYNEELTDLLAPEETKFADDKSKKPLALMEDGKGGV 227
Query: 193 EVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSHWQIFYIIVDDKKMVLHAV 252
V GL E V AD ++++L+ G+ R T N+ SSRSH IF + + K+
Sbjct: 228 FVRGLEEEIVSTADEIYKVLEKGSAKRRTAETLLNKQSSRSH-SIFSVTIHIKECTPEG- 285
Query: 253 WCG*LLWGKI*SMAREQKVTFG*LT*LAVSV*GKLKLKEKG*R--NLSS*ISLYHHLVML 310
E+ V G L V + G + G R + L+ L
Sbjct: 286 ---------------EEIVKSGKLN--LVDLAGSENISRSGAREGRAREAGEINKSLLTL 328
Query: 311 LQPL--LPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATR 368
+ + L ++ R SKLT +L+ SLGG KT + +SPS L ETL +L++A R
Sbjct: 329 GRVINALVEHSGHIPYRESKLTRLLRDSLGGKTKTCVIATVSPSVHCLEETLSTLDYAHR 388
Query: 369 VRGIESGPARKQVDL-----------TELLKYKQMAEKSKH-----------DEKEARKL 406
+ I++ P Q + E LK + A + K+ +E E + +
Sbjct: 389 AKHIKNKPEVNQKMMKSAIMKDLYSEIERLKQEVYAAREKNGIYIPKERYTQEEAEKKAM 448
Query: 407 QDNLQSVQMRLATREFMCRNLQD 429
D ++ +++ ++ +LQ+
Sbjct: 449 ADKIEQMEVEGEAKDKQIIDLQE 471
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,769,603
Number of Sequences: 26719
Number of extensions: 353109
Number of successful extensions: 1656
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 1389
Number of HSP's gapped (non-prelim): 142
length of query: 430
length of database: 11,318,596
effective HSP length: 102
effective length of query: 328
effective length of database: 8,593,258
effective search space: 2818588624
effective search space used: 2818588624
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135319.10


