

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135311.5 + phase: 0
(346 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
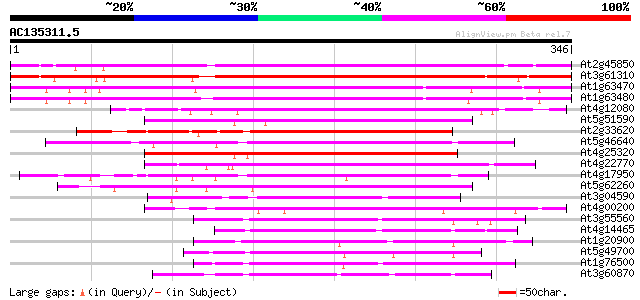
Score E
Sequences producing significant alignments: (bits) Value
At2g45850 putative AT-hook DNA-binding protein 301 5e-82
At3g61310 DNA-binding protein like 285 2e-77
At1g63470 DNA-binding like protein 217 9e-57
At1g63480 putative DNA-binding protein (At1g63480) 212 3e-55
At4g12080 putative DNA-binding protein 189 2e-48
At5g51590 unknown protein 187 6e-48
At2g33620 AT-hook DNA-binding protein (AHP1) 182 2e-46
At5g46640 unknown protein 180 1e-45
At4g25320 unknown protein 177 6e-45
At4g22770 putative DNA binding protein 176 2e-44
At4g17950 unknown protein 172 2e-43
At5g62260 unknown protein 165 3e-41
At3g04590 unknown protein 150 1e-36
At4g00200 putative transcription factor 146 2e-35
At3g55560 unknown protein 112 3e-25
At4g14465 unknown protein 107 7e-24
At1g20900 putative DNA-binding protein 101 7e-22
At5g49700 putative protein 100 2e-21
At1g76500 unknown protein 100 2e-21
At3g60870 putative protein 98 6e-21
>At2g45850 putative AT-hook DNA-binding protein
Length = 348
Score = 301 bits (770), Expect = 5e-82
Identities = 177/357 (49%), Positives = 216/357 (59%), Gaps = 20/357 (5%)
Query: 1 MDHGDHMALSNSASYYMQQRVLPGSGAQPELHVSPSFNQ----LSNPNLPFQSNIGGGGS 56
MD D M LS S SYY+ R L GSG P H SP Q L N N PF S G GS
Sbjct: 1 MDRRDAMGLSGSGSYYIH-RGLSGSGP-PTFHGSPQQQQGLRHLPNQNSPFGSGSTGFGS 58
Query: 57 -------NIGTTLPLESSAISSQGVNMSGHTGVPSGETVKRKRGRPRKYGADRVVSLALS 109
++ T + GVNM PS +KRKRGRPRKYG D VSLALS
Sbjct: 59 PSLHGDPSLATAAGGAGALPHHIGVNMIAPPPPPSETPMKRKRGRPRKYGQDGSVSLALS 118
Query: 110 PSPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGED 169
S + P KRGRGRPPGSGKKQ++AS GELM S+G F PHVI ++ GED
Sbjct: 119 SSSVSTITPNNSN----KRGRGRPPGSGKKQRMASVGELMPSSSGMSFTPHVIAVSIGED 174
Query: 170 IAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTEN 229
IA+K++ FSQ RA+CVLS+SG+VS+ + +PS S G +KYEG F I+++S Y+ +
Sbjct: 175 IASKVIAFSQQGPRAICVLSASGAVSTATLIQPSASPGAIKYEGRFEILALSTSYIVATD 234
Query: 230 GSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKESSED 289
GS RNR G LS+SL PDGR+ GGA+GGPL+AASPVQV++GSF+W K K+KK+E
Sbjct: 235 GSFRNRTGNLSVSLASPDGRVIGGAIGGPLIAASPVQVIVGSFIWAAPKIKSKKREEEAS 294
Query: 290 AEGTVESDHQGAHNPAALNSISPNQNLTPTSSLSPWSAASRQMDMGNSHADIDLMRG 346
DH N N+ISP P +L WS SRQMDM ++HADIDLMRG
Sbjct: 295 EVVQETDDHHVLDNNN--NTISPVPQQQPNQNLI-WSTGSRQMDMRHAHADIDLMRG 348
>At3g61310 DNA-binding protein like
Length = 354
Score = 285 bits (730), Expect = 2e-77
Identities = 176/366 (48%), Positives = 226/366 (61%), Gaps = 32/366 (8%)
Query: 1 MDHGDHMALSNSASYYMQQRVLPGSG-----AQPELHVSPSFNQLSNPNLPFQSNIG--- 52
MD D MALS S SYY+Q R +PGSG QP H S F+ +N PF SN
Sbjct: 1 MDRRDAMALSGSGSYYIQ-RGIPGSGPPPPQTQPTFHGSQGFHHFTNSISPFGSNPNPNP 59
Query: 53 --GGGSN--IGTTLPLESSAISSQGVNMSGHTGVPSGET-VKRKRGRPRKYGADR-VVSL 106
GG S + LP++SS S PSG+T VKRKRGRPRKYG D VSL
Sbjct: 60 NPGGVSTGFVSPPLPVDSSPADSSAAAAGALVAPPSGDTSVKRKRGRPRKYGQDGGSVSL 119
Query: 107 ALSPS---PTPSSNPGTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIE 163
ALSPS +P+SN KRGRGRPPGSGKKQ+L+S GE+M S G F PHVI
Sbjct: 120 ALSPSISNVSPNSN---------KRGRGRPPGSGKKQRLSSIGEMMPSSTGMSFTPHVIV 170
Query: 164 IASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGC 223
++ GEDIA+K+++FS RA+CVLS+SG+VS+ + +P+ S GT+ YEG F ++S+S
Sbjct: 171 VSIGEDIASKVISFSHQGPRAICVLSASGAVSTATLLQPAPSHGTIIYEGLFELISLSTS 230
Query: 224 YVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKK 283
Y+ T + NR G L++SL PDGR+ GG +GGPL+AAS VQV++GSF+W K K KK
Sbjct: 231 YLNTTDNDYPNRTGSLAVSLASPDGRVIGGGIGGPLIAASQVQVIVGSFIWAIPKGKIKK 290
Query: 284 K-ESSEDAEGTVESDHQGAHNPAALNSISP--NQNLTPTSSLSPWSAASRQMDMGNSHAD 340
+ E+SED + T ++ N AA + P +QN+ T + WS SR MDM + H D
Sbjct: 291 REETSEDVQDT-DALENNNDNTAATSPPVPQQSQNIVQT-PVGIWSTGSRSMDMHHPHMD 348
Query: 341 IDLMRG 346
IDLMRG
Sbjct: 349 IDLMRG 354
>At1g63470 DNA-binding like protein
Length = 378
Score = 217 bits (552), Expect = 9e-57
Identities = 149/380 (39%), Positives = 204/380 (53%), Gaps = 36/380 (9%)
Query: 1 MDHGDHMALSNSAS-YYMQQRV---LPGSGAQPELHVSP---SFNQLSNPNL--PFQSNI 51
MD + MA S S +Y+Q+ V L S LH P +SNPN+ P SN
Sbjct: 1 MDGREAMAFPGSHSQFYLQRGVFTNLTPSQVASGLHAPPPPPGMRPMSNPNIHHPQASNP 60
Query: 52 GGG-------GSNIGTTLPLESSAISSQGVNMSGHTGVPSGETVKRKRGRPRKYGADRVV 104
G S+ G ++ + ++ ++ + VK+KRGRPRKY D V
Sbjct: 61 GPPFSMAEHRHSDFGHSIHMGMASPAAVQPTLQLPPPPSEQPMVKKKRGRPRKYVPDGQV 120
Query: 105 SLALSPSPT---PSSNPGTMTQ-GGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPH 160
SL LSP P S + +M+ PKR RGRPPG+G+KQ+LA+ GE M+ SAG F PH
Sbjct: 121 SLGLSPMPCVSKKSKDSSSMSDPNAPKRARGRPPGTGRKQRLANLGEWMNTSAGLAFAPH 180
Query: 161 VIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSM 220
VI + SGEDI +K+L+FSQ R RALC++S +G+VSSV +REP+ + +L +EG F I+S+
Sbjct: 181 VISVGSGEDIVSKVLSFSQKRPRALCIMSGTGTVSSVTLREPASTTPSLTFEGRFEILSL 240
Query: 221 SGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAK 280
G Y+ E G S++R GGLS+SL GP+G + GG + G L+AAS VQV+ SF++G
Sbjct: 241 GGSYLVNEEGGSKSRTGGLSVSLSGPEGHVIGGGI-GMLIAASLVQVVACSFVYGASAKS 299
Query: 281 NKK---------KESSEDAEGTVESDHQGAHNPAALNSISPNQNLTPTSSLSPW----SA 327
N K E +E+ A AA QN P +S W S
Sbjct: 300 NNNNNKTIKQEIKPKQEPTNSEMETTPGSAPEAAASTGQHTPQNF-PAQGMSGWPVSGSG 358
Query: 328 ASRQMDMG-NSHADIDLMRG 346
+ R +D N DIDL RG
Sbjct: 359 SGRSLDSSRNPLTDIDLTRG 378
>At1g63480 putative DNA-binding protein (At1g63480)
Length = 361
Score = 212 bits (539), Expect = 3e-55
Identities = 149/370 (40%), Positives = 196/370 (52%), Gaps = 33/370 (8%)
Query: 1 MDHGDHMALSNSASYYMQQRV----LPGSGAQPELHVSP---SFNQLSNPNL--PFQSNI 51
MD + MA S S Y QR L S LH P +SNPN+ P +N
Sbjct: 1 MDGREAMAFPGSHSQYYLQRGAFTNLAPSQVASGLHAPPPHTGLRPMSNPNIHHPQANNP 60
Query: 52 GGGGSNIGTTLPLES-SAISSQGVNMSGHTGVPSGETVKRKRGRPRKYGADRVVSLALSP 110
G S+ G T+ + S+ S V P VKRKRGRPRKYG V + +
Sbjct: 61 GPPFSDFGHTIHMGVVSSASDADVQPPPPPPPPEEPMVKRKRGRPRKYGEPMVSNKSRDS 120
Query: 111 SPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDI 170
SP N PKR RGRPPG+G+KQ+LA+ GE M+ SAG F PHVI I +GEDI
Sbjct: 121 SPMSDPNE-------PKRARGRPPGTGRKQRLANLGEWMNTSAGLAFAPHVISIGAGEDI 173
Query: 171 AAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENG 230
AAK+L+FSQ R RALC++S +G++SSV + +P + L YEG F I+S G Y+ E G
Sbjct: 174 AAKVLSFSQQRPRALCIMSGTGTISSVTLCKPGSTDRHLTYEGPFEIISFGGSYLVNEEG 233
Query: 231 SSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWG-RLKAKN-------- 281
SR+R GGLS+SL PDG + G V L+AA+ VQV+ SF++G R K N
Sbjct: 234 GSRSRTGGLSVSLSRPDGSIIAGGV-DMLIAANLVQVVACSFVYGARAKTHNNNNKTIRQ 292
Query: 282 KKKESSEDAEGTVESDHQGAHNPAALNSISPNQNLTPTSSLSPW----SAASRQMDM-GN 336
+K+ + ED +E+ A PAA QN + + + W S + R +D+ N
Sbjct: 293 EKEPNEEDNNSEMETTPGSAAEPAASAGQQTPQNFS-SQGIRGWPGSGSGSGRSLDICRN 351
Query: 337 SHADIDLMRG 346
D DL RG
Sbjct: 352 PLTDFDLTRG 361
>At4g12080 putative DNA-binding protein
Length = 356
Score = 189 bits (479), Expect = 2e-48
Identities = 120/304 (39%), Positives = 173/304 (56%), Gaps = 38/304 (12%)
Query: 63 PLESSAISSQGVNMSGHTGVPSGETVKRKRGRPRKYGADRVVSLALSP-----SPTPSSN 117
PL+ S +++ + G+ SG +K+KRGRPRKYG D V +ALSP +P PS
Sbjct: 65 PLQISTVTTT-TTTAAMEGI-SGGLMKKKRGRPRKYGPDGTV-VALSPKPISSAPAPSHL 121
Query: 118 PGTMTQ-----GGPKRGRGRPPGSGKK----QQLASFGELMSGSAGTGFIPHVIEIASGE 168
P + KR + +P S + Q+ + GE S G F PH+I + +GE
Sbjct: 122 PPPSSHVIDFSASEKRSKVKPTNSFNRTKYHHQVENLGEWAPCSVGGNFTPHIITVNTGE 181
Query: 169 DIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTE 228
D+ KI++FSQ R++CVLS++G +SSV +R+P SGGTL YEG F I+S+SG ++P +
Sbjct: 182 DVTMKIISFSQQGPRSICVLSANGVISSVTLRQPDSSGGTLTYEGRFEILSLSGSFMPND 241
Query: 229 NGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKESSE 288
+G +R+R GG+S+SL PDGR+ GG + G LVAASPVQV++GSFL G K K++
Sbjct: 242 SGGTRSRTGGMSVSLASPDGRVVGGGLAGLLVAASPVQVVVGSFLAGTDHQDQKPKKNKH 301
Query: 289 D-------AEGTVES--DHQGAHNPAALNSISPNQNLTPTSSLSPWSAASRQMDMGNSHA 339
D A + S DH+ H +++S+ N N TS S D N H
Sbjct: 302 DFMLSSPTAAIPISSAADHRTIH---SVSSLPVNNNTWQTSLAS---------DPRNKHT 349
Query: 340 DIDL 343
DI++
Sbjct: 350 DINV 353
>At5g51590 unknown protein
Length = 419
Score = 187 bits (476), Expect = 6e-48
Identities = 98/234 (41%), Positives = 142/234 (59%), Gaps = 32/234 (13%)
Query: 84 SGETVKRKRGRPRKYGADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGSG------ 137
S +K+KRGRPRKY D +++ LSP P SS P T G KRGRGR G G
Sbjct: 73 SSSELKKKRGRPRKYNPDGSLAVTLSPMPISSSVPLTSEFGSRKRGRGRGRGRGRGRGRG 132
Query: 138 ------------------KKQQLASFGELMSGSAGTG--------FIPHVIEIASGEDIA 171
K Q+ F S G G F PHV+ + +GED+
Sbjct: 133 QGQGSREPNNNNNDNNWLKNPQMFEFNNNTPTSGGGGPAEIVSPSFTPHVLTVNAGEDVT 192
Query: 172 AKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGS 231
KI+TFSQ +RA+C+LS++G +S+V +R+ SGGTL YEGHF I+S++G ++P+E+G
Sbjct: 193 MKIMTFSQQGSRAICILSANGPISNVTLRQSMTSGGTLTYEGHFEILSLTGSFIPSESGG 252
Query: 232 SRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKE 285
+R+R GG+S+SL G DGR+FGG + G +AA PVQVM+GSF+ G+ +++ ++++
Sbjct: 253 TRSRAGGMSVSLAGQDGRVFGGGLAGLFIAAGPVQVMVGSFIAGQEESQQQQQQ 306
>At2g33620 AT-hook DNA-binding protein (AHP1)
Length = 351
Score = 182 bits (463), Expect = 2e-46
Identities = 106/236 (44%), Positives = 145/236 (60%), Gaps = 20/236 (8%)
Query: 42 NPNLPFQSNIGGGGSNIGTTLPLESSAISSQGVNMSGHTGVPSGETVKRKRGRPRKYGAD 101
+P +Q N G S + LP G G TG S E VK++RGRPRKYG D
Sbjct: 60 SPPQQYQPNSAGENSVLNMNLP---------GGESGGMTGTGS-EPVKKRRGRPRKYGPD 109
Query: 102 R-VVSLALSPSPTPS---SNPGTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGF 157
+SL L+P PS S P + GG K+ RGRPPGS K+ GS G GF
Sbjct: 110 SGEMSLGLNPG-APSFTVSQPSSGGDGGEKK-RGRPPGSSSKRLKLQ----ALGSTGIGF 163
Query: 158 IPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHI 217
PHV+ + +GED+++KI+ + RA+CVLS++G++S+V +R+ + SGGT+ YEG F I
Sbjct: 164 TPHVLTVLAGEDVSSKIMALTHNGPRAVCVLSANGAISNVTLRQSATSGGTVTYEGRFEI 223
Query: 218 MSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFL 273
+S+SG + EN R+R GGLS+SL PDG + GG+V G L+AASPVQ+++GSFL
Sbjct: 224 LSLSGSFHLLENNGQRSRTGGLSVSLSSPDGNVLGGSVAGLLIAASPVQIVVGSFL 279
>At5g46640 unknown protein
Length = 386
Score = 180 bits (456), Expect = 1e-45
Identities = 115/317 (36%), Positives = 167/317 (52%), Gaps = 40/317 (12%)
Query: 23 PGSGAQPELHVSPSFNQLSNPNLPFQSNIGGGGSNIGTTLPLESSAISSQGVNMSGHTGV 82
P + A P + + + + +P LPF + + ++ + S G G
Sbjct: 27 PNAAASPLMVPTSTSQPIQHPRLPFGNQQQSQTFHQQQQQQMDQKTLESLGFG----DGS 82
Query: 83 PSGET-------------VKRKRGRPRKYGADRVVSLALSP-SPTPSSNPGTMTQGGP-- 126
PS + VK+KRGRPRKY D ++L L+P SP S+ + +GG
Sbjct: 83 PSSQPMRFGIDDQNQQLQVKKKRGRPRKYTPDGSIALGLAPTSPLLSAASNSYGEGGVGD 142
Query: 127 ------------KRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKI 174
KR RGRPPGS KKQ + + G++G GF PHVIE+ +GEDIA+K+
Sbjct: 143 SGGNGNSVDPPVKRNRGRPPGSSKKQL-----DALGGTSGVGFTPHVIEVNTGEDIASKV 197
Query: 175 LTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRN 234
+ FS +R +C+LS+SG+VS V++R+ S S G + YEG F I+++SG + E S N
Sbjct: 198 MAFSDQGSRTICILSASGAVSRVMLRQASHSSGIVTYEGRFEIITLSGSVLNYEVNGSTN 257
Query: 235 RDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKESSEDAEGTV 294
R G LS++L GPDG + GG+V G LVAA+ VQV++GSF+ +AK K+ S A G
Sbjct: 258 RSGNLSVALAGPDGGIVGGSVVGNLVAATQVQVIVGSFV---AEAKKPKQSSVNIARGQN 314
Query: 295 ESDHQGAHNPAALNSIS 311
N S+S
Sbjct: 315 PEPASAPANMLNFGSVS 331
>At4g25320 unknown protein
Length = 404
Score = 177 bits (450), Expect = 6e-45
Identities = 92/205 (44%), Positives = 131/205 (63%), Gaps = 12/205 (5%)
Query: 84 SGETVKRKRGRPRKYGADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGSG--KKQQ 141
S E +K+KRGRPRKY D + + LSP P SS P T KRGRGR + KK Q
Sbjct: 81 SAEQLKKKRGRPRKYNPDGTLVVTLSPMPISSSVPLTSEFPPRKRGRGRGKSNRWLKKSQ 140
Query: 142 LASF----------GELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSS 191
+ F G + G F PHV+ + +GED+ KI+TFSQ +RA+C+LS++
Sbjct: 141 MFQFDRSPVDTNLAGVGTADFVGANFTPHVLIVNAGEDVTMKIMTFSQQGSRAICILSAN 200
Query: 192 GSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLF 251
G +S+V +R+ SGGTL YEG F I+S++G ++ ++G +R+R GG+S+ L GPDGR+F
Sbjct: 201 GPISNVTLRQSMTSGGTLTYEGRFEILSLTGSFMQNDSGGTRSRAGGMSVCLAGPDGRVF 260
Query: 252 GGAVGGPLVAASPVQVMIGSFLWGR 276
GG + G +AA PVQVM+G+F+ G+
Sbjct: 261 GGGLAGLFLAAGPVQVMVGTFIAGQ 285
>At4g22770 putative DNA binding protein
Length = 334
Score = 176 bits (445), Expect = 2e-44
Identities = 100/252 (39%), Positives = 152/252 (59%), Gaps = 14/252 (5%)
Query: 84 SGETVKRKRGRPRKYGADRVVSLALSPSPTPSSNPGT-----MTQGGPKRGRGRP----P 134
S +K++RGRPRKYG D ++ LSP+P S+ P T + KRG+ +P P
Sbjct: 67 SSGPIKKRRGRPRKYGHDGA-AVTLSPNPISSAAPTTSHVIDFSTTSEKRGKMKPATPTP 125
Query: 135 GS--GKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSG 192
S K Q+ + GE SA F PH+I + +GED+ +I++FSQ + A+CVL ++G
Sbjct: 126 SSFIRPKYQVENLGEWSPSSAAANFTPHIITVNAGEDVTKRIISFSQQGSLAICVLCANG 185
Query: 193 SVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFG 252
VSSV +R+P SGGTL YEG F I+S+SG ++P+++ +R+R GG+S+SL PDGR+ G
Sbjct: 186 VVSSVTLRQPDSSGGTLTYEGRFEILSLSGTFMPSDSDGTRSRTGGMSVSLASPDGRVVG 245
Query: 253 GAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAALNSISP 312
G V G LVAA+P+QV++G+FL G + + K + + + + N A +I P
Sbjct: 246 GGVAGLLVAATPIQVVVGTFLGGTNQQEQTPKPHNHNFMSSPLM--PTSSNVADHRTIRP 303
Query: 313 NQNLTPTSSLSP 324
+ P S+ +P
Sbjct: 304 MTSSLPISTWTP 315
>At4g17950 unknown protein
Length = 439
Score = 172 bits (436), Expect = 2e-43
Identities = 121/324 (37%), Positives = 177/324 (54%), Gaps = 58/324 (17%)
Query: 7 MALSNSASYYMQQRVLPGSGAQPELHVSPSFNQLSNPNLPFQ------SNIGGGGSNIGT 60
M + S S M QR+ G P P +Q +P Q ++G GS
Sbjct: 57 MGHNTSTSQAMHQRLPFGGSMSPH---QPQQHQYHHPQPQQQIDQKTLESLGFDGS---- 109
Query: 61 TLPLESSAISSQGVNMSGHTGVPSGETVKRKRGRPRKYGAD--------RVVSLALSPS- 111
SS ++Q +M G+ + VK+KRGRPRKY AD ++L L+P+
Sbjct: 110 ----PSSVAATQQHSM--RFGIDH-QQVKKKRGRPRKYAADGGGGGGGGSNIALGLAPTS 162
Query: 112 --PTPSSNPGTMTQGG----------------PKRGRGRPPGSGKKQQLASFGELMSGSA 153
P+ S++ G +GG KR RGRPPGSGKKQ + + G+
Sbjct: 163 PLPSASNSYGGGNEGGGGGDSAGANANSSDPPAKRNRGRPPGSGKKQL-----DALGGTG 217
Query: 154 GTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISG--GTLKY 211
G GF PHVIE+ +GEDIA KIL F+ RA+C+LS++G+V++V++R+ + S GT+KY
Sbjct: 218 GVGFTPHVIEVKTGEDIATKILAFTNQGPRAICILSATGAVTNVMLRQANNSNPTGTVKY 277
Query: 212 EGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGS 271
EG F I+S+SG ++ +E+ + + G LS+SL G +GR+ GG V G LVA S VQV++GS
Sbjct: 278 EGRFEIISLSGSFLNSESNGTVTKTGNLSVSLAGHEGRIVGGCVDGMLVAGSQVQVIVGS 337
Query: 272 FLWGRLKAKNKKKESSEDAEGTVE 295
F + K+K+S+ A+ T E
Sbjct: 338 F----VPDGRKQKQSAGRAQNTPE 357
>At5g62260 unknown protein
Length = 404
Score = 165 bits (418), Expect = 3e-41
Identities = 101/275 (36%), Positives = 149/275 (53%), Gaps = 31/275 (11%)
Query: 30 ELHVSPSFNQLSNPNLPFQSNIGGGGSNIGTTLP---LESSAISSQGVNMSGHTGVPSGE 86
E +PSF Q +P + + T LP SSA V T +
Sbjct: 26 EFQQNPSFLQFVSP------------TTVVTPLPPPPAPSSAPVPTTVTPGSATASTGSD 73
Query: 87 TVKRKRGRPRKYGAD-----RVVSLALSPSPTPSSNPGT---MTQGGPKRGRGRPPGSGK 138
K+KRGRPRKY D R + LSP+P SS P + + G + + +P K
Sbjct: 74 PTKKKRGRPRKYAPDGSLNPRFLRPTLSPTPISSSIPLSGDYQWKRGKAQQQHQPLEFVK 133
Query: 139 KQQLASFGEL--------MSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSS 190
K +G +S G F H + GED+ K++ +SQ +RA+C+LS+
Sbjct: 134 KSHKFEYGSPAPTPPLPGLSCYVGANFTTHQFTVNGGEDVTMKVMPYSQQGSRAICILSA 193
Query: 191 SGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRL 250
+GS+S+V + +P+ +GGTL YEG F I+S+SG ++PTENG ++ R GG+SISL GP+G +
Sbjct: 194 TGSISNVTLGQPTNAGGTLTYEGRFEILSLSGSFMPTENGGTKGRAGGMSISLAGPNGNI 253
Query: 251 FGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKE 285
FGG + G L+AA PVQV++GSF+ +N+KK+
Sbjct: 254 FGGGLAGMLIAAGPVQVVMGSFIVMHQAEQNQKKK 288
>At3g04590 unknown protein
Length = 411
Score = 150 bits (379), Expect = 1e-36
Identities = 87/200 (43%), Positives = 115/200 (57%), Gaps = 19/200 (9%)
Query: 86 ETVKRKRGRPRKY-------GADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGSGK 138
E VKRKRGRPRKY A ++ S A S S +T G G S K
Sbjct: 102 EPVKRKRGRPRKYVTPEQALAAKKLASSASSSSAKQRRELAAVTGGTVSTNSG----SSK 157
Query: 139 KQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVI 198
K QL S G+ G F PH++ IA GED+ KI+ F+ LCVLS+SG++S+
Sbjct: 158 KSQLGSVGK-----TGQCFTPHIVNIAPGEDVVQKIMMFANQSKHELCVLSASGTISNAS 212
Query: 199 IREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGP 258
+R+P+ SGG L YEG + I+S+SG Y+ TE G + GGLS+SL DG++ GGA+G
Sbjct: 213 LRQPAPSGGNLPYEGQYEILSLSGSYIRTEQG---GKSGGLSVSLSASDGQIIGGAIGSH 269
Query: 259 LVAASPVQVMIGSFLWGRLK 278
L AA PVQV++G+F R K
Sbjct: 270 LTAAGPVQVILGTFQLDRKK 289
>At4g00200 putative transcription factor
Length = 345
Score = 146 bits (368), Expect = 2e-35
Identities = 107/305 (35%), Positives = 157/305 (51%), Gaps = 59/305 (19%)
Query: 84 SGETV-KRKRGRPRKYGADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQL 142
SGE K++RGRPRKY A+ +P PSS+ + KR RG+ G K+
Sbjct: 50 SGEASGKKRRGRPRKYEANG--------APLPSSSVPLVK----KRVRGKLNGFDMKKMH 97
Query: 143 ASFGELMSGS-----------AGTGFIPHVIEIASGE-----------------DIAAKI 174
+ G SG G+ F PHVI + +GE DI +I
Sbjct: 98 KTIGFHSSGERFGVGGGVGGGVGSNFTPHVITVNTGEVCILEEKGPKLSLGRRFDITMRI 157
Query: 175 LTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRN 234
++FSQ RA+C+LS++G +S+V +R+P GGTL YEG F I+S+SG ++ TEN S+
Sbjct: 158 ISFSQQGPRAICILSANGVISNVTLRQPDSCGGTLTYEGRFEILSLSGSFMETENQGSKG 217
Query: 235 RDGGLSISLLGPDGRLFGGAVGGPLVAASPVQ----------VMIGSFLWGRLKAKNK-K 283
R GG+S+SL GPDGR+ GG V G L+AA+P+Q V++GSF+ + K +
Sbjct: 218 RSGGMSVSLAGPDGRVVGGGVAGLLIAATPIQVTHESNNNVYVVVGSFITSDQQDHQKPR 277
Query: 284 KESSEDAEGTVESDHQGAHNPAALNSI----SPNQNLTPTS-SLSPWSAASRQMDMGNSH 338
K+ E A V S P S+ +P++ P+S +S W+ + Q NS
Sbjct: 278 KQRVEHAPAAVMSVPPPPSPPPPAASVFSPTNPDREQPPSSFGISSWT--NGQDMPRNSA 335
Query: 339 ADIDL 343
DI++
Sbjct: 336 TDINI 340
>At3g55560 unknown protein
Length = 310
Score = 112 bits (280), Expect = 3e-25
Identities = 71/214 (33%), Positives = 115/214 (53%), Gaps = 14/214 (6%)
Query: 114 PSSNPGTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAK 173
P+ PG+ + +R RGRPPGS K + +++ + HV+EIA+G D+A
Sbjct: 75 PAVEPGSGSGSTGRRPRGRPPGSKNKPKSPV---VVTKESPNSLQSHVLEIATGADVAES 131
Query: 174 ILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSR 233
+ F++ R R + VLS SG V++V +R+P+ SGG + G F I+SM G ++PT S
Sbjct: 132 LNAFARRRGRGVSVLSGSGLVTNVTLRQPAASGGVVSLRGQFEILSMCGAFLPT--SGSP 189
Query: 234 NRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSF---LWGRLKAKNKKKESS--- 287
GL+I L G G++ GG V GPL+A+ PV V+ +F + RL + ++++
Sbjct: 190 AAAAGLTIYLAGAQGQVVGGGVAGPLIASGPVIVIAATFCNATYERLPIEEEQQQEQPLQ 249
Query: 288 -EDAEGTVE--SDHQGAHNPAALNSISPNQNLTP 318
ED + E D++ +N + P N+ P
Sbjct: 250 LEDGKKQKEENDDNESGNNGNEGSMQPPMYNMPP 283
>At4g14465 unknown protein
Length = 281
Score = 107 bits (268), Expect = 7e-24
Identities = 65/187 (34%), Positives = 101/187 (53%), Gaps = 9/187 (4%)
Query: 127 KRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALC 186
+R RGRPPGS K + F ++ + HV+EI+ G D+A I FS+ R R +C
Sbjct: 67 RRPRGRPPGSKNKPKAPIF---VTRDSPNALRSHVLEISDGSDVADTIAHFSRRRQRGVC 123
Query: 187 VLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGP 246
VLS +GSV++V +R+ + GG + +G F I+S++G ++P G S GL++ L G
Sbjct: 124 VLSGTGSVANVTLRQAAAPGGVVSLQGRFEILSLTGAFLP---GPSPPGSTGLTVYLAGV 180
Query: 247 DGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAA 306
G++ GG+V GPL+A V V+ +F + E ED G+ H G +P
Sbjct: 181 QGQVVGGSVVGPLLAIGSVMVIAATF--SNATYERLPMEEEEDGGGS-RQIHGGGDSPPR 237
Query: 307 LNSISPN 313
+ S P+
Sbjct: 238 IGSNLPD 244
>At1g20900 putative DNA-binding protein
Length = 311
Score = 101 bits (251), Expect = 7e-22
Identities = 71/221 (32%), Positives = 110/221 (49%), Gaps = 22/221 (9%)
Query: 114 PSSNPGTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAK 173
P S+P T + KR RGRPPGS K A +++ + HV+E++ G DI
Sbjct: 73 PDSDPNTSSSAPGKRPRGRPPGSKNK---AKPPIIVTRDSPNALRSHVLEVSPGADIVES 129
Query: 174 ILTFSQVRARALCVLSSSGSVSSVIIREP---------SISGGTLKYEGHFHIMSMSGCY 224
+ T+++ R R + VL +G+VS+V +R+P S GG + G F I+S++G
Sbjct: 130 VSTYARRRGRGVSVLGGNGTVSNVTLRQPVTPGNGGGVSGGGGVVTLHGRFEILSLTGTV 189
Query: 225 VPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSF---LWGRLKAKN 281
+P GGLSI L G G++ GG+V PL+A++PV +M SF ++ RL +
Sbjct: 190 LPPPAPPGA---GGLSIFLAGGQGQVVGGSVVAPLIASAPVILMAASFSNAVFERLPIEE 246
Query: 282 KKKESSEDAEGTVESDHQGAHNPAALNSISPNQNLTPTSSL 322
+++E G Q P+A SP +T L
Sbjct: 247 EEEEGGGGGGGGGGGPPQMQQAPSA----SPPSGVTGQGQL 283
>At5g49700 putative protein
Length = 276
Score = 99.8 bits (247), Expect = 2e-21
Identities = 63/191 (32%), Positives = 107/191 (55%), Gaps = 12/191 (6%)
Query: 108 LSPSPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASG 167
LS + TP+ + ++ +R RGRPPGS K + F ++ P+++E+ SG
Sbjct: 39 LSSTVTPTVDDSSIEV--VRRPRGRPPGSKNKPKPPVF---VTRDTDPPMSPYILEVPSG 93
Query: 168 EDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSIS--GGTLKYEGHFHIMSMSGCYV 225
D+ I F + ++ +CVLS SGSV++V +R+PS + G T+ + G F ++S+S ++
Sbjct: 94 NDVVEAINRFCRRKSIGVCVLSGSGSVANVTLRQPSPAALGSTITFHGKFDLLSVSATFL 153
Query: 226 PTENGSSRNR--DGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSF---LWGRLKAK 280
P +S + ++SL GP G++ GG V GPL++A V V+ SF + RL A+
Sbjct: 154 PPPPRTSLSPPVSNFFTVSLAGPQGQIIGGFVAGPLISAGTVYVIAASFNNPSYHRLPAE 213
Query: 281 NKKKESSEDAE 291
++K S+ E
Sbjct: 214 EEQKHSAGTGE 224
>At1g76500 unknown protein
Length = 302
Score = 99.8 bits (247), Expect = 2e-21
Identities = 70/209 (33%), Positives = 105/209 (49%), Gaps = 17/209 (8%)
Query: 114 PSSNPGTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAK 173
P S+P T G KR RGRPPGS K + +++ + HV+E++SG DI
Sbjct: 60 PGSDPVTSGSTG-KRPRGRPPGSKNKPKPPV---IVTRDSPNVLRSHVLEVSSGADIVES 115
Query: 174 ILTFSQVRARALCVLSSSGSVSSVIIREPSI---------SGGTLKYEGHFHIMSMSGCY 224
+ T+++ R R + +LS +G+V++V +R+P+ +GG + G F I+S++G
Sbjct: 116 VTTYARRRGRGVSILSGNGTVANVSLRQPATTAAHGANGGTGGVVALHGRFEILSLTGTV 175
Query: 225 VPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKK 284
+P + GGLSI L G G++ GG V PLVA+ PV +M SF + +
Sbjct: 176 LPP---PAPPGSGGLSIFLSGVQGQVIGGNVVAPLVASGPVILMAASFSNATFERLPLED 232
Query: 285 ESSEDAE-GTVESDHQGAHNPAALNSISP 312
E E E G V G P S SP
Sbjct: 233 EGGEGGEGGEVGEGGGGEGGPPPATSSSP 261
>At3g60870 putative protein
Length = 265
Score = 98.2 bits (243), Expect = 6e-21
Identities = 64/209 (30%), Positives = 107/209 (50%), Gaps = 16/209 (7%)
Query: 89 KRKRGRPRKYGADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQLASFGEL 148
++KRGR + + L+ P+ N +R RGRP GS K + +
Sbjct: 27 RQKRGREEEGVEPNNIGEDLATFPSGEENIKK------RRPRGRPAGSKNKPKAPI---I 77
Query: 149 MSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGT 208
++ + F HV+EI + D+ + F++ R R +CVL+ +G+V++V +R+P GG
Sbjct: 78 VTRDSANAFRCHVMEITNACDVMESLAVFARRRQRGVCVLTGNGAVTNVTVRQP--GGGV 135
Query: 209 LKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVM 268
+ G F I+S+SG ++P + + GL + L G G++ GG+V GPL A+SPV VM
Sbjct: 136 VSLHGRFEILSLSGSFLPPPAPPAAS---GLKVYLAGGQGQVIGGSVVGPLTASSPVVVM 192
Query: 269 IGSFLWGRLKAKNKKKESSEDAEGTVESD 297
SF G + E E+ E ++ +
Sbjct: 193 AASF--GNASYERLPLEEEEETEREIDGN 219
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.130 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,341,979
Number of Sequences: 26719
Number of extensions: 397148
Number of successful extensions: 1324
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 1186
Number of HSP's gapped (non-prelim): 92
length of query: 346
length of database: 11,318,596
effective HSP length: 100
effective length of query: 246
effective length of database: 8,646,696
effective search space: 2127087216
effective search space used: 2127087216
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135311.5


