

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135311.3 - phase: 1 /pseudo
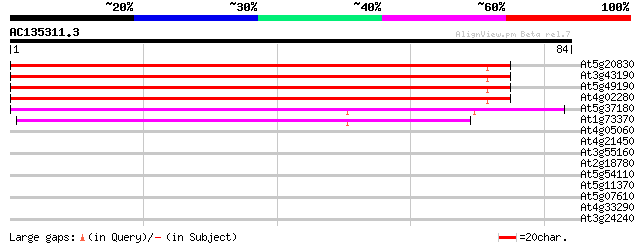
(84 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g20830 sucrose-UDP glucosyltransferase 115 3e-27
At3g43190 sucrose synthase like protein 104 7e-24
At5g49190 sucrose synthase 97 2e-21
At4g02280 putative sucrose synthetase 89 3e-19
At5g37180 sucrose synthase-like protein 50 2e-07
At1g73370 50 2e-07
At4g05060 unknown protein 26 4.0
At4g21450 unknown protein 25 5.2
At3g55160 putative protein 25 5.2
At2g18780 unknown protein 25 5.2
At5g54110 membrane associated protein 25 6.8
At5g11370 putative protein 25 6.8
At5g07610 putative protein 25 8.9
At4g33290 hypothetical protein 25 8.9
At3g24240 receptor kinase, putative 25 8.9
>At5g20830 sucrose-UDP glucosyltransferase
Length = 808
Score = 115 bits (289), Expect = 3e-27
Identities = 60/80 (75%), Positives = 63/80 (78%), Gaps = 5/80 (6%)
Query: 1 IVLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGSANVNYVLELDF 60
IVLPPWVALAVR PG+ EYLRVN+HALVVE LQPAEFL FKEELVDG N N+ LELDF
Sbjct: 80 IVLPPWVALAVRPRPGVWEYLRVNLHALVVEELQPAEFLHFKEELVDGVKNGNFTLELDF 139
Query: 61 EPFTASFPRP-----IGGGV 75
EPF AS PRP IG GV
Sbjct: 140 EPFNASIPRPTLHKYIGNGV 159
>At3g43190 sucrose synthase like protein
Length = 808
Score = 104 bits (260), Expect = 7e-24
Identities = 54/80 (67%), Positives = 63/80 (78%), Gaps = 5/80 (6%)
Query: 1 IVLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGSANVNYVLELDF 60
IVLPP+VALAVR PG+ EY+RVN+H LVVE LQ +E+L+FKEELVDG N N+ LELDF
Sbjct: 80 IVLPPFVALAVRPRPGVWEYVRVNLHDLVVEELQASEYLQFKEELVDGIKNGNFTLELDF 139
Query: 61 EPFTASFPRP-----IGGGV 75
EPF A+FPRP IG GV
Sbjct: 140 EPFNAAFPRPTLNKYIGDGV 159
>At5g49190 sucrose synthase
Length = 805
Score = 96.7 bits (239), Expect = 2e-21
Identities = 49/80 (61%), Positives = 63/80 (78%), Gaps = 5/80 (6%)
Query: 1 IVLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGSANVNYVLELDF 60
IVLPP+VALA+R PG+ EY+RVNV+ L V++L +E+L+FKEELV+G AN +Y+LELDF
Sbjct: 75 IVLPPFVALAIRPRPGVREYVRVNVYELSVDHLTVSEYLRFKEELVNGHANGDYLLELDF 134
Query: 61 EPFTASFPRP-----IGGGV 75
EPF A+ PRP IG GV
Sbjct: 135 EPFNATLPRPTRSSSIGNGV 154
>At4g02280 putative sucrose synthetase
Length = 809
Score = 89.4 bits (220), Expect = 3e-19
Identities = 48/80 (60%), Positives = 58/80 (72%), Gaps = 5/80 (6%)
Query: 1 IVLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGSANVNYVLELDF 60
IV+PP+VALAVR PG+ EY+RVNV L VE L +E+L+FKEELVDG + + LELDF
Sbjct: 80 IVVPPFVALAVRPRPGVWEYVRVNVFELSVEQLTVSEYLRFKEELVDGPNSDPFCLELDF 139
Query: 61 EPFTASFPRP-----IGGGV 75
EPF A+ PRP IG GV
Sbjct: 140 EPFNANVPRPSRSSSIGNGV 159
>At5g37180 sucrose synthase-like protein
Length = 843
Score = 50.1 bits (118), Expect = 2e-07
Identities = 35/89 (39%), Positives = 46/89 (51%), Gaps = 6/89 (6%)
Query: 1 IVLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGS-ANVNYVLELD 59
+V+PP VA AVR PG +Y++VN L VE L ++LK KE L D + AN LE+D
Sbjct: 75 VVIPPNVAFAVRGTPGNWQYVKVNSSNLSVEALSSTQYLKLKEFLFDENWANDENALEVD 134
Query: 60 FEPFTASFP-----RPIGGGVVVDEEKKG 83
F + P IG G+ K G
Sbjct: 135 FGALDFTLPWLSLSSSIGNGLSFVSSKLG 163
>At1g73370
Length = 942
Score = 50.1 bits (118), Expect = 2e-07
Identities = 28/69 (40%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Query: 2 VLPPWVALAVRLMPGI*EYLRVNVHALVVENLQPAEFLKFKEELVDGS-ANVNYVLELDF 60
V+PP+VALA R PG EY++VN L V+ + ++LK KE + D S + LE+DF
Sbjct: 84 VVPPFVALAARPNPGFWEYVKVNSGDLTVDEITATDYLKLKESVFDESWSKDENALEIDF 143
Query: 61 EPFTASFPR 69
+ PR
Sbjct: 144 GAIDFTSPR 152
>At4g05060 unknown protein
Length = 287
Score = 25.8 bits (55), Expect = 4.0
Identities = 10/17 (58%), Positives = 14/17 (81%)
Query: 68 PRPIGGGVVVDEEKKGR 84
P+PIG G+V+DE K+ R
Sbjct: 257 PKPIGEGLVIDEWKQRR 273
>At4g21450 unknown protein
Length = 295
Score = 25.4 bits (54), Expect = 5.2
Identities = 16/45 (35%), Positives = 24/45 (52%), Gaps = 4/45 (8%)
Query: 40 KFKEELVDGSANVNYVLELDFEPFTASFPRPIGGGVVVDEEKKGR 84
K K +L + A V E +P + P+ IG G+V+DE K+ R
Sbjct: 240 KLKRQLAEADAAV----EARKKPPEETGPKMIGEGLVIDEWKERR 280
>At3g55160 putative protein
Length = 2149
Score = 25.4 bits (54), Expect = 5.2
Identities = 14/37 (37%), Positives = 18/37 (47%)
Query: 35 PAEFLKFKEELVDGSANVNYVLELDFEPFTASFPRPI 71
P E+LK VD S V+ L P T+S P P+
Sbjct: 656 PIEWLKMALTHVDESVRVDAAETLFLNPKTSSLPSPL 692
>At2g18780 unknown protein
Length = 370
Score = 25.4 bits (54), Expect = 5.2
Identities = 12/27 (44%), Positives = 16/27 (58%)
Query: 56 LELDFEPFTASFPRPIGGGVVVDEEKK 82
L++D +P T + GG VDEEKK
Sbjct: 304 LKVDMKPLTGNSFSYSGGSFFVDEEKK 330
>At5g54110 membrane associated protein
Length = 266
Score = 25.0 bits (53), Expect = 6.8
Identities = 16/45 (35%), Positives = 24/45 (52%), Gaps = 4/45 (8%)
Query: 40 KFKEELVDGSANVNYVLELDFEPFTASFPRPIGGGVVVDEEKKGR 84
K K +L + A V E +P + PR +G G+V+DE K+ R
Sbjct: 210 KLKRQLDEAEAAV----EARKKPPPETGPRVVGEGLVIDEWKERR 250
>At5g11370 putative protein
Length = 311
Score = 25.0 bits (53), Expect = 6.8
Identities = 12/36 (33%), Positives = 19/36 (52%)
Query: 28 LVVENLQPAEFLKFKEELVDGSANVNYVLELDFEPF 63
L+V L P + L+ K EL+ + L+L F+ F
Sbjct: 272 LIVSKLDPQKELEMKNELLSSCGSTTCNLDLVFKQF 307
>At5g07610 putative protein
Length = 420
Score = 24.6 bits (52), Expect = 8.9
Identities = 14/48 (29%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Query: 24 NVHALVVENLQPAEFLKFKEELVDGSANVNYVLELDFEPFTASFPRPI 71
N++ + V N + ++ E S +V Y +D EP A+FP I
Sbjct: 294 NLYYIEVNNQSSSNLRVYEMESNSSSWSVKY--NVDLEPLAAAFPEMI 339
>At4g33290 hypothetical protein
Length = 430
Score = 24.6 bits (52), Expect = 8.9
Identities = 13/30 (43%), Positives = 15/30 (49%)
Query: 53 NYVLELDFEPFTASFPRPIGGGVVVDEEKK 82
N L +D EP I GG +DEEKK
Sbjct: 297 NKFLRIDIEPNNNIMVPFIYGGFFIDEEKK 326
>At3g24240 receptor kinase, putative
Length = 558
Score = 24.6 bits (52), Expect = 8.9
Identities = 14/32 (43%), Positives = 18/32 (55%), Gaps = 4/32 (12%)
Query: 53 NYVLELDFEPFTASFPRPIGGGVVVDEEKKGR 84
N ++ LDFEP+ A F G +VDE GR
Sbjct: 341 NILIGLDFEPYIADF----GLAKLVDEGDIGR 368
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.148 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,876,110
Number of Sequences: 26719
Number of extensions: 64508
Number of successful extensions: 174
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 164
Number of HSP's gapped (non-prelim): 15
length of query: 84
length of database: 11,318,596
effective HSP length: 60
effective length of query: 24
effective length of database: 9,715,456
effective search space: 233170944
effective search space used: 233170944
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC135311.3


