

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135232.5 + phase: 0
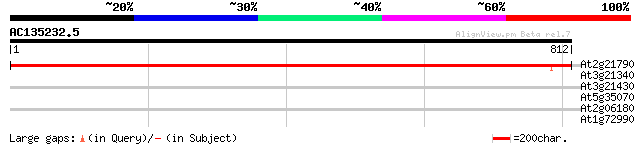
(812 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g21790 ribonucleoside-diphosphate reductase large subunit 1432 0.0
At3g21340 serine/threonine-specific protein kinase, putative 32 1.4
At3g21430 unknown protein 30 5.4
At5g35070 putative protein 29 9.3
At2g06180 hypothetical protein 29 9.3
At1g72990 unknown protein 29 9.3
>At2g21790 ribonucleoside-diphosphate reductase large subunit
Length = 816
Score = 1432 bits (3706), Expect = 0.0
Identities = 687/816 (84%), Positives = 753/816 (92%), Gaps = 4/816 (0%)
Query: 1 MYVVKRDGRQEAVHFDKITARLKKLSYGLSTEHCDPVLVAQKVCAGVYKGVTTSQLDELA 60
MYVVKRDGRQE VHFDKITARLKKLSYGLS++HCDPVLVAQKVCAGVYKGVTTSQLDELA
Sbjct: 1 MYVVKRDGRQETVHFDKITARLKKLSYGLSSDHCDPVLVAQKVCAGVYKGVTTSQLDELA 60
Query: 61 AETAAGMTANHPDYASLAARIAVSNLHKNTLKSFSETVKVMYNHFNHRSGKKAPLIADDV 120
AETAA MT NHPDYASLAARIAVSNLHKNT KSFSET+K M+ H N RSG K+PLIADDV
Sbjct: 61 AETAAAMTCNHPDYASLAARIAVSNLHKNTKKSFSETIKDMFYHVNDRSGLKSPLIADDV 120
Query: 121 YEIIIKNAARLDSEIIYDRDFDYDYFGFKTLERSYLLKVQGVVVERPQHMIMRVSVGIHK 180
+EII++NAARLDSEIIYDRDF+YDYFGFKTLERSYLLKVQG VVERPQHM+MRV+VGIHK
Sbjct: 121 FEIIMQNAARLDSEIIYDRDFEYDYFGFKTLERSYLLKVQGTVVERPQHMLMRVAVGIHK 180
Query: 181 DDVESAVKTYHFMSQRWFTHASPTLFNAGTPRPQLSSCFLVCMKEDSIEGIYDTLKECAV 240
DD++S ++TYH MSQRWFTHASPTLFNAGTPRPQLSSCFLVCMK+DSIEGIY+TLKECAV
Sbjct: 181 DDIDSVIQTYHLMSQRWFTHASPTLFNAGTPRPQLSSCFLVCMKDDSIEGIYETLKECAV 240
Query: 241 ISKSAGGIGVSVHNIRAHGSYIRGTNGTSNGIVPMLRVFNDTARYVDQGGGKRKGAFAVY 300
ISKSAGGIGVSVHNIRA GSYIRGTNGTSNGIVPMLRVFNDTARYVDQGGGKRKGAFAVY
Sbjct: 241 ISKSAGGIGVSVHNIRATGSYIRGTNGTSNGIVPMLRVFNDTARYVDQGGGKRKGAFAVY 300
Query: 301 LEPWHADIFEFLELKINHGKEEQRARDLFYALWVPDLFMERVQSNGEWSLFCPNEAPGLA 360
LEPWHAD++EFLEL+ NHGKEE RARDLFYALW+PDLFMERVQ+NG+WSLFCPNEAPGLA
Sbjct: 301 LEPWHADVYEFLELRKNHGKEEHRARDLFYALWLPDLFMERVQNNGQWSLFCPNEAPGLA 360
Query: 361 DCWGEEYDKLYTQYEKAGKAKKIVQAQSLWFAILKSQIETGNPYMLFKDACNRKSNQQNL 420
DCWG E++ LYT+YE+ GKAKK+VQAQ LW+ IL SQ+ETG PYMLFKD+CNRKSNQQNL
Sbjct: 361 DCWGAEFETLYTKYEREGKAKKVVQAQQLWYEILTSQVETGTPYMLFKDSCNRKSNQQNL 420
Query: 421 GTIKSSNLCTEIIEYTSPTETAVCNLASIALPRYVRDKGLPVESHQSKLVGSRGSSSRYF 480
GTIKSSNLCTEIIEYTSPTETAVCNLASIALPR+VR+KG+P++SH KL GS S +RYF
Sbjct: 421 GTIKSSNLCTEIIEYTSPTETAVCNLASIALPRFVREKGVPLDSHPPKLAGSLDSKNRYF 480
Query: 481 DFEKLGEVTAVVTTNLNKIIDVNYYPVETAERSNLRHRPIGIGVQGLADTFILLGMAFDS 540
DFEKL EVTA VT NLNKIIDVNYYPVETA+ SN+RHRPIGIGVQGLAD FILLGM FDS
Sbjct: 481 DFEKLAEVTATVTVNLNKIIDVNYYPVETAKTSNMRHRPIGIGVQGLADAFILLGMPFDS 540
Query: 541 PEAQQLNKDIFETIYYHALKTSCELAAKEGPYETYSGSPVSKGILQPDMWGVTPSNLWDW 600
PEAQQLNKDIFETIYYHALK S ELAA+ GPYETY+GSPVSKGILQPDMW V PS+ WDW
Sbjct: 541 PEAQQLNKDIFETIYYHALKASTELAARLGPYETYAGSPVSKGILQPDMWNVIPSDRWDW 600
Query: 601 GALREMISKTGVRNSLLVAPMPTASTSQILGNNECFEPYTSNIYNRRVLSGEFVVVNKHL 660
LR+MISK GVRNSLLVAPMPTASTSQILGNNECFEPYTSNIY+RRVLSGEFVVVNKHL
Sbjct: 601 AVLRDMISKNGVRNSLLVAPMPTASTSQILGNNECFEPYTSNIYSRRVLSGEFVVVNKHL 660
Query: 661 LHDLTEMGLWNPALKNKIIYDNGSVQNLSEIPAELKVIYKTVWEIKQKILVDMAVDRGCY 720
LHDLT+MGLW P LKNK+I +NGS+ N++EIP +LK IY+TVWEIKQ+ +VDMA DRGCY
Sbjct: 661 LHDLTDMGLWTPTLKNKLINENGSIVNVAEIPDDLKAIYRTVWEIKQRTVVDMAADRGCY 720
Query: 721 IDQSQSLNIHMDKPNFGKLTSLHFYAWSKGLKTGMYYLRTRAASDAIKFTVDTSAIKDQP 780
IDQSQSLNIHMDKPNF KLTSLHFY W KGLKTGMYYLR+RAA+DAIKFTVDT+ +K++P
Sbjct: 721 IDQSQSLNIHMDKPNFAKLTSLHFYTWKKGLKTGMYYLRSRAAADAIKFTVDTAMLKEKP 780
Query: 781 KV----KEADDEDDDTKMAQMVCSLTNRDECEACGS 812
V KE ++ED++TK+AQMVCSLTN +EC ACGS
Sbjct: 781 SVAEGDKEVEEEDNETKLAQMVCSLTNPEECLACGS 816
>At3g21340 serine/threonine-specific protein kinase, putative
Length = 880
Score = 32.0 bits (71), Expect = 1.4
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 2/76 (2%)
Query: 570 GPYETYSGSPVSKGILQPDMWGVTPSNLWDWGALREMISKTGVRNSLLVAPMPTASTSQI 629
G Y+ + P L P++W N G ++E+I KT + SL V + T ++S +
Sbjct: 116 GNYDGLNNPPSFDLYLGPNLWVTVDMNGRTNGTIQEIIHKT-ISKSLQVCLVKTGTSSPM 174
Query: 630 LGNNECFEPYTSNIYN 645
+ E P +N YN
Sbjct: 175 INTLE-LRPLKNNTYN 189
>At3g21430 unknown protein
Length = 961
Score = 30.0 bits (66), Expect = 5.4
Identities = 22/84 (26%), Positives = 35/84 (41%), Gaps = 3/84 (3%)
Query: 396 SQIETGNPYMLFKDACNRKSNQQNLGTIKSSNLCTEIIEYTSPTETAVCNLASIALPRYV 455
S + +P + K + R S Q+ G S++LC +E SP+ + + ALP V
Sbjct: 239 SDFKGTDPASMSKSSSLRNSKQRRYG---SNDLCNPELERKSPSSSLIQKRRQKALPAKV 295
Query: 456 RDKGLPVESHQSKLVGSRGSSSRY 479
L S + S+G Y
Sbjct: 296 LKDELAASSQVIEPCNSKGIGEEY 319
>At5g35070 putative protein
Length = 356
Score = 29.3 bits (64), Expect = 9.3
Identities = 18/64 (28%), Positives = 32/64 (49%), Gaps = 6/64 (9%)
Query: 439 TETAVCNLASIALPRYVRDKGL------PVESHQSKLVGSRGSSSRYFDFEKLGEVTAVV 492
T T + L+ + LP+ +RD G +++ +SKLV + YFD +L T ++
Sbjct: 18 TNTRLVTLSHVDLPKRIRDVGEHTEDSGEIDTGRSKLVIEEVCFADYFDHRELSFATILL 77
Query: 493 TTNL 496
+ L
Sbjct: 78 SVEL 81
>At2g06180 hypothetical protein
Length = 793
Score = 29.3 bits (64), Expect = 9.3
Identities = 17/62 (27%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query: 226 DSIEGIYDTLKECAVISKSAGGIGVSVHNIRAHGSYIRGTNGT--SNGIVPMLRVFNDTA 283
DS G YDT K AV + AGG+G+ + + +G + ++P LR+ ++ +
Sbjct: 128 DSGGGDYDTPKVAAVTQERAGGVGLCLRQFLREMWAVLPDSGVHPRDALLPRLRLSSNVS 187
Query: 284 RY 285
++
Sbjct: 188 KF 189
>At1g72990 unknown protein
Length = 697
Score = 29.3 bits (64), Expect = 9.3
Identities = 18/53 (33%), Positives = 25/53 (46%), Gaps = 1/53 (1%)
Query: 146 FGFKTLERSYLLKVQGVVVERPQ-HMIMRVSVGIHKDDVESAVKTYHFMSQRW 197
FGF E SY+ K G + P+ H +V V DV+ V Y ++RW
Sbjct: 452 FGFLLYESSYIAKKSGNTLRIPKVHDRAQVFVSCLSQDVDVGVLRYIGTTERW 504
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,218,531
Number of Sequences: 26719
Number of extensions: 789998
Number of successful extensions: 1863
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1860
Number of HSP's gapped (non-prelim): 6
length of query: 812
length of database: 11,318,596
effective HSP length: 107
effective length of query: 705
effective length of database: 8,459,663
effective search space: 5964062415
effective search space used: 5964062415
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC135232.5


