

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135230.12 + phase: 0
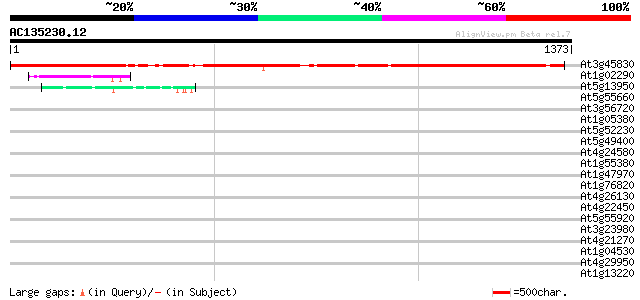
(1373 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g45830 DNA-binding protein - like 1149 0.0
At1g02290 unknown protein 131 2e-30
At5g13950 unknown protein 75 2e-13
At5g55660 putative protein 41 0.004
At3g56720 putative protein 38 0.046
At1g05380 unknown protein 37 0.060
At5g52230 unknown protein 37 0.079
At5g49400 putative protein 37 0.079
At4g24580 putative protein 37 0.10
At1g55380 hypothetical protein 36 0.13
At1g47970 Unknown protein (T2J15.12) 36 0.13
At1g76820 putative translation initiation factor IF-2 36 0.18
At4g26130 uncharacterized protein 35 0.30
At4g22450 hypothetical protein 35 0.30
At5g55920 nucleolar protein-like 35 0.39
At3g23980 unknown protein 35 0.39
At4g21270 kinesin-related protein katA 34 0.51
At1g04530 unknown protein 34 0.51
At4g29950 unknown protein 34 0.67
At1g13220 putative nuclear matrix constituent protein 34 0.67
>At3g45830 DNA-binding protein - like
Length = 1298
Score = 1149 bits (2971), Expect = 0.0
Identities = 678/1388 (48%), Positives = 865/1388 (61%), Gaps = 120/1388 (8%)
Query: 1 MAIEKNSFKVSRVDTECEPMSKESMSSGDEEDVQRRNSGNESDEDDDEFDDADSGAGSDD 60
MAIEK++ KVSR D E S +SMSS +E + +S+++DD+FD+ DSGAGSDD
Sbjct: 1 MAIEKSNVKVSRFDLEYSHGSGDSMSSYEERRKNSVVNNVDSEDEDDDFDEDDSGAGSDD 60
Query: 61 FDLLELGETGAEFCQIGNQTCSIPLELYDLSGLEDILSVDVWNDCLSEEERFELAKYLPD 120
FDLLEL ETGAEFCQ+GN TCSIP ELYDL LEDILSVDVWN+CL+E+ERF L+ YLPD
Sbjct: 61 FDLLELAETGAEFCQVGNVTCSIPFELYDLPSLEDILSVDVWNECLTEKERFSLSSYLPD 120
Query: 121 MDQETFVQTLKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYREGLNFVQKRQHYHLL 180
+DQ TF++TLKELF GCNF FGSPVKKLFDMLKGG CEPR LY EG + + +HYH L
Sbjct: 121 VDQLTFMRTLKELFEGCNFHFGSPVKKLFDMLKGGQCEPRNTLYLEGRSLFLRTKHYHSL 180
Query: 181 KKHQNTMVSNLCQMRDAWLNCRGYSIEERLRVLNIMTSQKSLMGEKMDDLEADSSE--ES 238
+K+ N MV NLCQ RDAW +C+GYSI+E+LRVLNI+ SQK+LM EK DD E DSSE E
Sbjct: 181 RKYHNDMVVNLCQTRDAWTSCKGYSIDEKLRVLNIVKSQKTLMREKKDDFEDDSSEKDEP 240
Query: 239 GEGMWSRKNKDKKNAQK-LGRFPFQGVGSGLDFHPREQSMVMEQEKYSKQNPKGILKLAG 297
+ W RK KD+K+ QK L R GV SGL+F PR Q +EQ+ Y K PK K
Sbjct: 241 FDKPWGRKGKDRKSTQKKLARHAGYGVDSGLEF-PRRQLAAVEQDLYGK--PKSKPKFPF 297
Query: 298 SKTHLAKDPTAHSSSVYHGLDMNPRLNGSAFAHPQHNISTGYDLGSIRRTRDQLWNGDNE 357
+KT + T Y+G MN N S+ ++ D D++
Sbjct: 298 AKTSVGPYATG-----YNGYGMNSAYNPSSLVRQRYGSEDNID-------------DDDQ 339
Query: 358 EEI----SYRDRNALRGSLMDMSSALRVGKRHDLLRGDEIEGGNLMGLSMSSKTDLRGYT 413
+ + S RDR + R GK+H R E + MG SS+ Y+
Sbjct: 340 DPLFGMGSRRDR--------EKPGYSRPGKKHKSSRDGEPISEHFMGPPYSSRQYHSNYS 391
Query: 414 RNPNQSSDMQLFAAKPPSKK-KGKYAENVQQFVGSRGSKLSHNVDSIHSPDPDDLFYNKR 472
++ ++++Q A K KG A+ RG H K
Sbjct: 392 KSSRYANNIQPHAFADQMKPVKGSLAD-------LRGDLYRHG---------------KN 429
Query: 473 PAQELGMSSLFKYEDWNPKSKKRKAERESPDLSYTAYRSSSPQVSNRLFSSDFRTKSSQE 532
+ + +D N KSKK K+ER+SPD S +YR+S Q++ R +SDF QE
Sbjct: 430 HGDGFSVDPRYISDDLNSKSKKLKSERDSPDTSLRSYRASMQQMNERFLNSDFGENHVQE 489
Query: 533 KIRGSFVQNGRKDMKPLRGSHMLARGEETESDSSEQWDDDDDNNPLLQSKFAYPIGKAAG 592
KIR + V N R + R S M ++TESDSS +DD+++ N L+++K + +G
Sbjct: 490 KIRVNVVPNARSGVAAFRDSRMFMGNDDTESDSSHGYDDEEERNRLMRNKSSVSVGGMNN 549
Query: 593 SLTKPLKSHLDPMKAKFSRTDMKATQS--------KKIGGFAEQGNMHGADNYLSKNAKK 644
S LKS D K+K + DM+ + K +G E G + + K+ +K
Sbjct: 550 SHFPILKSRQDIKKSKSRKKDMQENELLDGRSAYLKYLGVSGEHIYAPGTEKHSFKSKQK 609
Query: 645 SKIFNGSPVRNPAGKFMEENYPSVSDMLNGGHDDWRQLYKS-KNDQIRDEPVQR--FDMP 701
K+ + SP+ N + + E+ P S ++ ++ ++S +N Q R++ + R F P
Sbjct: 610 GKMRDRSPLENFSSRDFEDG-PITSLSEFQDRNNRKEFFRSNRNSQTREQMIDRTLFQRP 668
Query: 702 SSTSYAAEHKKKGRIGLDHSSMRSKYLHDYGNDEDDSLENRLLADENGVGQSRFWRKGQK 761
S+ + K+ +++D+S E R L + + R RK Q
Sbjct: 669 SAKPNLSGRKR------------------VFDEDDESHEMRTLVN----ARDRLSRKYQV 706
Query: 762 NVAHKDDRDERSEVPL-LGCNSAMKKRKMKFGAADFGERDEDANLLSSNPSKIDDLPAFS 820
+ + DE E L + CN+ KKRK + D R+++ +L ++
Sbjct: 707 SEDDGNSGDENLEARLFVSCNALSKKRKTRESLMDMERREDNGDLQLYPDIQLPVGDVTV 766
Query: 821 LKRKSKKKPGAEMVISEMENSELPLTHTVTADVEVETKPQKKPYILITPTVHTGFSFSIM 880
KRK KKK ++ ++E S++P ++ EVETKPQKKP++LITPTVHTGFSFSI+
Sbjct: 767 SKRKGKKKMEVDVGFLDLETSDIPKA----SEAEVETKPQKKPFVLITPTVHTGFSFSIV 822
Query: 881 HLLTAVRTAMISPPEVESLEAGKPVEQQNKAQEDSLNGVISSDKVDDKVAANVEPSDQK- 939
HLL+AVR AM S +SL+ K V +N E NG ++ +D N P
Sbjct: 823 HLLSAVRMAMTSLRPEDSLDVSKSVAVENAEHETGENGASVPEEAED----NKSPQQGNG 878
Query: 940 NVPSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAVYE 999
N+PSLTIQEIV+ V+SNPGDPCILETQEPLQDL+RGVLKIFSSKT+PLGAKGWK L +E
Sbjct: 879 NLPSLTIQEIVSCVKSNPGDPCILETQEPLQDLIRGVLKIFSSKTSPLGAKGWKPLVTFE 938
Query: 1000 KSTRSWSWIGPVLHNSSDHDPIEEVTSPEAWGLPHKMLVKLVDSFANWLKCGQDTLKQIG 1059
+ST+ WSWIGPVL SD + +EEVTSPEAW LPHKMLVKLVDSFANWLK GQ+TL+QIG
Sbjct: 939 RSTKCWSWIGPVL-GPSDQETVEEVTSPEAWSLPHKMLVKLVDSFANWLKTGQETLQQIG 997
Query: 1060 SLPAPPLELMQINLDEKERFRDLRAQKSLNTISPSSEEVRAYFRKEELLRYSIPDRAFSY 1119
SLP PPL LMQ NLDEKERF+DLRAQKSL+TI+ SSEE RAYFRKEE LRYSIPDRAF Y
Sbjct: 998 SLPEPPLSLMQCNLDEKERFKDLRAQKSLSTITQSSEEARAYFRKEEFLRYSIPDRAFVY 1057
Query: 1120 TAADGKKSIVAPLRRCGGKPTSKARDHFMLKRDRPPHVTILCLVRDAAARLPGSIGTRAD 1179
TAADGKKSIVAPLRR GGKPTSKARDHFMLKR+RPPHVTILCLVRDAAARLPGSIGTRAD
Sbjct: 1058 TAADGKKSIVAPLRRGGGKPTSKARDHFMLKRERPPHVTILCLVRDAAARLPGSIGTRAD 1117
Query: 1180 VCTLIRDSQYIVEDVSDEKINQVVSGALDRLHYERDPCVLFDQERKLWVYLHREREEEDF 1239
VCTLIRDSQYIVEDVSD ++NQVVSGALDRLHYERDPCV FD ERKLWVYLHR+RE+EDF
Sbjct: 1118 VCTLIRDSQYIVEDVSDSQVNQVVSGALDRLHYERDPCVQFDSERKLWVYLHRDREQEDF 1177
Query: 1240 DDDGTSSTKKWKRQKKDVADQS-DQAPVTVACNGTGEQSGYDLCSDLNV-DPPCIEDDKE 1297
+DDGTSSTKKWKR KK+ A+Q+ +Q VTVA G EQ+ ++ S+ +P ++ D+
Sbjct: 1178 EDDGTSSTKKWKRPKKEAAEQTEEQEAVTVAFLGNEEQTETEMGSEPKTGEPTGLDGDQG 1237
Query: 1298 AVQLLTTDTRPNAEDQVVVNPVSEVGNSCEDNSMT-WE---ALDLNPTRE---LCQENST 1350
A L +T AE+Q + N+ + N T WE A+ NP + +CQENS
Sbjct: 1238 ATDQLCNETEQAAEEQ-------DGENTAQGNEPTIWEPDPAVVSNPVEDNTFICQENSV 1290
Query: 1351 NEDFGDES 1358
N+DF DE+
Sbjct: 1291 NDDFDDET 1298
>At1g02290 unknown protein
Length = 443
Score = 131 bits (330), Expect = 2e-30
Identities = 87/263 (33%), Positives = 132/263 (50%), Gaps = 24/263 (9%)
Query: 46 DDEFDDADSGAGSDDFDLLELGETGAEFCQIGNQTCSIPLELYDLSGLEDILSVDVWNDC 105
D E DD SDD+D+ ++ E + Q C+IP ELYDL L ILSV+ WN
Sbjct: 10 DSEDDD------SDDYDIAQVN---CELALVEGQLCNIPYELYDLPDLTGILSVETWNSL 60
Query: 106 LSEEERFELAKYLPDMDQETFVQTLKELFTGCNFQFGSPVKKLFDMLKGGLCEPRVALYR 165
L+EEERF L+ +LPDMD +TF T++EL G N FG+P K + L GGL P+VA ++
Sbjct: 61 LTEEERFFLSCFLPDMDPQTFSLTMQELLDGANLYFGNPEDKFYKNLLGGLFTPKVACFK 120
Query: 166 EGLNFVQKRQHYHLLKKHQNTMVSNLCQMRDAWLNCRGYSIEERLRVLNIMTSQKSLMGE 225
EG+ FV++R++Y+ LK + ++ +M+ W+ G + R+L I + +
Sbjct: 121 EGVMFVKRRKYYYSLKFYHEKLIRTFTEMQRVWVQ-YGNKLGNYSRLL-IWSGRTQTGNL 178
Query: 226 KMDDLEADSSEESGEGMWSRKNKD-----KKNAQKLGRFPFQGVGSGL--------DFHP 272
K+ DL S+E K + ++N K FP G
Sbjct: 179 KLLDLNRVPSKEMDSATCRFKTPNVVKPVERNRSKSLTFPRSGSSKNSLKIKITKEGVFR 238
Query: 273 REQSMVMEQEKYSKQNPKGILKL 295
+ S ++ + + PKG+LKL
Sbjct: 239 YQGSSLVSAGHHHQTLPKGVLKL 261
>At5g13950 unknown protein
Length = 939
Score = 75.5 bits (184), Expect = 2e-13
Identities = 94/424 (22%), Positives = 168/424 (39%), Gaps = 67/424 (15%)
Query: 79 QTCSIPLELYDLSGLEDILSVDVWNDCLSEEERFELAKYLPD-MDQETFVQTLKELFTGC 137
Q C +P E + L L ++LS +VW CLS+ ER L ++LP+ +D E VQ L + G
Sbjct: 85 QVCPVPHETFQLENLSEVLSNEVWRSCLSDGERNYLRQFLPEGVDVEQVVQALLD---GE 141
Query: 138 NFQFGSPVKKLFDMLKGGLCEPRVALYREGLNFVQKRQHYHLLKKHQNTMVSNLCQMRDA 197
NF FG+P + G P + RE KR++Y L+K+ ++ L +++
Sbjct: 142 NFHFGNPSLDWGTAVCSGKAHPDQIVSREECLRADKRRYYSNLEKYHQDIIDYLQTLKEK 201
Query: 198 WLNCRGYSIEERLRVLNIMTSQKSLMGEKMDDLEADSSEESGEGMWSRKNKDKKN----- 252
W +C+ + ++ +M + +++ + SG W+ +K +
Sbjct: 202 WESCK----DPEKDIVKMMWGRSRGGNAQVNGSCQGLTAASGSSSWNEDDKPDSSDNMIS 257
Query: 253 -AQKLGRFPFQGVGSGLDFHPREQSMVMEQEKYSKQN--PKGILKLAGSKTHLAKDPTAH 309
+ G + SGL+ + + V K K+N PK ++ +++
Sbjct: 258 PVVRCGEVQRRSKRSGLEKEKTQNNGVNVGGKVRKKNVLPKDSIQQTDGAKYMS---YLK 314
Query: 310 SSSVYHGLDMNPRLNGSAFAHPQHNISTGYDLGSIRRTRDQLWNGDNEEEISYRDRNALR 369
S H + + + +G + + + G+I Q + EEE + + L
Sbjct: 315 ISKKQHQIVTSMKQSGKSI----QSRALNRIFGNIDSLDVQPYGVFVEEEQKKLNAHWLH 370
Query: 370 GSLMDMSSALRVGKRHDLLRGDEIEGGNLMGLSMSSKTDL-------------------- 409
+ D+ +A + KR L + D I MG + K +L
Sbjct: 371 -LVKDLPAAYAIWKRLQLQKRDIISS---MGRELKEKCNLWMEDKQQQYAAENPLQKHDV 426
Query: 410 ---RGYTRNPNQSSDMQ-------LFAA-----KPPSKKKGKYAENVQQ-----FVGSRG 449
G + NPNQS DM +F+ PSK Y + + VG+
Sbjct: 427 QYNNGESLNPNQSGDMAPDGEDSGIFSQISGKNHSPSKDSSSYGDQITDSGRCLLVGTHS 486
Query: 450 SKLS 453
S++S
Sbjct: 487 SQVS 490
>At5g55660 putative protein
Length = 759
Score = 41.2 bits (95), Expect = 0.004
Identities = 42/188 (22%), Positives = 70/188 (36%), Gaps = 11/188 (5%)
Query: 414 RNPNQSSDMQLFAAKPPSKKKGKYAENVQQFVGSRGSKLSHNVDSIHSPDPDDLFYNKRP 473
R P +SS A SK+ K + ++ + ++H+ D DD K
Sbjct: 468 RTPKKSSPA---AGSSSSKRSAKSQKKTEEATRTNKKSVAHSDDESEEEKEDDEEEEKEQ 524
Query: 474 AQELGMSSLFKYEDWNPKSKKRKAERESPDLSYTAYRSSSPQVSNRLFSSDFR-TKSSQE 532
E + E+ N K+E E+P LS + S + S R +++S +
Sbjct: 525 EVE-------EEEEENENGIPDKSEDEAPQLSESEENVESEEESEEETKKKKRGSRTSSD 577
Query: 533 KIRGSFVQNGRKDMKPLRGSHMLARGEETESDSSEQWDDDDDNNPLLQSKFAYPIGKAAG 592
K + +K P + S ++ + ++ DDD D +P SK A
Sbjct: 578 KKESAGKSRSKKTAVPTKSSPPKKATQKRSAGKRKKSDDDSDTSPKASSKRKKTEKPAKE 637
Query: 593 SLTKPLKS 600
PLKS
Sbjct: 638 QAAAPLKS 645
>At3g56720 putative protein
Length = 390
Score = 37.7 bits (86), Expect = 0.046
Identities = 38/158 (24%), Positives = 67/158 (42%), Gaps = 13/158 (8%)
Query: 495 RKAERESPDLSYTAYRSSSPQVSNRLFSSDFRTKSSQEKIRGSFVQNGRKDMKPLRGSHM 554
R ++R + Y+ R+ S + S+ +S+ S EK G + +GR+ H
Sbjct: 126 RHSDRRRVETEYSRLRNDSDRSSHDKYSNSGHRVKSNEK--GEDLSSGRR--------HS 175
Query: 555 LARGEETESDSSEQWDDDDDNNPLLQSKFAYPIGKAAGSLTKPLKSHLDPMKAKFSRTDM 614
+R E+ E S +W D ++ + + + P + + +KS D K T+
Sbjct: 176 DSRVEDNEKRGS-RWGFGDRHSRVERKEHEDP--EISKEKEVHVKSSRDRSDGKCLATED 232
Query: 615 KATQSKKIGGFAEQGNMHGADNYLSKNAKKSKIFNGSP 652
+ T SKK+ GF G N + K++ I SP
Sbjct: 233 RDTHSKKLKGFISDKFTTGNTNDAAAEEKQTSILKPSP 270
>At1g05380 unknown protein
Length = 1138
Score = 37.4 bits (85), Expect = 0.060
Identities = 86/385 (22%), Positives = 142/385 (36%), Gaps = 90/385 (23%)
Query: 353 NGDNEEEISYRDRNALRGSLMDMSSALRVGKRHDLLRGDEIEGGN-LMGLSMSSKTDLRG 411
+G E + R RN SS D ++G N ++ + +++KT+
Sbjct: 156 SGSGREGLFDRRRNTFVNGTCSASSQEDSSSESDSDEPMRVQGINGVLKVKVNNKTNTLA 215
Query: 412 YTRNPNQSSDMQLFAAKPPSKKKGKYAENVQQFVGSRGSKLSHNVDSIHSPDPDDLFYNK 471
+ NP D +++ +PPS +K + ENV V + S+NVD N
Sbjct: 216 ASINPR---DAEIYE-RPPSSRKAQRRENV---VVKPPFRKSNNVD------------NN 256
Query: 472 RPAQELGMSSLFKYEDWNPKSKKRKAERESPDLSY-TAYRSSSPQVSNRLFSSDFRTKSS 530
++E MS KSK++K+E P + T +S+ P++ N + R +
Sbjct: 257 SESEESDMSR---------KSKRKKSEYSKPKKEFNTKSKSTFPELVNPDVREERRGRRG 307
Query: 531 --------QEKIRGSFVQNG-RKDMKPLRGSHMLARGEETESDSSEQWDDDDDNNPLLQS 581
+E+I+G G D KP R N L +
Sbjct: 308 GGTDKQRLRERIKGMLTDAGWTIDYKPRR------------------------NQSYLDA 343
Query: 582 KFAYPIGKAAGSLTKPLKSHLDPMKAKFSRTDMKATQSKKIGGFAEQGNMHGADNYLSKN 641
+ P G A S+ K + L +K + + A K A ++ ++K
Sbjct: 344 VYVNPSGTAYWSIIKAYDALLKQLKDE----GVDARPRKDTAAVASV-----SEEIVNKL 394
Query: 642 AKKSKIFNGSPVRNPAGKFMEENYPSVSDMLNGGHDDWRQLYKSKNDQIRDEPVQRFDMP 701
A+K+K R+ K ++N S SD N KS+ D +R +
Sbjct: 395 ARKAK-----KTRSEMTKKWKQN-SSGSDSEN----------KSEGGAYTDTSEER--IR 436
Query: 702 SSTSYAAEHKKKGRIGLDHSSMRSK 726
SS + KKGR G D + K
Sbjct: 437 SSIKLGGKSTKKGRNGADWDELHKK 461
>At5g52230 unknown protein
Length = 746
Score = 37.0 bits (84), Expect = 0.079
Identities = 60/269 (22%), Positives = 109/269 (40%), Gaps = 27/269 (10%)
Query: 705 SYAAEHKKKGRIGLDHSSMRSKYLHDYGNDED--DSLENRLLADENGVGQSRFWRKGQ-- 760
S + EH K ++ D S + S+ L D G E+ D +E +L+A Q++ +
Sbjct: 143 SSSDEHSKNCKMTSDLSIVTSQVLEDLGKKEEVKDPIEKQLIAKRVTRSQTKASTTEEVV 202
Query: 761 ----KNVAHKDDRDERSEVPLLGCNSAMKKRKMKFGAADFGERDEDANLLSSNPSKID-- 814
+N++ + + E+ V NS+++ +K K A E ++D++ SK++
Sbjct: 203 VDLKRNLSSSNAKSEKDSV-----NSSVRSQKPKKEAVMKEEEEQDSSEKRITRSKVEEK 257
Query: 815 --DLPAFSLKRKSKKKPGAEMVISEMENSELPLTHTVTADVE--VETKPQKKPYILITPT 870
+L +R SK+ G E+ + + + V D E E K + K ++ P
Sbjct: 258 KNELSNSVARRTSKRLAGIELEPTPELKTRAKVQRIVPLDDEPTPELKTRTKVQRVVPPD 317
Query: 871 VHTGFSFSIMHLLTAVRTAMISPPEVE-SLEAGKPVEQQNKAQEDSLNGVISSDKVDDKV 929
L T + I PP+ E +LE + Q D + + K KV
Sbjct: 318 -----DEPTPELKTRTKIQRIVPPDDEPTLELKTRTKVQRILPPD--DELTPELKSRTKV 370
Query: 930 AANVEPSDQKNVPSLTIQEIVNRVRSNPG 958
V P D+ T ++ R+ + G
Sbjct: 371 QRIVPPDDELTPEFKTRTKVQQRIPPDDG 399
>At5g49400 putative protein
Length = 280
Score = 37.0 bits (84), Expect = 0.079
Identities = 41/208 (19%), Positives = 82/208 (38%), Gaps = 17/208 (8%)
Query: 372 LMDMSSALRVGKRHDLLRGDEIEGGNLMGLSMSSKTDLRGYTRNPNQSSDMQLFAAKPPS 431
LM A+R+G+R D G + S K G+ + + +++ ++P
Sbjct: 73 LMSTEKAVRLGERKDKTMSASAVGSSASAASQCQKCFQAGHWTY--ECKNERVYISRPSR 130
Query: 432 KKKGKYAENVQQFVGSRGSKLSHNVDSIHSPDPDDLFYNKRPAQELGMSSLFKYEDWNPK 491
++ K + + +VD + D DD +RP G + + K + +
Sbjct: 131 TQQLKNPK----------LRTKPSVDDLDGSDDDD--EEERPDATNGKAEVEKRSKKSKR 178
Query: 492 SKKRKAERESPD---LSYTAYRSSSPQVSNRLFSSDFRTKSSQEKIRGSFVQNGRKDMKP 548
+ K++ ES + T SS + S+ SS + + + + +K K
Sbjct: 179 KHRSKSDSESDSEASVFETDSDGSSGESSSEYSSSSDSEDERRRRRKAKKSKKKQKQRKE 238
Query: 549 LRGSHMLARGEETESDSSEQWDDDDDNN 576
R + + E +ES+S+ D D+D +
Sbjct: 239 RRRRYSSSSSESSESESASDSDSDEDRS 266
>At4g24580 putative protein
Length = 790
Score = 36.6 bits (83), Expect = 0.10
Identities = 17/43 (39%), Positives = 26/43 (59%), Gaps = 1/43 (2%)
Query: 20 MSKESMSSGDEEDVQRRNSGNESDEDDDEFDDADSGA-GSDDF 61
+ + S+S G D + SG E DD+E+DD D G+ GS+D+
Sbjct: 346 LQEGSLSPGLYSDSEESGSGTEEGSDDEEYDDDDDGSQGSEDY 388
>At1g55380 hypothetical protein
Length = 661
Score = 36.2 bits (82), Expect = 0.13
Identities = 22/80 (27%), Positives = 39/80 (48%), Gaps = 1/80 (1%)
Query: 8 FKVSRVDTECEPMSKESMSSGDEEDVQRRNSGN-ESDEDDDEFDDADSGAGSDDFDLLEL 66
F V+++ S S S D++D ++ E D DDD+ D +DS ++FD
Sbjct: 95 FDVTKIPKNWMDSSSSSDESEDDDDDEKSIKEECEDDGDDDDGDSSDSDDDDNNFDANND 154
Query: 67 GETGAEFCQIGNQTCSIPLE 86
G + ++ + + C +PLE
Sbjct: 155 GASLSDGNHLKCKCCQVPLE 174
>At1g47970 Unknown protein (T2J15.12)
Length = 198
Score = 36.2 bits (82), Expect = 0.13
Identities = 17/49 (34%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Query: 25 MSSGDEEDVQRRNSGNESDEDDDEFDDADSGAGSDDFDLLELGETGAEF 73
+ S ++ED + GN D+DDD+ DD D +D D+ + G+ G E+
Sbjct: 78 VQSAEDEDEEGDEDGN-GDDDDDDGDDDDDDDDDEDEDVEDEGDLGTEY 125
>At1g76820 putative translation initiation factor IF-2
Length = 1146
Score = 35.8 bits (81), Expect = 0.18
Identities = 57/245 (23%), Positives = 93/245 (37%), Gaps = 41/245 (16%)
Query: 711 KKKGRIGLDHSSMRSKYLHDYGNDEDDSLENRLLADENGVGQSRFWRKGQKNVAHKD--- 767
KKKG+ R+K D+DD ++ +E V + F +K+ KD
Sbjct: 57 KKKGK-------KRNKKGTQQQQDDDDDFADKFAVEEEVVPDNAFVGNNKKSKGKKDGGS 109
Query: 768 -------DRDERSE-----VPLLGCNSAMKKRKMKFGAADFGERDEDANLLSSNPSKIDD 815
D D R E + G + KK+ A G DE+++ ++ SK +
Sbjct: 110 VSFALLADEDNREEAEDNDITFSGRKKSSKKKSSSV-LASIG--DEESSQSKTSDSKSVE 166
Query: 816 LPAFSLKRKSKKKPGAEMVISEMEN-----SELPLTHTVTADVEVETKPQKKPYILITPT 870
L + +K KKK + + E ++ +EL T E K Q +P + P
Sbjct: 167 LVESARSKKHKKKNKSGRTLQEDDDLDKLLAELGETPAAGKPASEEEKDQAQPE-PVAPV 225
Query: 871 VHTGFSFSIMHLLTAVRTAMISPPEVE-------SLEAGKPVEQQNKAQEDSLNGVISSD 923
+TG ++ TA E E + A VE + + QE+S+ +
Sbjct: 226 ENTGEEETVE---TAAAKKKKKKKEKEKEKKAAAAAAATSSVEAKEEKQEESVTEPLQPR 282
Query: 924 KVDDK 928
K D K
Sbjct: 283 KKDAK 287
>At4g26130 uncharacterized protein
Length = 286
Score = 35.0 bits (79), Expect = 0.30
Identities = 42/192 (21%), Positives = 71/192 (36%), Gaps = 30/192 (15%)
Query: 289 PKGILKLAGSKTHLAKDPTAHSSSVYHGLDMNPR-----------LNGSAFAHPQHNIST 337
P I ++ HL P+ S Y G D NP +N F PQHN
Sbjct: 63 PSLIDRVKSINFHLYNSPSPESEIHYSGSDPNPNPPPSLLQRVKSINMPYFKFPQHNSEG 122
Query: 338 GYDLGSIRRTRDQLWNGDNEEEISYRDRNALRGSLMDMSSALRVGKRHDLLRGDEIEGGN 397
Y + D+ D ++I D+++ R G L R I+ +
Sbjct: 123 DYAAYELMTQPDETNRVDPIDKIPED----------DVTTEPRFGAPSLLQRVKSIKLPS 172
Query: 398 LMGLSMSSKTDLRGYTRNPNQSSDMQLFAAKPPSKKKGKYAENVQQFVGSR--GSKLSHN 455
L +++ +TR ++SS KP +KKK K + + + R G +
Sbjct: 173 LYRSDPDPTPEVQTHTRTKSESS-------KPATKKKKKATKKMMKSASERHIGREEEET 225
Query: 456 VDSIHSPDPDDL 467
V+++ P+ +
Sbjct: 226 VEAVEKRRPETM 237
>At4g22450 hypothetical protein
Length = 457
Score = 35.0 bits (79), Expect = 0.30
Identities = 56/266 (21%), Positives = 105/266 (39%), Gaps = 26/266 (9%)
Query: 765 HKDDRDERSEVPLLGCNSAMKKRKMKFGAADFGERDEDANLLSSNPSKIDDLPAFSLKRK 824
H+DD + +++P+ + + R++ D DE ++L + S + + P++S K K
Sbjct: 154 HQDDDGDSTQIPITQDDDG-ESRQIPLNLDD----DEATHILPTQ-SNVGNGPSYSSKSK 207
Query: 825 SKKKPGAEMVISEMENSELPLTHTVTADVEVETKPQKKPYILITPTVHTGFS------FS 878
S KK S E + + ++ K LI +H FS
Sbjct: 208 SSKKR------SRSVQDEQGVAEVIKDSIKSRDKILSHKNQLIE--MHPKFSCCQLRAMQ 259
Query: 879 IMHLLTAVRT-AMISPPEVESLEAGKPVEQQNKAQEDSLNGVISSDKVDDKVAANVEPSD 937
I+H L A+R + + ++ L+ Q ED N V+ ++ + A+++ S
Sbjct: 260 ILHSLPAIRVWSPLYKAAIQHLKKDITNRQTFLFYEDDENKVLY---LEFETGADLDSSK 316
Query: 938 QKNVPSLTIQEIVNRVRSNPGDPCILETQEPLQDLVRGVLKIFSSKTAPLGAKGWKVLAV 997
Q S+ +QE V+ + N G P L + L+ +T + W++L
Sbjct: 317 QSTSQSVVLQENVSLMLFNRGGP--PTNSRELFNRRHSSLRSVIERTFGVWKAKWRILDR 374
Query: 998 YEKSTRSWSWIGPVLHNSSDHDPIEE 1023
WI V + H+ I +
Sbjct: 375 KHPKYEVKKWIKIVTSTMALHNYIRD 400
>At5g55920 nucleolar protein-like
Length = 682
Score = 34.7 bits (78), Expect = 0.39
Identities = 31/103 (30%), Positives = 48/103 (46%), Gaps = 10/103 (9%)
Query: 27 SGDEEDVQRRNSGNE-----SDEDDDEFDDADSGAGSDDFDLLELGETGAEFCQIGNQTC 81
S DEE+ G++ DED++ DADS A SDD D+ + ++ A + Q
Sbjct: 111 SDDEEENDEETLGDDFLEGSGDEDEEGSLDADSDADSDDDDI--VAKSDAIDRDLAMQKK 168
Query: 82 SIPLELYDLSGLEDILSVDVWNDCL---SEEERFELAKYLPDM 121
EL D +D+ + +D +EEE E A+ PD+
Sbjct: 169 DAAAELEDFIKQDDVHDEEPEHDAFRLPTEEELEEEARGPPDL 211
>At3g23980 unknown protein
Length = 741
Score = 34.7 bits (78), Expect = 0.39
Identities = 63/290 (21%), Positives = 112/290 (37%), Gaps = 35/290 (12%)
Query: 507 TAYRSSSPQVSNRLFSSDFRTKSSQE---KIRGSFVQNGRKDMKPLRGSHMLARGEETES 563
T+ R S Q + R+ S +Q+ ++ GS VQ KP S L +T S
Sbjct: 177 TSSRKDSLQPTTRMAGSFDEVAKNQQGSGELGGSIVQ------KPTLSSSYLFNSPDTSS 230
Query: 564 DSSEQWD---DDDDNNPLLQSKFAYPIGKAAGSLTKPLKSHLDPMKAKFSRTDMKATQSK 620
SE D + ++PL +K + ++ S L P + ++ +++A
Sbjct: 231 RPSEPSDFSVNITSSSPLNSAKSEATVKRSRPSFLDSLNISRAP-ETQYQHPEIQADLVT 289
Query: 621 KIGGFAEQGNMHGADNYLS----KNAKKSKIFNGSPVRNPAGKFMEENYPSVSDMLNG-- 674
G + G +Y+S N S S NP KF YP+ + ++ G
Sbjct: 290 SSGSQLSGSDGFGP-SYISGRRDSNGPSSLTSGASDYPNPFEKFRSSLYPAANGVMPGFT 348
Query: 675 ------GHDDWRQLYKSKNDQIRDEPVQRFDMPSSTSYAAEHKKKGRIGLDHSSMRSKYL 728
+DD+ L + D +++ + D+ +S + A + ++SSM Y
Sbjct: 349 DFSMPKQNDDFTALEQHIEDLTQEKFSLQRDLDASRALAES------LASENSSMTDTYN 402
Query: 729 HDYG--NDEDDSLENRLLADENGVGQSRFWRKGQKNVAHK-DDRDERSEV 775
G N D +E + +G+ R N + + DERS++
Sbjct: 403 QQRGLVNQLKDDMERLYQQIQAQMGELESVRVEYANAQLECNAADERSQI 452
>At4g21270 kinesin-related protein katA
Length = 793
Score = 34.3 bits (77), Expect = 0.51
Identities = 49/216 (22%), Positives = 87/216 (39%), Gaps = 22/216 (10%)
Query: 229 DLEADSSEESGEGMWSRKNKDKKNAQKLGRFPFQGVGSGLDFHPREQSMVMEQEKYSKQN 288
+LEA + EE + S+ ++ + + ++ H RE+ + EK
Sbjct: 138 ELEARTKEEELQATISKLEENVVSLHEKLAKEESSTQDAIECHRREKEARVAAEKVQASL 197
Query: 289 PKGILKLAGSKTHLAKDPTAHSSSVYHGLDMNPRLNGSAFAHPQHNISTGYDLGSIRRTR 348
+ + K+ K + T+ DM RL + Q+N DL ++R
Sbjct: 198 GEELDKVKEEKMAAKQKVTSLE-------DMYKRLQEYNTSLQQYNSKLQTDLETVRAA- 249
Query: 349 DQLWNGDNEEEISYRDRNALRG---SLMDMSSALRVGKRHDLLRGDEIEGGNLMGLSMSS 405
L + E+ + + LRG SL D S+ RV L+ D I+ + + +S
Sbjct: 250 --LTRAEKEKSSILENLSTLRGHSKSLQDQLSSSRV------LQDDAIKQKDSL---LSE 298
Query: 406 KTDLRGYTRNPNQSSDMQLFAAKPPSKKKGKYAENV 441
T+LR + D Q+ ++ S++ KY ENV
Sbjct: 299 VTNLRNELQQVRDDRDRQVVQSQKLSEEIRKYQENV 334
>At1g04530 unknown protein
Length = 310
Score = 34.3 bits (77), Expect = 0.51
Identities = 13/25 (52%), Positives = 18/25 (72%)
Query: 38 SGNESDEDDDEFDDADSGAGSDDFD 62
+ ++ DEDDDE DD SG G D+F+
Sbjct: 219 NADDDDEDDDEDDDESSGQGKDEFE 243
>At4g29950 unknown protein
Length = 828
Score = 33.9 bits (76), Expect = 0.67
Identities = 40/172 (23%), Positives = 60/172 (34%), Gaps = 25/172 (14%)
Query: 1184 IRDSQYIVEDVSDEKINQVVSGALDRLHY----ERDPCVLFDQE--RKLWVYLHREREEE 1237
+R S + D D+ I+ V + + P ++ Q W LH+ EEE
Sbjct: 436 VRSSALSINDGFDQSISPAVPARTNSFPSGSTSPKSPLIITPQSYWEDQWRVLHKAAEEE 495
Query: 1238 DFDDDGTSSTKKWKRQKKDVADQSDQAPVTVACNGTGEQSGYDLCSDLNVDPPCIEDDKE 1297
K W R K+ +S+ + NG E ++
Sbjct: 496 KKSPSPIQKKKPWFRVKRLFRAESEPTHSAKSPNGKSE----------------VKISSV 539
Query: 1298 AVQLLTTDTRPNAEDQVVVNPVSEVGNSCEDNSMTWEALDLNPTRELCQENS 1349
A LL R + V NP+ V N ED+S+ E D+N E E S
Sbjct: 540 ARNLLEDFNRQLVSEPVEANPIDVVNN--EDSSIR-ETEDINTDFETAAEES 588
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 33.9 bits (76), Expect = 0.67
Identities = 12/34 (35%), Positives = 24/34 (70%)
Query: 20 MSKESMSSGDEEDVQRRNSGNESDEDDDEFDDAD 53
+++++ GDEE+ + ++ NE ++DDD+ DD D
Sbjct: 1073 VNEDTNEDGDEEEDEAQDDDNEENQDDDDDDDGD 1106
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.131 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,597,015
Number of Sequences: 26719
Number of extensions: 1611839
Number of successful extensions: 6403
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 5745
Number of HSP's gapped (non-prelim): 359
length of query: 1373
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1262
effective length of database: 8,352,787
effective search space: 10541217194
effective search space used: 10541217194
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC135230.12


