

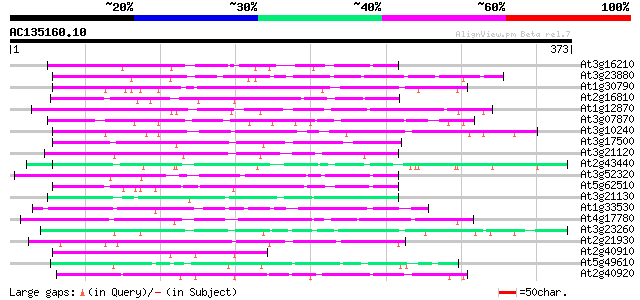
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135160.10 + phase: 0
(373 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g16210 hypothetical protein 88 7e-18
At3g23880 unknown protein 83 3e-16
At1g30790 hypothetical protein 83 3e-16
At2g16810 unknown protein 80 2e-15
At1g12870 hypothetical protein 75 6e-14
At3g07870 unknown protein 74 1e-13
At3g10240 hypothetical protein 74 2e-13
At3g17500 hypothetical protein 73 3e-13
At3g21120 hypothetical protein 72 5e-13
At2g43440 hypothetical protein 72 6e-13
At3g52320 putative protein 68 9e-12
At5g62510 putative protein 67 1e-11
At3g21130 hypothetical protein 67 2e-11
At1g33530 hypothetical protein 65 5e-11
At4g17780 MYB transcription factor like protein 65 8e-11
At3g23260 unknown protein 65 8e-11
At2g21930 unknown protein 62 4e-10
At2g40910 unknown protein 62 5e-10
At5g49610 unknown protein 62 7e-10
At2g40920 unknown protein 61 9e-10
>At3g16210 hypothetical protein
Length = 360
Score = 88.2 bits (217), Expect = 7e-18
Identities = 73/246 (29%), Positives = 115/246 (46%), Gaps = 41/246 (16%)
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQ--NAPNMKFIFC 83
L EEL IEIL+RL K L+RF+CV K+W ++I P F + + + + F
Sbjct: 5 LPEELAIEILVRLSMKDLARFRCVCKTWRDLINDPGFTETYRDMSPAKFVSFYDKNFYML 64
Query: 84 DGGNDQKSIPIKSLFPQDVARIE---IYGSCDGVFCLKGISSCITRHDQLILWNPTTKEV 140
D I K FP D + I+ CDG C+ ++ L++WNP +K+
Sbjct: 65 DVEGKHPVITNKLDFPLDQSMIDESTCVLHCDGTLCV------TLKNHTLMVWNPFSKQF 118
Query: 141 HLIPRAPSLGNHYSDESLYGFG--AVNDDFKVV----KLNISNSNRMAKINSLLKADIYD 194
++P P + Y D ++ GFG V+DD+KVV +L++S A +++
Sbjct: 119 KIVPN-PGI---YQDSNILGFGYDPVHDDYKVVTFIDRLDVST------------AHVFE 162
Query: 195 LSTKSW--TPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFRDNQF 252
T SW + +S+P R T ++ YWI +D ILCF+ +++
Sbjct: 163 FRTGSWGESLRISYPDWH-----YRDRRGTFLDQYLYWIAYRSSADRF-ILCFNLSTHEY 216
Query: 253 RKLEAP 258
RKL P
Sbjct: 217 RKLPLP 222
>At3g23880 unknown protein
Length = 364
Score = 82.8 bits (203), Expect = 3e-16
Identities = 91/328 (27%), Positives = 134/328 (40%), Gaps = 51/328 (15%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMK----FIFCD 84
E+ EILLRLP KSL+RFKCV SW ++I FA + +L A +
Sbjct: 17 EMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTSTKSPYGVITT 76
Query: 85 GGNDQKSIPIKSLFPQDVARI-------------EIYGSCDGVFCLKGISSCITRHDQLI 131
KS I SL+ + ++ G+C G+ C + L
Sbjct: 77 SRYHLKSCCIHSLYNASTVYVSEHDGELLGRDYYQVVGTCHGLVCFH-----VDYDKSLY 131
Query: 132 LWNPTTKEVHLIPRAPSLGNHYSDES---LYGFG--AVNDDFKVVKLNISNSNRMAKINS 186
LWNPT K L R S SD+ YGFG DD+KVV L KI
Sbjct: 132 LWNPTIK---LQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVAL--LQQRHQVKI-- 184
Query: 187 LLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFD 246
+ IY K W S P +V SR +NG W +S S + I+ +D
Sbjct: 185 --ETKIYSTRQKLWRSNTSFPSGVVVA--DKSRSGIYINGTLNWAATS-SSSSWTIISYD 239
Query: 247 FRDNQFRKLEAPK-LGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTLEQNG----W 301
++F++L P G F + +++G L V C+ ++W +++ G W
Sbjct: 240 MSRDEFKELPGPVCCGR--GCFTMTLGDLRGCLSMVCY--CKGANADVWVMKEFGEVYSW 295
Query: 302 AKKYNIDTKMSIFHIYGLW-NDGAEILV 328
+K +I + LW +DG +L+
Sbjct: 296 SKLLSIPGLTDF--VRPLWISDGLVVLL 321
>At1g30790 hypothetical protein
Length = 399
Score = 82.8 bits (203), Expect = 3e-16
Identities = 87/306 (28%), Positives = 139/306 (44%), Gaps = 43/306 (14%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF--------ATRRNRLLILQNA--PNM 78
+L +EIL RLP KSL +FKCV K W++II + F +TR ++ N P+
Sbjct: 12 DLTVEILTRLPAKSLMKFKCVSKLWSSIIHNQSFIDSFYSISSTRPRFIVAFSNGSFPSD 71
Query: 79 K----FIFCDG--GNDQKSIPIKSL---FPQDVARIEIYGSCDGVFCLKGISSCITRHDQ 129
K FIF G++ S I +L P + C V G +C + + +
Sbjct: 72 KEKRLFIFSSSHEGHESSSSVITNLDTTIPSLTVSNNLASRCISV---NGFIAC-SLYTR 127
Query: 130 LILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNIS-NSNRMAKINSLL 188
+ NP+T++V ++P PS + G+ V+D FK + L S N+ + + L+
Sbjct: 128 FTICNPSTRQVIVLPILPSGRAPDMRSTCIGYDPVDDQFKALALISSCIPNKDSTVEHLV 187
Query: 189 KADIYDLSTKSWTPLVSH---PPITMVTRIQPSRYNTLVNG-VYYWITSSDGSDAARILC 244
D SW + + PP + VT +NG VYY + S A I+C
Sbjct: 188 LTLKGDKKNYSWRQIQGNNNIPPYSPVT------MRVCINGVVYYGAWTPRQSMNAVIVC 241
Query: 245 FDFRDNQFRKLEAPKLGHYIPFFCDD--VFEIKGYLGYVVQYR-CRIVWLEIWTL---EQ 298
FD R + ++ PK + +C+D + E KG L +V+ R R ++W L E+
Sbjct: 242 FDVRSEKITFIKTPK---DVVRWCNDSILMEYKGKLASIVRNRYSRFDTFDLWVLEDIEK 298
Query: 299 NGWAKK 304
W+K+
Sbjct: 299 QEWSKQ 304
>At2g16810 unknown protein
Length = 295
Score = 79.7 bits (195), Expect = 2e-15
Identities = 69/250 (27%), Positives = 121/250 (47%), Gaps = 33/250 (13%)
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFC--DG 85
++L IEIL RLP KS+ RFKCV K W++++ S YF NR LI+ + P C D
Sbjct: 43 QDLLIEILTRLPPKSVMRFKCVSKFWSSLLSSRYFC---NRFLIVPSQPQPSLYMCLLDR 99
Query: 86 GNDQKSI---------PIKSLFPQDVARIEIYGSCDGVFCLKGISSCI--TRHDQLILWN 134
N KS+ P +F QD+ ++ G F L+ + I TR+ + ++N
Sbjct: 100 YNYSKSLILSSAPSTSPYSFVFDQDLTIRKM-----GGFFLRILRGFIFFTRNLKARIYN 154
Query: 135 PTTKEVHLIPRAPS----LGNHYSDESLYGFGAVNDDFKVV-KLNISNSNRMAKINSLLK 189
PTT+++ ++P G Y+ VND +K++ ++ ++ N + + S L
Sbjct: 155 PTTRQLVILPTIKESDIIAGPPYNILYFICHDPVNDRYKLLCTVSYASDNDLQNLKSELW 214
Query: 190 ADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFRD 249
+ + + SW + + P + PS + +NGV Y++ +D ++ FD R
Sbjct: 215 IFVLE-AGGSWKRVANEFPHHV-----PSHLDLNMNGVLYFLAWTD-PHTCMLVSFDVRS 267
Query: 250 NQFRKLEAPK 259
+F ++ P+
Sbjct: 268 EEFNTMQVPR 277
>At1g12870 hypothetical protein
Length = 416
Score = 75.1 bits (183), Expect = 6e-14
Identities = 84/346 (24%), Positives = 141/346 (40%), Gaps = 62/346 (17%)
Query: 15 RKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQN 74
R KL I L +++ EI L+LP K+L RFK + K W + ++S YF+ R ++ +
Sbjct: 22 RYKKLMIASSSLPDDVVEEIFLKLPVKALMRFKSLSKQWRSTLESCYFSQRHLKIAERSH 81
Query: 75 APNMK-FIFCDGGNDQKSIPIKSLFPQDVARIE-------------IYG--SCDGVFCLK 118
+ K I + N I +++ + V+ + IY SCDG+FC+
Sbjct: 82 VDHPKVMIITEKWNPDIEISFRTISLESVSFLSSALFNFPRGFHHPIYASESCDGIFCIH 141
Query: 119 GISSCITRHDQLILWNPTTKEVHLIPRA----------PSLGNHYSDESLYGFGAVN--- 165
+ + + NP T+ +P A P+L ++L VN
Sbjct: 142 S-----PKTQDIYVVNPATRWFRQLPPARFQIFMHKLNPTL------DTLRDMIPVNHLA 190
Query: 166 ----DDFKVVKLNISNSNRMAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYN 221
D+K+V L S+++R+ K +++D +W L P RI +
Sbjct: 191 FVKATDYKLVWLYNSDASRVT------KCEVFDFKANAWRYLTCIPSY----RIYHDQKP 240
Query: 222 TLVNGVYYWITSSDGSDAARILCFDFRDNQFRKLEAPKL-GHYIPFFCDDVFEIKGYLGY 280
NG YW T + ++ +++ D FR L P L P D Y
Sbjct: 241 ASANGTLYWFTETYNAE-IKVIALDIHTEIFRLLPKPSLIASSEPSHIDMCIIDNSLCMY 299
Query: 281 VVQYRCRIVWLEIWTLE--QNGWAKKYNIDTKMSI---FHIYGLWN 321
+ +I+ EIW L+ ++ W K Y I+ S FH+ +N
Sbjct: 300 ETEGDKKII-QEIWRLKSSEDAWEKIYTINLLSSSYCHFHVLDGYN 344
>At3g07870 unknown protein
Length = 417
Score = 74.3 bits (181), Expect = 1e-13
Identities = 85/330 (25%), Positives = 140/330 (41%), Gaps = 73/330 (22%)
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFI---- 81
L E++ +I RLP S++R V +SW +++ T+ RL ++P +
Sbjct: 28 LPEDIIADIFSRLPISSIARLMFVCRSWRSVL------TQHGRLSSSSSSPTKPCLLLHC 81
Query: 82 ---------FCDGGNDQKSIPIKSL---FPQDVARIEIYGSCDGVFCLKGISSCITRHDQ 129
F D ++K I K F + ++ GSC+G+ CL S +D
Sbjct: 82 DSPIRNGLHFLDLSEEEKRIKTKKFTLRFASSMPEFDVVGSCNGLLCL----SDSLYNDS 137
Query: 130 LILWNP-TTKEVHLIPRAPSLGNHYSDESL---YGFGAVNDDFKVVKL-----NISNSNR 180
L L+NP TT + L P N Y D+ L +GF + ++KV+K+ + SN+N
Sbjct: 138 LYLYNPFTTNSLEL----PECSNKYHDQELVFGFGFHEMTKEYKVLKIVYFRGSSSNNNG 193
Query: 181 MAKINSLL-----KADIYDLSTK------SWTPLVSHPPITMVTRIQPSRYNTLVNGVYY 229
+ + + + I LS+K SW L P V R LVNG +
Sbjct: 194 IYRGRGRIQYKQSEVQILTLSSKTTDQSLSWRSL-GKAPYKFVKRSS----EALVNGRLH 248
Query: 230 WITSSDGSDAAR-ILCFDFRDNQFRKLEAPKLGHYIPFFCDDVFEIKGYLGYVVQYRCRI 288
++T R + FD D +F+++ P G + + +KG L C +
Sbjct: 249 FVTRPRRHVPDRKFVSFDLEDEEFKEIPKPDCGG-LNRTNHRLVNLKGCL-------CAV 300
Query: 289 VW-----LEIWTLEQNG----WAKKYNIDT 309
V+ L+IW ++ G W K+Y+I T
Sbjct: 301 VYGNYGKLDIWVMKTYGVKESWGKEYSIGT 330
>At3g10240 hypothetical protein
Length = 389
Score = 73.6 bits (179), Expect = 2e-13
Identities = 89/353 (25%), Positives = 147/353 (41%), Gaps = 55/353 (15%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF----ATRRNRLLILQNAPNMKFIFCD 84
+L EILLRLP KS++RF+CV K W++I PYF TR RLL+ A F+
Sbjct: 30 DLVSEILLRLPEKSVARFRCVSKPWSSITTEPYFINLLTTRSPRLLLCFKANEKFFVSSI 89
Query: 85 GGNDQ------KSIPIKSL-------FPQDVARIEIYGSCDGVFCLKGISSCITRHDQLI 131
+ Q KS L F +++ S +G+ C + +LI
Sbjct: 90 PQHRQTFETWNKSHSYSQLIDRYHMEFSEEMNYFPPTESVNGLICFQ-------ESARLI 142
Query: 132 LWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNISNSNRMAKINSLLKAD 191
+WNP+T+++ ++P+ GN G+ V KV+ + S + ++ +L A
Sbjct: 143 VWNPSTRQLLILPKPN--GNSNDLTIFLGYDPVEGKHKVMCMEFSATYDTCRVLTLGSAQ 200
Query: 192 IYDLSTKSWTPLVSHPPITMVTRIQPSRYNT--LVNGVYYWITSSDGSDAARILCFDFRD 249
K W + +H + + Y++ +NGV Y I ++ FD R
Sbjct: 201 ------KLWRTVKTH------NKHRSDYYDSGRCINGVVYHIAYVKDMCVWVLMSFDVRS 248
Query: 250 NQFRKLEAPKLGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTLEQ-NGWAKK---- 304
F +E P + D + + G L V + + +W LE+ N W+ K
Sbjct: 249 EIFDMIELPSSDVH----KDVLIDYNGRLACVGREIIEKNGIRLWILEKHNKWSSKDFLA 304
Query: 305 --YNIDTKMSI--FHIYGLWNDGAEILVGEFGQR--QLTSCDHHGNVLRQFQL 351
+ID +S F + G + G I V + ++ D N R+F+L
Sbjct: 305 PLVHIDKSLSTNKFLLKGFTHAGEIIYVESMFHKSAKIFFYDPVRNTSRRFEL 357
>At3g17500 hypothetical protein
Length = 438
Score = 72.8 bits (177), Expect = 3e-13
Identities = 64/244 (26%), Positives = 104/244 (42%), Gaps = 30/244 (12%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDGGND 88
+L EIL R+P SL R + KSWNN K F + + ++ + + D
Sbjct: 7 DLVEEILSRVPATSLKRLRSTCKSWNNCYKDQRFTEKHS---VIAAKQFLVLMLKDCRVS 63
Query: 89 QKSIPIKSLFPQDVA-RIEIYG------SCDGVFCLKGISSCITRHDQLILWNPTTKEVH 141
S+ + + A IE+ G G+F G+ C T+ D+L +WNP T +
Sbjct: 64 SVSVNLNEIHNNIAAPSIELKGVVGPQMQISGIFHCDGLLLCTTKDDRLEVWNPCTGQTR 123
Query: 142 LIPRAPSLGNHYSDESLYGFGAVNDD----FKVVKL-NISNSNRMAKINSLLKADIYDLS 196
+ + HY S + G VN++ +K+++ N SN + +IYD S
Sbjct: 124 RVQHS----IHYKTNSEFVLGYVNNNSRHSYKILRYWNFYMSNYRVS-----EFEIYDFS 174
Query: 197 TKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFRDNQFRKLE 256
+ SW + P + + NT YW+ S + D IL FDF ++++L
Sbjct: 175 SDSWRFIDEVNPYCLTEGEVSLKGNT------YWLASDEKRDIDLILRFDFSIERYQRLN 228
Query: 257 APKL 260
P L
Sbjct: 229 LPIL 232
>At3g21120 hypothetical protein
Length = 367
Score = 72.0 bits (175), Expect = 5e-13
Identities = 65/258 (25%), Positives = 112/258 (43%), Gaps = 39/258 (15%)
Query: 24 MKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNR---------LLILQN 74
M L E+L +EIL ++P SL+RF+ + WN ++ FA + +++++
Sbjct: 1 MHLPEDLVLEILSKVPAVSLARFRSTCRRWNALVVDGSFAKKHYAYGPRQYPIVIMLIEF 60
Query: 75 APNMKFIFCDGGNDQKSIPIKSLFPQDVARIEIYGSCDGV-----FCLKGISSCITRHDQ 129
+ I G N+ P L Q + + S + V F G+ C T+ +
Sbjct: 61 RVYLVSIDLHGINNNNGAPSAKLTGQFSLKDPLSNSSEEVDIRNAFHCDGLLLCCTKDRR 120
Query: 130 LILWNPTTKEVHLI-PRAPSLGNHYSDESLYGFGAVN--DDFKVVKLN-ISNSNRMAKIN 185
L++WNP + E I PR N Y + LY G N +K+++++ + N
Sbjct: 121 LVVWNPCSGETKWIQPR-----NSYKESDLYALGYDNRSSSYKILRMHPVGNPFH----- 170
Query: 186 SLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSR-YNTLVNGVYYWITSS----DGSDAA 240
+++++YD ++ SW + +T IQ + Y V G YW S D
Sbjct: 171 --IESEVYDFASHSWRSV----GVTTDFHIQTNESYGMNVKGTTYWFALSKDWWSSDDRR 224
Query: 241 RILCFDFRDNQFRKLEAP 258
+L FDF +F+ L P
Sbjct: 225 FLLSFDFSRERFQCLPLP 242
>At2g43440 hypothetical protein
Length = 779
Score = 71.6 bits (174), Expect = 6e-13
Identities = 91/370 (24%), Positives = 147/370 (39%), Gaps = 61/370 (16%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDGGN- 87
EL +I LRLP KS+ +FK V + W +I++S F RR L + K + N
Sbjct: 16 ELLEDIFLRLPLKSILKFKTVSRQWRSILESKLFVERRGNL----QKHHRKILAAYNCNY 71
Query: 88 -DQKSIPIKSLFPQDVARIEIY-------GSCDGVFCLKGISSCITRHDQLILWNPTTKE 139
+ SI +S F D + ++ +CDG+ CIT + NP+ +
Sbjct: 72 FMRPSIFPESRFEGDEEIVYLHCDAAQPSMTCDGLL-------CITEPGWFNVLNPSAGQ 124
Query: 140 VHLIPRAPSLGNHYSDESLYGFGA--VNDDFKVVKLNISNSNRMAKINSLLKADIYDLST 197
+ P P + L GFG V +K+V++ + + I D+ T
Sbjct: 125 LRRFPPGPGPVKGPQENWLLGFGRDNVTGRYKIVRM---------CFHDCYEFGILDIET 175
Query: 198 KSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFRDNQFRKLEA 257
W+ L S PP M+ P + VNG YW+ S G IL D + + +
Sbjct: 176 GVWSKLRS-PPHNML----PGSKSVCVNGSIYWLQISAG---YIILAMDLHEESYHGV-- 225
Query: 258 PKLGHYIPFFC----DDVFEIKGYLGYVVQYRCRIVW-LEIWT--LEQNGWAKKYNIDTK 310
H++P + ++ L + W LEIW+ +E GW+K Y+
Sbjct: 226 ----HHLPATWVTQETQLVNLEDRLAMAMTTNVGPEWILEIWSMDIEGKGWSKGYSWSKS 281
Query: 311 MSIFHIYGL-----WNDGAEILVGEFGQRQLTSCDHHGNVLRQF----QLDTLENACFWY 361
SI +G+ W V Q L D+H + + + ++ L +
Sbjct: 282 YSISLAHGVGFSWPWQKRWFTPVWVSKQGNLLFYDNHKRLFKYYSGTDEIRCLSSNICVI 341
Query: 362 YEYVPSMAPL 371
YV ++APL
Sbjct: 342 SSYVENLAPL 351
Score = 60.1 bits (144), Expect = 2e-09
Identities = 87/384 (22%), Positives = 150/384 (38%), Gaps = 69/384 (17%)
Query: 12 SRLRKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLI 71
S++ + + + + + EL EI L LP KS+ +FK V K W +I++S F RR L
Sbjct: 385 SKMEEERENTNSIYIVSELLEEIFLGLPLKSILKFKTVSKQWRSILESNLFVERRRTLQ- 443
Query: 72 LQNAPNMKFIF-CDGGNDQKSIPIKSLFPQDVARIEIYG-------SCDGVFCLKGISSC 123
+N P + + CD +P KS F D + ++ +CDG+ C
Sbjct: 444 -KNHPKILAAYNCDYCTRPGILP-KSQFEGDEEIVYLHTDATQPSMTCDGLVC------- 494
Query: 124 ITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNISNSNRMAK 183
IT + N +T ++ P V +K+V++ +
Sbjct: 495 ITEPGWFNVLNVSTGQLRRFLPGPDPDK------------VTGKYKIVRMCFHDCYEFG- 541
Query: 184 INSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARIL 243
I D+ + W+ L+S P I V + VNG YW+ S + IL
Sbjct: 542 --------ILDIESGEWSKLMSPPHIMRV-----GSKSVCVNGSIYWLQISV---SYIIL 585
Query: 244 CFDFRDNQFRKLEAPKLGHYIP--FFCDD--VFEIKGYLGYVVQYRCRIVW-LEIWTL-- 296
D F + +++P + D + ++ L + + W LEIW++
Sbjct: 586 ALDLHQETFNGV------YHLPATWVTQDTQLVNLEDRLAMAMTTKVGPEWILEIWSMDI 639
Query: 297 EQNGWAKKYNIDTKMSIFHIYGL-----WNDGAEILVGEFGQRQLTSCDHHGNVLRQF-- 349
E+ GW+K+Y SI + + W V Q L D+H + + +
Sbjct: 640 EEKGWSKRYTWSKAYSISLAHRVVVSWPWQKRWFTPVSVSKQGNLVFYDNHKRLFKYYSG 699
Query: 350 --QLDTLENACFWYYEYVPSMAPL 371
++ L + YV ++APL
Sbjct: 700 TDEIRCLSSNINVISSYVENLAPL 723
>At3g52320 putative protein
Length = 390
Score = 67.8 bits (164), Expect = 9e-12
Identities = 65/278 (23%), Positives = 120/278 (42%), Gaps = 45/278 (16%)
Query: 4 LIVHNKKFSRLRKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFA 63
L V +K + ++ K + ++ EE+ I+IL+RLP KSL RFKCV K W ++I S YF
Sbjct: 5 LCVAVRKKEKQKQKKTVVFLPEIPEEMLIDILIRLPAKSLMRFKCVSKLWLSLITSRYFT 64
Query: 64 TR-------------------RNRLLILQNAPNMKFIFCDGG----NDQKSIPIKSLFPQ 100
R +++ L+LQ++ + + D + +IPI +
Sbjct: 65 NRFFKPSSPSCLFAYLVDRENQSKYLLLQSSSSSRHDHSDTSVSVIDQHSTIPIMGGYLV 124
Query: 101 DVARIEIYGSCDGVFCLKGISSCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYG 160
+ AR G+ C + ++ + NP+T+++ +P S N + + +G
Sbjct: 125 NAAR--------GLLCYR-------TGRRVKVCNPSTRQIVELPIMRSKTNVW---NWFG 166
Query: 161 FGAVNDDFKVVKLNISNSNRMAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRY 220
+D++KV+ L + + S + + + SW SH T P
Sbjct: 167 HDPFHDEYKVLSLFWEVTKEQTVVRSEHQVLVLGVGA-SWRNTKSHH--TPHRPFHPYSR 223
Query: 221 NTLVNGVYYWITSSDGSDAARILCFDFRDNQFRKLEAP 258
++GV Y+ +D + ++ FD +F +E P
Sbjct: 224 GMTIDGVLYYSARTDANRCV-LMSFDLSSEEFNLIELP 260
>At5g62510 putative protein
Length = 420
Score = 67.4 bits (163), Expect = 1e-11
Identities = 74/251 (29%), Positives = 121/251 (47%), Gaps = 35/251 (13%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQN--APNMKFIF---- 82
+L IEIL RLP KSL RFKCV K W+++I+S +F+ NR L + + P+ +I
Sbjct: 47 DLLIEILTRLPHKSLMRFKCVSKQWSSLIRSRFFS---NRYLTVASPLRPHRLYISLVDH 103
Query: 83 -CDG----GNDQKSIPIK----SLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLILW 133
CD + ++S+ + S F QD+ ++ G V L+G+ I + L+
Sbjct: 104 KCDSREVCHSPRESVLLSFSSPSSFDQDLTTMQGMGGLHMV-TLRGLILYIV-CGKACLY 161
Query: 134 NPTTKEVHLIPRAP----SLGNHYSDESLYGFGAVNDDFKVVKLNISNSNR-MAKINSLL 188
NPTT++ +P GN +S G V D +KVV +S+S++ + I S
Sbjct: 162 NPTTRQSVTLPAIKFNIFVQGNEHSLLYFLGHDPVLDQYKVVCTFVSSSSQDLETIISEH 221
Query: 189 KADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWIT-SSDGSDAARILCFDF 247
+ ++ SW + P T P+R + GV Y++ +S D ++ FD
Sbjct: 222 WVFVLEVG-GSWKRIEFDQPHT------PTRSGLCIGGVIYYLAFTSMFQDI--VVTFDV 272
Query: 248 RDNQFRKLEAP 258
R +F ++AP
Sbjct: 273 RSEEFNIIQAP 283
>At3g21130 hypothetical protein
Length = 367
Score = 66.6 bits (161), Expect = 2e-11
Identities = 60/254 (23%), Positives = 103/254 (39%), Gaps = 35/254 (13%)
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDG 85
L E+L +EIL R+ SL+R + K WN ++K A + + AP +
Sbjct: 10 LSEDLIVEILSRVSAVSLARLRTTSKRWNALVKDERLAKKHSAY-----APRQSLVIT-- 62
Query: 86 GNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLK-------------------GISSCITR 126
D + + + ++++ G F LK G+ C T+
Sbjct: 63 LIDSRVYLMNVSLQYGIEKVDLSAKLTGQFSLKDPLSNSLEEVDIRNVFHCDGLLLCSTK 122
Query: 127 HDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNISNSNRMAKINS 186
++L++WNP + E I P SD G+ + K+++++ S +I
Sbjct: 123 DNRLVVWNPCSGETRWI--QPRSSYKVSDIYALGYDNTSSCHKILRMDRSED----RIPI 176
Query: 187 LLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWIT-SSDGSDAAR-ILC 244
+ +YD ++KSW + + I R V G YW+ + DG +LC
Sbjct: 177 QTEYQVYDFTSKSWL-VDGVAGGLFIPSIGTRRRGLSVKGNTYWLALTEDGPPFDMFLLC 235
Query: 245 FDFRDNQFRKLEAP 258
FDF + FR+L P
Sbjct: 236 FDFSTDGFRRLSLP 249
>At1g33530 hypothetical protein
Length = 441
Score = 65.5 bits (158), Expect = 5e-11
Identities = 75/274 (27%), Positives = 115/274 (41%), Gaps = 44/274 (16%)
Query: 16 KTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNA 75
+T L++E L + L EIL RLP K L R K + K W ++I+S + A + RL L+
Sbjct: 90 ETTLAVE---LPDVLVEEILQRLPVKYLVRLKSISKGWKSLIESDHLAEKHLRL--LEKK 144
Query: 76 PNMKFI-FCDGGNDQKSIPIK---------SLFPQDVARIEIYGSCDGVFCLKGISSCIT 125
+K I + KSI IK ++ + + GSC+G+ C+ + S
Sbjct: 145 YGLKEIKITVERSTSKSICIKFFSRRSGMNAINSDSDDLLRVPGSCNGLVCVYELDSV-- 202
Query: 126 RHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNISNSNRMAKIN 185
+ L NP T + P G S +G V +KV+ L +R+ +
Sbjct: 203 ---YIYLLNPMTGVTRTL--TPPRGTKLSVG--FGIDVVTGTYKVMVL--YGFDRVGTV- 252
Query: 186 SLLKADIYDLSTKSWTP-LVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILC 244
++DL T W + P+ + P R VNG +W+ +SD S+ IL
Sbjct: 253 ------VFDLDTNKWRQRYKTAGPMPLSCIPTPERNPVFVNGSLFWLLASDFSE---ILV 303
Query: 245 FDFRDNQFRKLEAPKLGHYIPFFCDDVFEIKGYL 278
D +FR L P DDV GY+
Sbjct: 304 MDLHTEKFRTLSQPN-------DMDDVDVSSGYI 330
>At4g17780 MYB transcription factor like protein
Length = 745
Score = 64.7 bits (156), Expect = 8e-11
Identities = 69/311 (22%), Positives = 129/311 (41%), Gaps = 36/311 (11%)
Query: 8 NKKFSRLRKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRN 67
NK+ R+ + + + + + +L +I LRLP KS+ K V K W +I++S F RR
Sbjct: 357 NKQSPRMEEEEKNPSSIYIVADLLEDIFLRLPLKSILISKSVSKRWRSILESKTFVERR- 415
Query: 68 RLLILQNAPNMKFIF-CDGGNDQKSIPIKSLFPQDVARIEIY-------GSCDGVFCLKG 119
+ LQ + + C G + + +P S + + ++ +CDG+
Sbjct: 416 --MSLQKKRKILAAYNCKCGWEPRLLPGSSQCKGNEEIVYLHCNAAQPSFTCDGLV---- 469
Query: 120 ISSCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNISNSN 179
CI + + NP T+++ + +++G V +KVVK+ + + +
Sbjct: 470 ---CILEPRWIDVLNPWTRQLRRY--------GFGFGTIFGVDKVTGSYKVVKMCLISFS 518
Query: 180 RMAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDA 239
+ + ++ + D+ T W ++S PP ++ R + NG YW+ A
Sbjct: 519 EICARDPEVEYSVLDVETGEWR-MLSPPPY----KVFEVRKSECANGSIYWL-HKPTERA 572
Query: 240 ARILCFDFRDNQFRKLEAPKLGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTL--E 297
IL D + + P + F + ++ L Y LEIW++ E
Sbjct: 573 WTILALDLHKEELHNISVPDMSVTQETF--QIVNLEDRLAIANTYTKTEWKLEIWSMDTE 630
Query: 298 QNGWAKKYNID 308
W K Y+ID
Sbjct: 631 VETWTKTYSID 641
>At3g23260 unknown protein
Length = 362
Score = 64.7 bits (156), Expect = 8e-11
Identities = 90/388 (23%), Positives = 153/388 (39%), Gaps = 64/388 (16%)
Query: 21 IERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNR--------LLIL 72
+E L EL+ EIL R+P K L+R + K WN + K+ FA + + +++
Sbjct: 1 MEWRSLPVELQEEILSRVPAKYLARLRSTSKQWNALSKTGSFAKKHSANATKEPLIIMLK 60
Query: 73 QNAPNMKFIFCDG--GNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQL 130
+ + + G N +S + S I CDG+ L C + + L
Sbjct: 61 DSRVYLASVNLHGVHNNVAQSFELGSRLYLKDPHISNVFHCDGLLLL-----CSIKENTL 115
Query: 131 ILWNPTTKEVHLI-PRAPSLGNHYSDESLYGFGAVNDD----FKVVKLNISNSNRMAKIN 185
+WNP + E LI PR ++Y + Y G N +KV+++ IS +
Sbjct: 116 EVWNPCSGEAKLIKPR----HSYYKESDFYALGYDNKSSCKKYKVLRV-ISQVHVQGDFK 170
Query: 186 SLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCF 245
++ +IYD + SW H T ++ Q ++ V G YW+ + L F
Sbjct: 171 --IEYEIYDFTNDSWR---VHGATTELSIRQ--KHPVSVKGSTYWVVRNRYFPYKYFLSF 223
Query: 246 DFRDNQFRKLEAPKLGHYIPFFCDDVFEIK-------GYLGYVVQYRCRIVWLEIWTLEQ 298
DF +F+ L P+ P+ D+ ++ GY + VW+
Sbjct: 224 DFSTERFQSLSLPQ---PFPYLVTDLSVVREEQLCLFGYYNWSTTSEDLNVWVTTSLGSV 280
Query: 299 NGWAKKYNID---TKMSIFHIYG---LWNDGAEILVGEFGQR---------QLTSCDHHG 343
W+K I ++ +F YG L ++ + LV Q+ + D HG
Sbjct: 281 VSWSKFLTIQIIKPRVDMFD-YGMSFLVDEQNKSLVCWISQKVLHIVGEIYHIQDLDDHG 339
Query: 344 NVLRQFQLDTLENACFWYYEYVPSMAPL 371
+ L ++C YVPS+A +
Sbjct: 340 ------RDSALRSSCSVLMNYVPSLAQI 361
>At2g21930 unknown protein
Length = 396
Score = 62.4 bits (150), Expect = 4e-10
Identities = 69/281 (24%), Positives = 122/281 (42%), Gaps = 40/281 (14%)
Query: 13 RLRKTKLSIERMKL-GEELEI------EILLRLPTKSLSRFKCVQKSWNNIIKSPYF--- 62
R +KTK+S + + G ++I EIL RLP K+L+RF V K + +II++ F
Sbjct: 5 RSKKTKISDDLITCSGNSVQIPFDLIPEILKRLPVKTLARFLSVSKEYTSIIRNRDFMKS 64
Query: 63 -----ATRRNRLL--ILQNAPNMKFIFCDGGNDQKSIPIKSLFPQDVARIEIYG-SCDGV 114
+TR L+ I + F D G S L +++ + S G+
Sbjct: 65 YLINSSTRPQSLIFTIAGGGIHCFFSLIDQGESTSSSKPTYLMNCPHLQLKTFAPSVHGL 124
Query: 115 FCLKGISSCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVV--- 171
C S+ I +LI+ NP+T+ ++P+ + NH G+ ++ D+KV+
Sbjct: 125 ICHGPPSTLIVSSPRLIVSNPSTRRSIILPKIDA--NHECIYHHMGYDPIDGDYKVLCMM 182
Query: 172 -KLNISNSNRMAKINSLLKADIYDL-STKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYY 229
+++ +AK + ++ L SW + PP + P + +NGV Y
Sbjct: 183 KGMHVYQRRYLAK-----ELQVFTLRKGNSWRMVEDFPPHCLCHEDTP---DLCINGVLY 234
Query: 230 WITSSDGSDAARILCFDFRDNQFRKLEA-------PKLGHY 263
++ D + ++ FD R +F ++ PKL Y
Sbjct: 235 YVAMLDTASNHAVMSFDVRSEKFDLIKGGPDGDLNPKLTRY 275
>At2g40910 unknown protein
Length = 449
Score = 62.0 bits (149), Expect = 5e-10
Identities = 41/153 (26%), Positives = 74/153 (47%), Gaps = 10/153 (6%)
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDGGND 88
+L +EI++RLP KS+ RFKC+ K W+++I YF + + P++ D G
Sbjct: 72 DLLMEIVMRLPAKSMVRFKCISKQWSSLISCRYFCNSLFTSVTRKKQPHIHMCLVDHGGQ 131
Query: 89 QKSIPIKSLFPQDVARI---EIYGSCDGVFCLKGISS--CITRHDQLILWNPTTKEVHLI 143
+ + + S P + + ++ + G F L + C + ++ ++NPTT++ +
Sbjct: 132 RVLLSLSSTSPDNTCYVVDQDLSLTGMGGFFLNLVRGLLCFSVREKACIYNPTTRQRLTL 191
Query: 144 PRAPS----LGNHYSDESLY-GFGAVNDDFKVV 171
P S L + D Y G VND +K+V
Sbjct: 192 PAIKSDIIALKDERKDIRYYIGHDPVNDQYKLV 224
>At5g49610 unknown protein
Length = 359
Score = 61.6 bits (148), Expect = 7e-10
Identities = 72/292 (24%), Positives = 118/292 (39%), Gaps = 45/292 (15%)
Query: 28 EELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRN------RLLILQNAPNMKFI 81
+E+ ++IL RLP KSL RFK V KSW + YF + N +LL+ + + + I
Sbjct: 11 DEVILQILARLPVKSLFRFKSVCKSWYRLPSDKYFTSLFNQLSVKEQLLVAEVSDSSSLI 70
Query: 82 FCDGGNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLILWNPTTKEVH 141
D + + F +D RI + S +G+ C S I + NP+T+E
Sbjct: 71 CVDNLRGVSELSLD--FVRDRVRIRV--SSNGLLC----CSSIPEKGVYYVCNPSTREYR 122
Query: 142 LIP--RAPSLGNHYSDESLYGFGAVND----DFKVVKLNISNSNRMAKINSLLKADIYDL 195
+P R + Y D G D F VV S S + ++D
Sbjct: 123 KLPKSRERPVTRFYPDGEATLVGLACDLSKNKFNVVLAGYHRSFGQRPDGSFI-CLVFDS 181
Query: 196 STKSWTPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFRDNQFRKL 255
+ W VS T + ++ VNG+ +W+ S IL D + +RK+
Sbjct: 182 ESNKWRKFVSVLEECSFTHMSKNQV-VFVNGMLHWLMSG----LCYILALDVEHDVWRKI 236
Query: 256 EAP---KLGH------YIPFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTLEQ 298
P ++G+ Y+ E G+L + + VW++IW + +
Sbjct: 237 SLPDEIRIGNGGGNRVYL-------LESDGFLSVI---QLSDVWMKIWKMSE 278
>At2g40920 unknown protein
Length = 436
Score = 61.2 bits (147), Expect = 9e-10
Identities = 71/303 (23%), Positives = 128/303 (41%), Gaps = 42/303 (13%)
Query: 32 IEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLLILQNAPNMKFIFCDGGNDQKS 91
+EIL+R P SL+RFKCV K W+++I S YF + Q P + D G +
Sbjct: 60 MEILMRFPLTSLTRFKCVSKQWSSLISSRYFCNLLYTTVTRQQ-PRLYMCLKDDGGHRVL 118
Query: 92 IPIKSLFPQDVARIEIYGSCD----GVFCLKGISS--CITRHDQLILWNPTTKEVHLIPR 145
+ I S + + + + G F L + C +R + ++NP+TK++ +P
Sbjct: 119 LSISSPSRGNTSFVVVEQDLSIPGMGGFFLNVVRGLMCFSRRKKARIYNPSTKQLLTLPA 178
Query: 146 APS-----LGNHYSDESLY-GFGAVNDDFKVVKLNISNSNRMAKINSLLKADIYDLSTK- 198
S G Y G V+D +K+V ++ S+ + ++ + LK++ + + +
Sbjct: 179 IKSDIVAQQGQTKHHPRYYIGHDPVSDQYKLV-CTVAISSLLPRLGN-LKSEHWVFALEA 236
Query: 199 --SWTPLVSHPPITMVTRIQPSRYNTLVNG-VYYWITSSDGSDAARILCFDFRDNQFRKL 255
SW +V P+ PS +G V ++ D + ++ FD R Q +
Sbjct: 237 GGSWKKVV---PLENYRHHAPSTEGRSTSGSVVRYMAWPDNYNCV-VVSFDIRSEQLTII 292
Query: 256 EAPKLGHYIPFFCDDVFEIKGYLGYVVQYRCRIVWL-----------EIWTLEQNG---W 301
P+ H D+V + +++Y +I ++W LE G W
Sbjct: 293 PVPREIH-----LDEVVPAVTMMADLIEYGGKIAIFYHTNLKDEGSADLWVLEDTGKSEW 347
Query: 302 AKK 304
+KK
Sbjct: 348 SKK 350
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.140 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,918,287
Number of Sequences: 26719
Number of extensions: 386186
Number of successful extensions: 1243
Number of sequences better than 10.0: 254
Number of HSP's better than 10.0 without gapping: 189
Number of HSP's successfully gapped in prelim test: 65
Number of HSP's that attempted gapping in prelim test: 882
Number of HSP's gapped (non-prelim): 310
length of query: 373
length of database: 11,318,596
effective HSP length: 101
effective length of query: 272
effective length of database: 8,619,977
effective search space: 2344633744
effective search space used: 2344633744
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC135160.10


