

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134967.11 - phase: 0
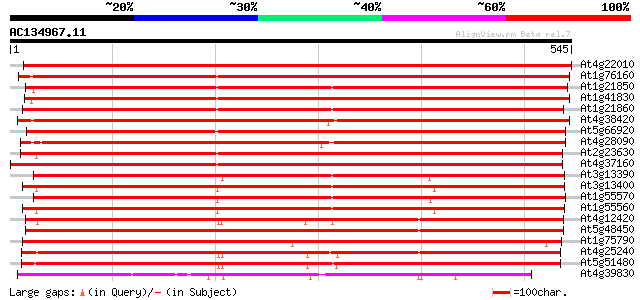
(545 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g22010 pectinesterase like protein 894 0.0
At1g76160 unknown protein 776 0.0
At1g21850 pectinesterase, putative 739 0.0
At1g41830 pectinesterase like protein 734 0.0
At1g21860 pectinesterase, putative 730 0.0
At4g38420 pectinesterase like protein 697 0.0
At5g66920 pectinesterase like protein 659 0.0
At4g28090 pectinesterase like protein 630 0.0
At2g23630 putative pectinesterase 625 e-179
At4g37160 pectinesterase like protein 622 e-178
At3g13390 L-ascorbate oxidase precursor, putative 592 e-169
At3g13400 pollen specific protein, putative 584 e-167
At1g55570 overlap with bases 100,099-109,160 of 'IGF' clone F20N... 582 e-166
At1g55560 unknown protein 577 e-165
At4g12420 pollen-specific protein - like predicted GPI-anchored ... 513 e-145
At5g48450 putative protein 510 e-145
At1g75790 polymer protein, putative 506 e-143
At4g25240 Pollen-specific protein SKU5 homologue (SKS1) 500 e-142
At5g51480 predicted GPI-anchored protein 484 e-137
At4g39830 putative L-ascorbate oxidase 216 2e-56
>At4g22010 pectinesterase like protein
Length = 541
Score = 894 bits (2310), Expect = 0.0
Identities = 419/533 (78%), Positives = 467/533 (87%), Gaps = 1/533 (0%)
Query: 14 VTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLII 73
+ +LL+ V ++PYRFFTWKITYGDIYPLGVKQQGILINGQFPGP IDA+TN+N+II
Sbjct: 9 IVLLLVLINGVLGDNPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPHIDAITNDNIII 68
Query: 74 SVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPSL 133
SV+NYL EPFLISWNG+Q R+NSWQDGV GT CPIPPGKNFTY +QVKDQIG+++YFPSL
Sbjct: 69 SVFNYLKEPFLISWNGVQQRKNSWQDGVVGTTCPIPPGKNFTYVIQVKDQIGSFYYFPSL 128
Query: 134 GMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFPD 193
HKAAGAFG IR+WSRPRIPVPF PP GDF LLAGDW K H LRR+LE G NLP PD
Sbjct: 129 AFHKAAGAFGAIRVWSRPRIPVPFSPPDGDFWLLAGDWYKTNHYVLRRLLEAGRNLPNPD 188
Query: 194 GLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYS 253
G+LINGRGW GNTFTV GKTYRFRISNVG+ATS+NFRIQGH++KLVEVEGSHT+QN Y+
Sbjct: 189 GVLINGRGWGGNTFTVQPGKTYRFRISNVGVATSLNFRIQGHTMKLVEVEGSHTVQNIYT 248
Query: 254 SLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPGPT 313
SLDIHLGQSYSVLV ANQ +DYYIV+S+RFTR+VLTTTSILHYS SR GVSGPVP GPT
Sbjct: 249 SLDIHLGQSYSVLVTANQAPQDYYIVISSRFTRKVLTTTSILHYSNSRKGVSGPVPNGPT 308
Query: 314 LDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAVNS 373
LD+ SS++QARTIR NLTASGPRPNPQGSYHYGLIKP RTI+LANSAP+INGKQRYAVN
Sbjct: 309 LDIASSLYQARTIRRNLTASGPRPNPQGSYHYGLIKPGRTIILANSAPWINGKQRYAVNG 368
Query: 374 VSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGN-AYLQTAVMGANFHEYVEIVFQNWE 432
S++APDTPLKLADYF IPGVF +G I TSP+GGN YLQ++VM ANF E++E+VFQNWE
Sbjct: 369 ASFVAPDTPLKLADYFKIPGVFNLGSIPTSPSGGNGGYLQSSVMAANFREFIEVVFQNWE 428
Query: 433 NSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVG 492
NSVQSWH+ GYSFFVVG GQWTP SR +YNLRD V+R T QVYPR+WTAIY+ALDNVG
Sbjct: 429 NSVQSWHVSGYSFFVVGMDGGQWTPGSRAKYNLRDAVSRSTVQVYPRAWTAIYIALDNVG 488
Query: 493 MWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASGRVTRPF 545
MWNIRSENW RQYLGQQFYLRVYT S S RDEYP PKNAL+CGRA GR TRPF
Sbjct: 489 MWNIRSENWARQYLGQQFYLRVYTSSTSYRDEYPPPKNALMCGRAKGRHTRPF 541
>At1g76160 unknown protein
Length = 541
Score = 776 bits (2004), Expect = 0.0
Identities = 374/536 (69%), Positives = 432/536 (79%), Gaps = 3/536 (0%)
Query: 9 SSLFLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTN 68
++ + + LLF +V EDPYRFF W ITYGDIYPLGV+QQGILING FPGP I +VTN
Sbjct: 8 AAALFIGLSLLF--AVTAEDPYRFFEWNITYGDIYPLGVRQQGILINGAFPGPDIHSVTN 65
Query: 69 ENLIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYF 128
+NLII+VYN L EPFL+SWNGIQ RRNS+ DGV GT CPIPPGKN+TY LQ+KDQIG+++
Sbjct: 66 DNLIINVYNSLDEPFLLSWNGIQQRRNSFVDGVYGTTCPIPPGKNYTYILQMKDQIGSFY 125
Query: 129 YFPSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHN 188
YFPSLG HKAAG FGGIRI SRPRIPVPFP PAGD T+L GDW K H LR L+NG
Sbjct: 126 YFPSLGFHKAAGGFGGIRILSRPRIPVPFPDPAGDTTVLIGDWYKANHTDLRAQLDNGKK 185
Query: 189 LPFPDGLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTL 248
LP PDG+LINGR +G T V+QGKTYRFRISNVGL S+NFRIQ H +K+VEVEG+HTL
Sbjct: 186 LPLPDGILINGRS-SGATLNVEQGKTYRFRISNVGLQDSLNFRIQDHKMKVVEVEGTHTL 244
Query: 249 QNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPV 308
Q T+SSLD+H+GQSYSVLV A+Q +DYY+VVS+RFT VLTTT I YS S GVSGP+
Sbjct: 245 QTTFSSLDVHVGQSYSVLVTADQTPRDYYVVVSSRFTSNVLTTTGIFRYSNSAGGVSGPI 304
Query: 309 PPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQR 368
P GPT+ + S+ QAR IR NL+ASGPRPNPQGSYHYG+I TRTI LA+SA ++GKQR
Sbjct: 305 PGGPTIQIDWSLNQARAIRTNLSASGPRPNPQGSYHYGMINTTRTIRLASSAGQVDGKQR 364
Query: 369 YAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGANFHEYVEIVF 428
YAVNSVS+ DTPLK+ADYF I GV+ G I PTGG YL T+VM ++ +VEI+F
Sbjct: 365 YAVNSVSFKPADTPLKIADYFKIDGVYRSGSIQYQPTGGGIYLDTSVMQVDYRTFVEIIF 424
Query: 429 QNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMAL 488
+N E+ VQSWH+DGYSF+VVG GQW+P+SR YNLRD VARCT QVYP SWTAI +AL
Sbjct: 425 ENSEDIVQSWHLDGYSFWVVGMDGGQWSPDSRNEYNLRDAVARCTVQVYPSSWTAILIAL 484
Query: 489 DNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASGRVTRP 544
DNVGMWN+RSE W RQYLGQQ YLRVYTPS SLRDEYP+PKNALLCGRASGR TRP
Sbjct: 485 DNVGMWNLRSEFWARQYLGQQLYLRVYTPSTSLRDEYPIPKNALLCGRASGRSTRP 540
>At1g21850 pectinesterase, putative
Length = 551
Score = 739 bits (1908), Expect = 0.0
Identities = 359/529 (67%), Positives = 422/529 (78%), Gaps = 4/529 (0%)
Query: 16 ILLLFF--TSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLII 73
IL LFF + V EDPY+FF W +TYG+I PL V QQGILING+FPGP I AVTN+NLII
Sbjct: 12 ILGLFFLISFVAAEDPYKFFEWHVTYGNISPLKVAQQGILINGKFPGPDIAAVTNDNLII 71
Query: 74 SVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPSL 133
+V+N+L EPFLISW+GI++ RNS+QDGV GT CPIPPGKN+TY LQVKDQIG+++YFPSL
Sbjct: 72 NVFNHLDEPFLISWSGIRNWRNSYQDGVYGTTCPIPPGKNYTYALQVKDQIGSFYYFPSL 131
Query: 134 GMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFPD 193
G HKAAG FG IRI SRPRIPVPFP PAGD+T+L GDW K H+ LR L+NG LPFPD
Sbjct: 132 GFHKAAGGFGAIRISSRPRIPVPFPAPAGDYTVLIGDWYKTNHKDLRAQLDNGGKLPFPD 191
Query: 194 GLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYS 253
G+LINGRG +G T ++ GKTYR RISNVGL S+NFRIQ H +KLVEVEG+HT+Q +S
Sbjct: 192 GILINGRG-SGATLNIEPGKTYRLRISNVGLQNSLNFRIQNHKMKLVEVEGTHTIQTPFS 250
Query: 254 SLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPGPT 313
SLD+H+GQSYSVL+ A+QP KDYYIVVS+RFT ++L T +LHYS S VSGP+P P
Sbjct: 251 SLDVHVGQSYSVLITADQPAKDYYIVVSSRFTSKILITAGVLHYSNSAGPVSGPIPEAP- 309
Query: 314 LDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAVNS 373
+ + S QAR I+ NL ASGPRPNPQG+YHYG IK TRTI LA+SA INGKQRYAVNS
Sbjct: 310 IQLRWSFDQARAIKTNLAASGPRPNPQGTYHYGKIKVTRTIKLASSAGNINGKQRYAVNS 369
Query: 374 VSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGANFHEYVEIVFQNWEN 433
S+ DTPLKLADYF I GV+ G I PT G Y T+VM ++ +VEIVF+NWE+
Sbjct: 370 ASFYPTDTPLKLADYFKIAGVYNPGSIPDQPTHGAIYPVTSVMQTDYKAFVEIVFENWED 429
Query: 434 SVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGM 493
VQ+WH+DGYSFFVVG G+W+ SR YNL D V+RCT QVYPRSWTAIY++LDNVGM
Sbjct: 430 IVQTWHLDGYSFFVVGMELGKWSAASRKVYNLNDAVSRCTVQVYPRSWTAIYVSLDNVGM 489
Query: 494 WNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASGRVT 542
WN+RSE WERQYLGQQFY+RVYTPS SLRDEY +PKNALLCGRA+G T
Sbjct: 490 WNLRSELWERQYLGQQFYMRVYTPSTSLRDEYLIPKNALLCGRATGHHT 538
>At1g41830 pectinesterase like protein
Length = 542
Score = 734 bits (1895), Expect = 0.0
Identities = 353/533 (66%), Positives = 418/533 (78%), Gaps = 4/533 (0%)
Query: 15 TILLL---FFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENL 71
TILL FF +V E PYRFF W +TYGDIYPLGV+QQGILINGQFPGP I +VTN+NL
Sbjct: 10 TILLFCLSFFAAVTAESPYRFFDWNVTYGDIYPLGVRQQGILINGQFPGPDIHSVTNDNL 69
Query: 72 IISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFP 131
II+V+N L EPFLISWNG+Q+RRNS+ DG+ GT CPIPP N+TY LQVKDQIG+++YFP
Sbjct: 70 IINVHNSLDEPFLISWNGVQNRRNSYVDGMYGTTCPIPPRSNYTYILQVKDQIGSFYYFP 129
Query: 132 SLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPF 191
SL HKAAG FGGIRI SRP IPVPF PAGD+T+L GDW K H L+ L+ G LP
Sbjct: 130 SLAFHKAAGGFGGIRILSRPGIPVPFADPAGDYTVLIGDWYKFNHTDLKSRLDRGRKLPS 189
Query: 192 PDGLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNT 251
PDG+LINGR NG T V+QGKTYR RISNVGL S+NFRIQ H +KLVEVEG+HTLQ
Sbjct: 190 PDGILINGRS-NGATLNVEQGKTYRLRISNVGLQDSLNFRIQNHRMKLVEVEGTHTLQTM 248
Query: 252 YSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPG 311
+SSLD+H+GQSYSVL+ A+Q +DYY+VVS+RFT +++TTT +L YS S SGP+P G
Sbjct: 249 FSSLDVHVGQSYSVLITADQSPRDYYVVVSSRFTDKIITTTGVLRYSGSSTPASGPIPGG 308
Query: 312 PTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAV 371
PT+ V S+ QAR IR NLTASGPRPNPQGSYHYGLI RTI+ +SA INGKQRY V
Sbjct: 309 PTIQVDWSLNQARAIRTNLTASGPRPNPQGSYHYGLIPLIRTIVFGSSAGQINGKQRYGV 368
Query: 372 NSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGANFHEYVEIVFQNW 431
NSVS++ DTPLKLAD+F I GV+ + IS PT G YL T+V+ ++ ++EIVF+N
Sbjct: 369 NSVSFVPADTPLKLADFFKISGVYKINSISDKPTYGGLYLDTSVLQVDYRTFIEIVFENQ 428
Query: 432 ENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNV 491
E+ VQS+H++GYSF+VVG GQW SR YNLRD V+R T QVYP+SWTAIY+ALDNV
Sbjct: 429 EDIVQSYHLNGYSFWVVGMDGGQWKTGSRNGYNLRDAVSRSTVQVYPKSWTAIYIALDNV 488
Query: 492 GMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASGRVTRP 544
GMWN+RSE W RQYLGQQ YLRV+T S SLRDEYP+PKN+ LCGRA GR TRP
Sbjct: 489 GMWNLRSEFWARQYLGQQLYLRVFTSSTSLRDEYPIPKNSRLCGRARGRHTRP 541
>At1g21860 pectinesterase, putative
Length = 538
Score = 730 bits (1884), Expect = 0.0
Identities = 349/526 (66%), Positives = 418/526 (79%), Gaps = 2/526 (0%)
Query: 13 LVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLI 72
++T LL + EDPYRFF W +TYG+I PLGV QQGILING+FPGP I ++TN+NLI
Sbjct: 11 MITTLLFLISLAFAEDPYRFFEWHVTYGNISPLGVAQQGILINGKFPGPDIISITNDNLI 70
Query: 73 ISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPS 132
I+V+N+L EPFL+SWNGI++ +NS+QDGV GT CPIPPGKN+TY LQVKDQIG+++YFPS
Sbjct: 71 INVFNHLDEPFLLSWNGIRNWKNSFQDGVYGTMCPIPPGKNYTYALQVKDQIGSFYYFPS 130
Query: 133 LGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFP 192
LG HKAAG FGGIRI SR IPVPFP PA D+TLL GDW K H+ L+ L+NG LP P
Sbjct: 131 LGFHKAAGGFGGIRISSRALIPVPFPTPADDYTLLVGDWYKTNHKDLKAQLDNGGKLPLP 190
Query: 193 DGLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTY 252
DG+LINGR +G T ++ GKTYR RISNVGL S+NFRIQ H++KLVEVEG +T+QN +
Sbjct: 191 DGILINGRS-SGATLNIEPGKTYRLRISNVGLQNSLNFRIQNHTMKLVEVEGRYTIQNLF 249
Query: 253 SSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPGP 312
SSLD+H+GQSYSVL+ A+QP KDYY+VVS+RFT ++LTTT +LHYS S VSGP+P GP
Sbjct: 250 SSLDVHVGQSYSVLITADQPAKDYYVVVSSRFTSKILTTTGVLHYSNSVAPVSGPIPDGP 309
Query: 313 TLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAVN 372
+ + S QAR IR NLTASGPRPNPQGSY YG+I TRTI LAN+ +I GKQRYAVN
Sbjct: 310 -IKLSWSFNQARAIRTNLTASGPRPNPQGSYRYGVINITRTIRLANNLGHIEGKQRYAVN 368
Query: 373 SVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGANFHEYVEIVFQNWE 432
S S+ DTPLKL DYF I GV+ G IS PT G + T+VM A+F +VE++F+N E
Sbjct: 369 SASFYPADTPLKLVDYFKIDGVYKPGSISDQPTNGAIFPTTSVMQADFRAFVEVIFENSE 428
Query: 433 NSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVG 492
+ VQSWH+DGYSF+VVG G+W+P SR YNL D + RCT QVYPRSWTAIY+ALDNVG
Sbjct: 429 DIVQSWHLDGYSFYVVGMELGKWSPASRKVYNLNDAILRCTIQVYPRSWTAIYIALDNVG 488
Query: 493 MWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRAS 538
MWN+RSE WERQYLGQQFY+RVYT S SLRDEY +PKNALLCGRAS
Sbjct: 489 MWNMRSEIWERQYLGQQFYMRVYTTSTSLRDEYLIPKNALLCGRAS 534
>At4g38420 pectinesterase like protein
Length = 549
Score = 697 bits (1798), Expect = 0.0
Identities = 341/545 (62%), Positives = 424/545 (77%), Gaps = 12/545 (2%)
Query: 8 VSSLFLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVT 67
V ++ ++TI ++ F V+ +DPYRFF W++TYG+I PLG+ Q+GILINGQ+PGP I +VT
Sbjct: 9 VWTMMMMTISIISF--VQADDPYRFFDWRVTYGNISPLGIPQRGILINGQYPGPDIYSVT 66
Query: 68 NENLIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTY 127
N+NLII+V+N L EPFL+SWNG+Q R+NS+QDGV GT CPIPPGKN+TY +QVKDQIG++
Sbjct: 67 NDNLIINVHNDLDEPFLLSWNGVQLRKNSYQDGVYGTTCPIPPGKNYTYAIQVKDQIGSF 126
Query: 128 FYFPSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGH 187
FYFPSL +HKAAG FGG RI SRPRIPVPFP PAGDFT L GDW K H+ L+ +L+ GH
Sbjct: 127 FYFPSLAVHKAAGGFGGFRILSRPRIPVPFPEPAGDFTFLIGDWFKHDHKVLKAILDRGH 186
Query: 188 NLPFPDGLLINGRGWN-GNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSH 246
LP P G+LING+G + ++ TV +GKTYRFRISNVGL ++NFRIQGH +KLVEVEG+H
Sbjct: 187 KLPLPQGVLINGQGVSYMSSITVHKGKTYRFRISNVGLQHTLNFRIQGHQMKLVEVEGTH 246
Query: 247 TLQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSG 306
T+Q+ Y+SLDIH+GQSYSVLV +QP +DY IVVST+F + L +S +HYS SR S
Sbjct: 247 TVQSMYTSLDIHVGQSYSVLVTMDQPDQDYDIVVSTKFVAKKLLVSSTIHYSNSRHSHSS 306
Query: 307 PV------PPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSA 360
P LD S+ QAR+IR NLTASGPRPNPQGSYHYG IK +RT++L +SA
Sbjct: 307 SANSVHVQQPADELD--WSIKQARSIRTNLTASGPRPNPQGSYHYGRIKISRTLILESSA 364
Query: 361 PYINGKQRYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAY-LQTAVMGAN 419
+ KQRYA+N VS++ DTPLKLADYF I GVF +G I P G ++T+VMGA+
Sbjct: 365 ALVKRKQRYAINGVSFVPGDTPLKLADYFKIKGVFKMGSIPDKPRRGRGMRMETSVMGAH 424
Query: 420 FHEYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPR 479
+++EI+FQN E VQS+H+DGYSF+VVG G W+ SR YNLRD ++R TTQVYP
Sbjct: 425 HRDFLEIIFQNREKIVQSYHLDGYSFWVVGTDRGTWSKASRREYNLRDAISRSTTQVYPE 484
Query: 480 SWTAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASG 539
SWTA+Y+ALDNVGMWN+RSE W RQYLGQQFYLRVY+P+ SLRDEY +PKNALLCGRAS
Sbjct: 485 SWTAVYVALDNVGMWNLRSEYWARQYLGQQFYLRVYSPTHSLRDEYLLPKNALLCGRASN 544
Query: 540 RVTRP 544
+ T P
Sbjct: 545 KHTTP 549
>At5g66920 pectinesterase like protein
Length = 546
Score = 659 bits (1700), Expect = 0.0
Identities = 313/524 (59%), Positives = 390/524 (73%), Gaps = 2/524 (0%)
Query: 17 LLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLIISVY 76
LL VK E PY+F+TW +TYG I PLGV QQ ILINGQFPGP+++ VTN+N+I+++
Sbjct: 23 LLSSLVIVKGESPYKFYTWTVTYGIISPLGVPQQVILINGQFPGPKLEVVTNDNIILNLI 82
Query: 77 NYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPSLGMH 136
N L +PFL++WNGI+ R+NSWQDGV GTNCPI P NFTY Q KDQIGT+ YFPS H
Sbjct: 83 NKLDQPFLLTWNGIKQRKNSWQDGVLGTNCPIQPNSNFTYKFQTKDQIGTFNYFPSTAFH 142
Query: 137 KAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFPDGLL 196
KAAG FG I +++RP IP+P+P P DFTLL GDW K H+ L++ L++G LPFPDG+L
Sbjct: 143 KAAGGFGAINVYARPGIPIPYPLPTADFTLLVGDWFKTNHKTLQQRLDSGGVLPFPDGML 202
Query: 197 INGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYSSLD 256
ING+ +TF+ DQGKTY RISNVGL+++ NFRIQGH++K+VEVEGSH +Q Y SLD
Sbjct: 203 INGQ--TQSTFSGDQGKTYMLRISNVGLSSTFNFRIQGHTMKVVEVEGSHVIQTDYDSLD 260
Query: 257 IHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPGPTLDV 316
IH+GQS +VLV NQ KDYYIV STRF R L+ +L YS SR+ SG P P ++
Sbjct: 261 IHVGQSLAVLVTLNQSPKDYYIVASTRFIRSKLSVMGLLRYSNSRVPASGDPPALPPGEL 320
Query: 317 VSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAVNSVSY 376
V S+ QART RWNLTA+ RPNPQGS+HYG+I PT+T + +NSAP INGKQRYAVN VSY
Sbjct: 321 VWSMRQARTFRWNLTANAARPNPQGSFHYGMISPTKTFVFSNSAPLINGKQRYAVNGVSY 380
Query: 377 IAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGANFHEYVEIVFQNWENSVQ 436
+ +TPLKLAD+F I GVF I + P+ + T+V+ + H+++EIVFQN E S+Q
Sbjct: 381 VKSETPLKLADHFGISGVFSTNAIQSVPSNSPPTVATSVVQTSHHDFLEIVFQNNEKSMQ 440
Query: 437 SWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGMWNI 496
SWH+DGY F+VVGFGSGQWTP R +NL D + R TTQVYP SWT I ++LDN GMWN+
Sbjct: 441 SWHLDGYDFWVVGFGSGQWTPAKRSLHNLVDALTRHTTQVYPESWTTILVSLDNQGMWNM 500
Query: 497 RSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASGR 540
RS WERQY GQQFYL+V+ +SL +EY P N LCG+A GR
Sbjct: 501 RSAIWERQYSGQQFYLKVWNSVQSLANEYNPPDNLQLCGKAVGR 544
>At4g28090 pectinesterase like protein
Length = 547
Score = 630 bits (1626), Expect = 0.0
Identities = 311/537 (57%), Positives = 402/537 (73%), Gaps = 14/537 (2%)
Query: 11 LFLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNEN 70
+ ++T ++ F VK ED F+ W++TYG I + ++GILINGQFPGP+I ++TN+N
Sbjct: 11 MMMMTTTIISF--VKAEDTL-FYNWRVTYGKIALDTLPRRGILINGQFPGPEIRSLTNDN 67
Query: 71 LIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYF 130
L+I+V N L +PFL+SWNG+ R+NS+QDGV GTNCPIPPGKN+TY QVKDQ+G+YFYF
Sbjct: 68 LVINVQNDLDDPFLLSWNGVHMRKNSYQDGVYGTNCPIPPGKNYTYDFQVKDQVGSYFYF 127
Query: 131 PSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLP 190
PSL + KAAG +G +RI+S PRIPVPFP PA DFT L DW + H L+++L+ G LP
Sbjct: 128 PSLAVQKAAGGYGSLRIYSLPRIPVPFPEPAEDFTFLVNDWYRRNHTTLKKILDGGRKLP 187
Query: 191 -FPDGLLINGRGWNG-NTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTL 248
PDG++ING+G + + TVD+GKTYRFR+SNVGL TS+N I GH LKL+EVEG+HT+
Sbjct: 188 LMPDGVMINGQGVSTVYSITVDKGKTYRFRVSNVGLQTSLNLEILGHQLKLIEVEGTHTV 247
Query: 249 QNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSR----IGV 304
Q Y+SLDIH+GQ+YS LV +QP ++Y IVVSTRF + + LHYS S+ I
Sbjct: 248 QTMYTSLDIHVGQTYSFLVTMDQPPQNYSIVVSTRFINAEVVIRATLHYSNSKGHKIITA 307
Query: 305 SGPVPPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYIN 364
P P DV S+ QA++IR NLTASGPR NPQGSYHYG +K +RT++L +SA +
Sbjct: 308 RRPDPD----DVEWSIKQAQSIRTNLTASGPRTNPQGSYHYGKMKISRTLILESSAALVK 363
Query: 365 GKQRYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSP-TGGNAYLQTAVMGANFHEY 423
KQRYA+N VS++ DTPLKLAD+F I VF VG I P GG L TAVMGA+ + +
Sbjct: 364 RKQRYAINGVSFVPSDTPLKLADHFKIKDVFKVGTIPDKPRRGGGIRLDTAVMGAHHNAF 423
Query: 424 VEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTA 483
+EI+FQN E VQS+H+DGY+F+VVG G W+ SR YNL+D ++R TTQVYP+SWTA
Sbjct: 424 LEIIFQNREKIVQSYHLDGYNFWVVGINKGIWSRASRREYNLKDAISRSTTQVYPKSWTA 483
Query: 484 IYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASGR 540
+Y+ALDNVGMWN+RS+ W RQYLGQQFYLRV++P+ S +DEYP+PKNALLCGRAS +
Sbjct: 484 VYVALDNVGMWNLRSQFWARQYLGQQFYLRVHSPNHSPKDEYPLPKNALLCGRASNK 540
>At2g23630 putative pectinesterase
Length = 541
Score = 625 bits (1613), Expect = e-179
Identities = 316/530 (59%), Positives = 374/530 (69%), Gaps = 4/530 (0%)
Query: 11 LFLVTILLLFFTSV---KCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVT 67
L L +L F+SV EDPY FFTW +TYG PLGV QQ ILINGQFPGP I+ VT
Sbjct: 7 LLLGFLLAYCFSSVFVINAEDPYLFFTWTVTYGTRSPLGVPQQVILINGQFPGPPIEGVT 66
Query: 68 NENLIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTY 127
N N++++V N L EPFLI+WNGI+ R+ SWQDGV GTNCPI P ++TY Q+KDQIGTY
Sbjct: 67 NNNIVVNVINKLDEPFLITWNGIKQRKMSWQDGVLGTNCPIQPKSSWTYHFQLKDQIGTY 126
Query: 128 FYFPSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGH 187
YF S MH+A+GAFG + + R I VP+P P DFTLL DW K+GH+ L+R L++
Sbjct: 127 AYFASTSMHRASGAFGALNVNQRSVIFVPYPKPDADFTLLVSDWYKMGHKELQRRLDSSR 186
Query: 188 NLPFPDGLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHT 247
LP PDGLLING G FT GK YRFRISNVG++TSINFRIQGH + LVEVEGSHT
Sbjct: 187 ALPPPDGLLINGAS-KGLVFTGQHGKIYRFRISNVGISTSINFRIQGHMMTLVEVEGSHT 245
Query: 248 LQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGP 307
LQ Y SLDIH+GQS +VLV PVKDY+IV STRFT+ +LTTT IL Y S+I S P
Sbjct: 246 LQEVYESLDIHVGQSVTVLVTLKAPVKDYFIVASTRFTKPILTTTGILSYQGSKIRPSHP 305
Query: 308 VPPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQ 367
+P GPT + S+ QARTIR NLTA+ RPNPQGS+HYG I RT +LAN INGK
Sbjct: 306 LPIGPTYHIHWSMKQARTIRLNLTANAARPNPQGSFHYGTIPINRTFVLANGRAMINGKL 365
Query: 368 RYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGANFHEYVEIV 427
RY VN VSY+ P TPLKLAD+FNIPGVF I PT G + L T+V HEYVE V
Sbjct: 366 RYTVNRVSYVNPATPLKLADWFNIPGVFNFKTIMNIPTPGPSILGTSVFDVALHEYVEFV 425
Query: 428 FQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMA 487
FQN E S+QSWH+DG S +VVG+GSG W R YNL D V+R T QVYP SWT+I ++
Sbjct: 426 FQNNEGSIQSWHLDGTSAYVVGYGSGTWNMAKRRGYNLVDAVSRHTFQVYPMSWTSILVS 485
Query: 488 LDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRA 537
LDN GMWN+RS+ W R+YLGQ+ Y+RV+ KSL E P N L CG+A
Sbjct: 486 LDNKGMWNLRSQIWSRRYLGQELYVRVWNNEKSLYTESEPPVNVLFCGKA 535
>At4g37160 pectinesterase like protein
Length = 541
Score = 622 bits (1603), Expect = e-178
Identities = 308/538 (57%), Positives = 380/538 (70%), Gaps = 2/538 (0%)
Query: 1 MKSYYNVVSSLFLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPG 60
MK +V LF+ + L V EDPY F+TW +TYG PLGV QQ ILINGQFPG
Sbjct: 1 MKQTNLLVCKLFIGALFWLGSVLVNAEDPYMFYTWTVTYGTRSPLGVPQQVILINGQFPG 60
Query: 61 PQIDAVTNENLIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQV 120
P I+AVTN N+++++ N L EPFLI+WNG++ RR SWQDGV GTNCPI P N+TY Q+
Sbjct: 61 PAIEAVTNNNIVVNLINKLDEPFLITWNGVKQRRTSWQDGVLGTNCPIQPNSNWTYQFQL 120
Query: 121 KDQIGTYFYFPSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDW-SKLGHRRL 179
KDQIGTY YF S +H+A+GAFG + I R I P+P P GDFTLL DW S + H+ L
Sbjct: 121 KDQIGTYTYFASTSLHRASGAFGALNINQRSVITTPYPTPDGDFTLLVSDWFSNMTHKDL 180
Query: 180 RRVLENGHNLPFPDGLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKL 239
R+ L+ G LP PD LLING G FT QGKTY+FR+SNVG+ATSINFRIQ H++ L
Sbjct: 181 RKSLDAGSALPLPDALLINGVS-KGLIFTGQQGKTYKFRVSNVGIATSINFRIQNHTMSL 239
Query: 240 VEVEGSHTLQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSY 299
+EVEG+HTLQ +Y SLD+H+GQS +VLV V+DY+IV STRFT+ VLTTT+ L Y
Sbjct: 240 IEVEGAHTLQESYESLDVHVGQSMTVLVTLKASVRDYFIVASTRFTKPVLTTTASLRYQG 299
Query: 300 SRIGVSGPVPPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANS 359
S+ GP+P GPT + S+ QARTIR NLTA+ RPNPQGS+HYG I RT++LAN+
Sbjct: 300 SKNAAYGPLPIGPTYHIHWSMKQARTIRMNLTANAARPNPQGSFHYGTIPINRTLVLANA 359
Query: 360 APYINGKQRYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGAN 419
A I GK RY VN +SYI P TPLKLAD++NI GVF I ++PT G A++ T+V+
Sbjct: 360 ATLIYGKLRYTVNRISYINPTTPLKLADWYNISGVFDFKTIISTPTTGPAHIGTSVIDVE 419
Query: 420 FHEYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPR 479
HE+VEIVFQN E S+QSWH+DG S + VG+GSG W R RYNL D V R T QVYP
Sbjct: 420 LHEFVEIVFQNDERSIQSWHMDGTSAYAVGYGSGTWNVTMRKRYNLVDAVPRHTFQVYPL 479
Query: 480 SWTAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRA 537
SWT I ++LDN GMWN+RS+ W R+YLGQ+ Y+RV+ KSL E P N L CG+A
Sbjct: 480 SWTTILVSLDNKGMWNLRSQIWSRRYLGQELYVRVWNDEKSLYTEAEPPLNVLYCGKA 537
>At3g13390 L-ascorbate oxidase precursor, putative
Length = 554
Score = 592 bits (1526), Expect = e-169
Identities = 286/525 (54%), Positives = 361/525 (68%), Gaps = 10/525 (1%)
Query: 24 VKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLIISVYNYLTEPF 83
V+ EDPY W +TYG + PLGV QQ ILINGQFPGP +++ +N N+II+V+N L EPF
Sbjct: 21 VRAEDPYFHHVWNVTYGTVSPLGVPQQVILINGQFPGPNVNSTSNNNVIINVFNNLDEPF 80
Query: 84 LISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPSLGMHKAAGAFG 143
L++WNGIQHR+N WQDG GT CPI PG N+TY Q KDQIG+YFY+PS MH++AG FG
Sbjct: 81 LLTWNGIQHRKNCWQDGTPGTMCPIMPGTNYTYHFQPKDQIGSYFYYPSTAMHRSAGGFG 140
Query: 144 GIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFPDGLLINGRGWN 203
G+R+ SR IPVP+ P D+T+L GDW H +L++ L++G L PDG+LING+
Sbjct: 141 GLRVNSRLLIPVPYADPEDDYTVLIGDWYTKSHTQLKKFLDSGRTLGRPDGILINGKSGK 200
Query: 204 GN-----TFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYSSLDIH 258
G+ FT+ GKTYR RI NVGL TS+NFRIQ H LKLVE+EGSH LQN Y SLD+H
Sbjct: 201 GDGSDAPLFTLKPGKTYRVRICNVGLKTSLNFRIQNHKLKLVEMEGSHVLQNDYDSLDVH 260
Query: 259 LGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPGPTLDVVS 318
+GQ Y ++ ANQ KDYY+V S+RF + V+TTT +L Y + S +PPGP +
Sbjct: 261 VGQCYGTILTANQEAKDYYMVASSRFLKSVITTTGLLRYEGGKGPASSQLPPGP-VGWAW 319
Query: 319 SVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAVNSVSYIA 378
S+ Q R+ RWNLTAS RPNPQGSYHYG I TRTI L N+ ++GK RYA+N VS+
Sbjct: 320 SLNQFRSFRWNLTASAARPNPQGSYHYGKINITRTIKLVNTQGKVDGKLRYALNGVSHTD 379
Query: 379 PDTPLKLADYFNIPG-VFYVGGISTSPTG---GNAYLQTAVMGANFHEYVEIVFQNWENS 434
P+TPLKLA+YF + VF I+ +PT + + V+ ++E+VF+N E S
Sbjct: 380 PETPLKLAEYFGVADKVFKYDSITDNPTPEQIKSIKIVPNVLNITHRTFIEVVFENHEKS 439
Query: 435 VQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGMW 494
VQSWH+DGYSFF V G WTP R YNL D V+R T QVYP+ W AI + DN GMW
Sbjct: 440 VQSWHLDGYSFFAVAVEPGTWTPEKRKNYNLLDAVSRHTVQVYPKCWAAILLTFDNCGMW 499
Query: 495 NIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASG 539
N+RSEN ER+YLGQQ Y V +P KSLRDEY +P+ +L CG G
Sbjct: 500 NVRSENSERRYLGQQLYASVLSPEKSLRDEYNMPETSLQCGLVKG 544
>At3g13400 pollen specific protein, putative
Length = 551
Score = 584 bits (1506), Expect = e-167
Identities = 282/538 (52%), Positives = 370/538 (68%), Gaps = 12/538 (2%)
Query: 13 LVTILLLFFTSV---KCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNE 69
L+T+L+ ++V DPY ++TW +TYG PLG+ QQ ILINGQFPGP +++ +N
Sbjct: 6 LLTVLVCLASTVALVSAGDPYFYYTWNVTYGTAAPLGIPQQVILINGQFPGPNLNSTSNN 65
Query: 70 NLIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFY 129
N++I+V+N L EPFL++W+G+QHR+NSWQDGV+GT+CPIP G NFTY Q KDQIG+YFY
Sbjct: 66 NVVINVFNNLDEPFLLTWSGLQHRKNSWQDGVTGTSCPIPAGTNFTYHFQPKDQIGSYFY 125
Query: 130 FPSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNL 189
+PS +H+ AG FGG+R+ SR IPVP+ P D T+L DW H L+ L++G L
Sbjct: 126 YPSTALHRFAGGFGGLRVNSRLLIPVPYADPEDDRTILINDWYAKSHTALKNFLDSGRTL 185
Query: 190 PFPDGLLINGR-----GWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEG 244
PDG+LING+ G N FT+ GKTY++RI NVG +++NFRIQGH +KLVE+EG
Sbjct: 186 GSPDGVLINGKSGKLGGNNAPLFTMKPGKTYKYRICNVGFKSTLNFRIQGHKMKLVEMEG 245
Query: 245 SHTLQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGV 304
SH LQN Y SLD+H+GQ ++VLV A+Q K+YY+V STRF ++ ++T ++ Y S +
Sbjct: 246 SHVLQNDYDSLDVHVGQCFAVLVTADQVAKNYYMVASTRFLKKEVSTVGVMSYEGSNVQA 305
Query: 305 SGPVPPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYIN 364
S +P P + S+ Q R+ RWNLTAS RPNPQGSYHYG I TRTI LAN+ +N
Sbjct: 306 SSDIPKAP-VGWAWSLNQFRSFRWNLTASAARPNPQGSYHYGKINITRTIKLANTKNLVN 364
Query: 365 GKQRYAVNSVSYIAPDTPLKLADYFNI-PGVFYVGGISTSPTGGNAYL--QTAVMGANFH 421
GK R+ N VS++ +TPLKLA+YF + VF I P L + V+ F
Sbjct: 365 GKVRFGFNGVSHVDTETPLKLAEYFGMSEKVFKYNVIKDEPAAKITTLTVEPNVLNITFR 424
Query: 422 EYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSW 481
+VE+VF+N E S+QS+H+DGYSFF V G+WTP R YNL D V+R T QVYP+SW
Sbjct: 425 TFVEVVFENHEKSMQSFHLDGYSFFAVASEPGRWTPEKRNNYNLLDAVSRHTVQVYPKSW 484
Query: 482 TAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASG 539
+AI + DN GMWNIRSENWER+YLGQQ Y+ V +P KSLRDEY +P N LCG G
Sbjct: 485 SAILLTFDNAGMWNIRSENWERRYLGQQLYVSVLSPEKSLRDEYNIPLNTNLCGIVKG 542
>At1g55570 overlap with bases 100,099-109,160 of 'IGF' clone F20N2,
gb|AC002328. This region is annotated in the F20N2
entry, gb|AC002328
Length = 555
Score = 582 bits (1500), Expect = e-166
Identities = 283/526 (53%), Positives = 360/526 (67%), Gaps = 10/526 (1%)
Query: 24 VKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLIISVYNYLTEPF 83
V+ EDPY W +TYG PLGV QQ ILINGQFPGP I++ +N N+I++V+N L EPF
Sbjct: 22 VQAEDPYFHHVWNVTYGTASPLGVPQQVILINGQFPGPNINSTSNNNVIVNVFNNLDEPF 81
Query: 84 LISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPSLGMHKAAGAFG 143
LI+W GIQHR+N WQDG +GT CPIPPG+NFTY Q KDQIG+YFY+P+ MH+AAG FG
Sbjct: 82 LITWAGIQHRKNCWQDGTAGTMCPIPPGQNFTYHFQPKDQIGSYFYYPTTAMHRAAGGFG 141
Query: 144 GIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFPDGLLINGR--- 200
G+R+ SR IPVP+ P D+T+L DW H +L++ L++G + PDG+LING+
Sbjct: 142 GLRVNSRLLIPVPYADPEDDYTILINDWYTKSHTQLKKFLDSGRTIGRPDGILINGKSGK 201
Query: 201 --GWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYSSLDIH 258
G + FT+ GKTYR RI NVGL S+NFRIQ H +KLVE+EGSH LQN Y SLD+H
Sbjct: 202 TDGSDKPLFTLKPGKTYRVRICNVGLKASLNFRIQNHKMKLVEMEGSHVLQNDYDSLDVH 261
Query: 259 LGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPGPTLDVVS 318
+GQ + V+V A+Q KDYY++ STRF ++ LTTT +L Y + S +P P +
Sbjct: 262 VGQCFGVIVTADQEPKDYYMIASTRFLKKPLTTTGLLRYEGGKGPASSQLPAAP-VGWAW 320
Query: 319 SVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAVNSVSYIA 378
S+ Q R+ RWNLTAS RPNPQGSYHYG I TRTI L N+ ++GK RYA++ VS+
Sbjct: 321 SLNQYRSFRWNLTASAARPNPQGSYHYGKINITRTIKLVNTQGKVDGKLRYALSGVSHTD 380
Query: 379 PDTPLKLADYFNIPG-VFYVGGISTSPTGG---NAYLQTAVMGANFHEYVEIVFQNWENS 434
P+TPLKLA+YF + VF IS +P N ++ V+ ++E+VF+N E S
Sbjct: 381 PETPLKLAEYFGVADKVFKYDTISDNPNPDQIKNIKIEPNVLNITHRTFIEVVFENHERS 440
Query: 435 VQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGMW 494
VQSWH+DGYSFF V G WTP R YNL D V+R T QVYP+ W AI + DN GMW
Sbjct: 441 VQSWHLDGYSFFAVAVEPGTWTPEKRKNYNLLDAVSRHTVQVYPKCWAAILLTFDNCGMW 500
Query: 495 NIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASGR 540
NIRSEN ER+YLGQQ Y V +P KSLRDEY +P+ +L CG G+
Sbjct: 501 NIRSENAERRYLGQQLYASVLSPEKSLRDEYNMPETSLQCGLVKGK 546
>At1g55560 unknown protein
Length = 549
Score = 577 bits (1488), Expect = e-165
Identities = 278/538 (51%), Positives = 370/538 (68%), Gaps = 12/538 (2%)
Query: 13 LVTILLLFFTSV---KCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNE 69
L+T+L+ ++V DPY F TW +TYG PLGV Q+ ILINGQFPGP +++ +N
Sbjct: 5 LLTVLVCLVSTVAIVNAGDPYFFHTWNVTYGTASPLGVPQKVILINGQFPGPNLNSTSNN 64
Query: 70 NLIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFY 129
N++I+V+N+L EPFL++W+GIQHR+N WQDGV+GT+CPIP G+NFTY Q KDQIG+YFY
Sbjct: 65 NVVINVFNHLDEPFLLTWSGIQHRKNCWQDGVAGTSCPIPAGQNFTYHFQPKDQIGSYFY 124
Query: 130 FPSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNL 189
+P+ +H+ AG FGG+R+ SR IPVP+ P D+T+L GDW GH L+ L++G L
Sbjct: 125 YPTTSLHRFAGGFGGLRVNSRLLIPVPYADPEDDYTVLLGDWYTAGHTALKNFLDSGRTL 184
Query: 190 PFPDGLLINGR-----GWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEG 244
P+G+LING+ G N FT+ GKTY++R+ NVG +++NFRIQ H +KLVE+EG
Sbjct: 185 GLPNGVLINGKSGKVGGKNEPLFTMKPGKTYKYRLCNVGFKSTLNFRIQNHKMKLVEMEG 244
Query: 245 SHTLQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGV 304
SH +QN Y SLD+H+GQ +SVLV ANQ KDYY+V STRF ++ L+T ++ Y S +
Sbjct: 245 SHVIQNDYDSLDVHVGQCFSVLVTANQAAKDYYMVASTRFLKKELSTVGVIRYEGSNVQA 304
Query: 305 SGPVPPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYIN 364
S +P P + S+ Q R+ RWNLT++ RPNPQGSYHYG I TR+I L NS ++
Sbjct: 305 STELPKAP-VGWAWSLNQFRSFRWNLTSNAARPNPQGSYHYGKINITRSIKLVNSKSVVD 363
Query: 365 GKQRYAVNSVSYIAPDTPLKLADYFNI-PGVFYVGGISTSPTGGNAYL--QTAVMGANFH 421
GK R+ N VS++ +TPLKLA+YF + VF I P L Q V+ F
Sbjct: 364 GKVRFGFNGVSHVDTETPLKLAEYFQMSEKVFKYNVIKDEPAAKITALTVQPNVLNITFR 423
Query: 422 EYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSW 481
+VEI+F+N E ++QS+H+DGYSFF V G+WTP R YNL D V+R T QVYP+SW
Sbjct: 424 TFVEIIFENHEKTMQSFHLDGYSFFAVASEPGRWTPEKRENYNLLDAVSRHTVQVYPKSW 483
Query: 482 TAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASG 539
+AI + DN GMWNIRSEN ER+YLG+Q Y+ V +P KSLRDEY +P N LCG G
Sbjct: 484 SAILLTFDNAGMWNIRSENLERKYLGEQLYVSVLSPEKSLRDEYNIPLNTNLCGIVKG 541
>At4g12420 pollen-specific protein - like predicted GPI-anchored
protein
Length = 587
Score = 513 bits (1320), Expect = e-145
Identities = 266/546 (48%), Positives = 353/546 (63%), Gaps = 24/546 (4%)
Query: 16 ILLLFFTSVK---CEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLI 72
+LL+FF ++ DPY F+ ++++Y PLGV QQ I ING+FPGP I+ TNENL+
Sbjct: 7 LLLVFFVNISFCFAADPYSFYNFEVSYITASPLGVPQQVIAINGKFPGPTINVTTNENLV 66
Query: 73 ISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPS 132
++V N L E L+ WNGIQ RR SWQDGV GTNCPIPP N+TY QVKDQIG++FYFPS
Sbjct: 67 VNVRNKLDEGLLLHWNGIQQRRVSWQDGVLGTNCPIPPKWNWTYEFQVKDQIGSFFYFPS 126
Query: 133 LGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFP 192
L +A+G FG + R IPVPF P GD T+ GDW H LR+ L++G +L P
Sbjct: 127 LHFQRASGGFGSFVVNPRAIIPVPFSTPDGDITVTIGDWYIRNHTALRKALDDGKDLGMP 186
Query: 193 DGLLINGRG---WNG---------NTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLV 240
DG+LING+G +N T TV GKTYR R+SNVG++TS+NFRIQGH+L L
Sbjct: 187 DGVLINGKGPYRYNDTLVADGIDFETITVHPGKTYRLRVSNVGISTSLNFRIQGHNLVLA 246
Query: 241 EVEGSHTLQNTYSSLDIHLGQSYSVLVMANQPV-KDYYIVVSTRFTR----RVLTTTSIL 295
E EGS+T+Q Y+SLDIH+GQSYS LV +Q DYYIV S R R +T IL
Sbjct: 247 ESEGSYTVQQNYTSLDIHVGQSYSFLVTMDQNASSDYYIVASARVVNETIWRRVTGVGIL 306
Query: 296 HYSYSRIGVSGPVPPGP--TLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRT 353
Y+ S+ G +PPGP D S+ QAR+IRWN++ASG RPNPQGS+ YG I T
Sbjct: 307 KYTNSKGKAKGQLPPGPQDEFDKTFSMNQARSIRWNVSASGARPNPQGSFKYGSINVTDV 366
Query: 354 IMLANSAPY-INGKQRYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQ 412
+L N P I+GK+R +N +S+ P TP++LAD + V+ + P G A +
Sbjct: 367 YVLRNMPPVTISGKRRTTLNGISFKNPSTPIRLADKLKVKDVYKL-DFPKRPLTGPAKVA 425
Query: 413 TAVMGANFHEYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARC 472
T+++ + ++E+V QN + +QS+H+ GY+FFVVG G+WT NSRG YN D +AR
Sbjct: 426 TSIINGTYRGFMEVVLQNNDTKMQSYHMSGYAFFVVGMDYGEWTENSRGTYNKWDGIARS 485
Query: 473 TTQVYPRSWTAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNAL 532
T QVYP +W+AI ++LDN G WN+R+EN + YLGQ+ Y+RV P ++ + E+ P N L
Sbjct: 486 TIQVYPGAWSAILISLDNPGAWNLRTENLDSWYLGQETYVRVVNPDENNKTEFGHPDNVL 545
Query: 533 LCGRAS 538
CG S
Sbjct: 546 YCGALS 551
>At5g48450 putative protein
Length = 544
Score = 510 bits (1314), Expect = e-145
Identities = 260/528 (49%), Positives = 347/528 (65%), Gaps = 6/528 (1%)
Query: 16 ILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLIISV 75
++ L T DPY FF W ++Y PLG +QQ I INGQFPGP ++ TN N++++V
Sbjct: 14 VVFLSVTGALAADPYVFFDWTVSYLSASPLGTRQQVIGINGQFPGPILNVTTNWNVVMNV 73
Query: 76 YNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPSLGM 135
N L EP L++WNGIQHR+NSWQDGV GTNCPIP G N+TY QVKDQIG++FYFPS
Sbjct: 74 KNNLDEPLLLTWNGIQHRKNSWQDGVLGTNCPIPSGWNWTYEFQVKDQIGSFFYFPSTNF 133
Query: 136 HKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFPDGL 195
+A+G +GGI + +R IPVPF P GD TL DW H+ LR+ +E+ + L PDG+
Sbjct: 134 QRASGGYGGIIVNNRAIIPVPFALPDGDVTLFISDWYTKSHKVLRKDVESKNGLRPPDGI 193
Query: 196 LINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYSSL 255
+ING G + T G+TYRFR+ N G+ATS+NFRIQ H+L LVE EGS+T+Q Y+++
Sbjct: 194 VINGFGPFASNGTYLLGRTYRFRVHNSGIATSLNFRIQNHNLLLVETEGSYTIQQNYTNM 253
Query: 256 DIHLGQSYSVLVMANQP-VKDYYIVVSTRFTRRVLTT-TSILHYSYSRIGVSGPVPPGP- 312
DIH+GQS+S LV +Q DYYIV S RF + + ++L YS S+ SGP+P P
Sbjct: 254 DIHVGQSFSFLVTMDQSGSNDYYIVASPRFATSIKASGVAVLRYSNSQGPASGPLPDPPI 313
Query: 313 TLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAP-YINGKQRYAV 371
LD S+ QAR++R NL++ RPNPQGS+ YG I T ++ N P I G+ R +
Sbjct: 314 ELDTFFSMNQARSLRLNLSSGAARPNPQGSFKYGQITVTDVYVIVNRPPEMIEGRLRATL 373
Query: 372 NSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGANFHEYVEIVFQNW 431
N +SY+ P TPLKLA +NI GV+ + P + + T+V+ F +VEI+FQN
Sbjct: 374 NGISYLPPATPLKLAQQYNISGVYKL-DFPKRPMNRHPRVDTSVINGTFKGFVEIIFQNS 432
Query: 432 ENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNV 491
+ +V+S+H+DGY+FFVVG G WT NSR YN D VAR TTQV+P +WTA+ ++LDN
Sbjct: 433 DTTVKSYHLDGYAFFVVGMDFGLWTENSRSTYNKGDAVARSTTQVFPGAWTAVLVSLDNA 492
Query: 492 GMWNIRSENWERQYLGQQFYLRVYTPSKSL-RDEYPVPKNALLCGRAS 538
GMWN+R +N YLGQ+ YL V P + E VPKN++ CGR S
Sbjct: 493 GMWNLRIDNLASWYLGQELYLSVVNPEIDIDSSENSVPKNSIYCGRLS 540
>At1g75790 polymer protein, putative
Length = 545
Score = 506 bits (1304), Expect = e-143
Identities = 256/535 (47%), Positives = 343/535 (63%), Gaps = 11/535 (2%)
Query: 13 LVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENLI 72
LV I L+ P + W ++Y + LG +Q I+IN FPGP ++A N+ ++
Sbjct: 9 LVLISLVILELSYAFAPISSYQWVVSYSQRFILGGNKQVIVINDMFPGPILNATANDIIV 68
Query: 73 ISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPS 132
++++N L EPFL++WNG+Q R+NSWQDGV GTNCPI PG N+TY QVKDQIG+YFYFP+
Sbjct: 69 VNIFNNLPEPFLMTWNGLQLRKNSWQDGVRGTNCPILPGTNWTYRFQVKDQIGSYFYFPT 128
Query: 133 LGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFP 192
L + KAAG +G IRI+ +PVPFP P ++ +L GDW L H +R L+ GH+LP P
Sbjct: 129 LLLQKAAGGYGAIRIYPPELVPVPFPKPDEEYDILIGDWFYLDHTVMRASLDAGHSLPNP 188
Query: 193 DGLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTY 252
DG+L NGRG F + GKTYR RISNVGL T +NFRIQ H + LVE EG++ + Y
Sbjct: 189 DGILFNGRGPEETFFAFEPGKTYRLRISNVGLKTCLNFRIQDHDMLLVETEGTYVQKRVY 248
Query: 253 SSLDIHLGQSYSVLVMA-NQPV---KDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPV 308
SSLDIH+GQSYS+LV A PV + YYI + RFT L +++ Y S + G
Sbjct: 249 SSLDIHVGQSYSILVTAKTDPVGIYRSYYIFATARFTDSYLGGIALIRYPGSPLDPVGQG 308
Query: 309 PPGPTL-DVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQ 367
P P L D SSV QA +IR +L R NPQGSYHYG I TRTI+L N +GK
Sbjct: 309 PLAPALQDFGSSVEQALSIRMDLNVGAARSNPQGSYHYGRINVTRTIILHNDVMLSSGKL 368
Query: 368 RYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYLQTAVMGANFHEYVEIV 427
RY +N VS++ P+TPLKL D+F + G P+ L T+V+ ++ +++ IV
Sbjct: 369 RYTINGVSFVYPETPLKLVDHFQLNDTIIPGMFPVYPSNKTPTLGTSVVDIHYKDFIHIV 428
Query: 428 FQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMA 487
FQN ++S+HIDGY+FFVVG+G G W+ + + YNL D V+R T QVYP SWTAI +A
Sbjct: 429 FQNPLFGLESYHIDGYNFFVVGYGFGAWSESKKAGYNLVDAVSRSTVQVYPYSWTAILIA 488
Query: 488 LDNVGMWNIRSENWERQYLGQQFYLRVYTPSKS------LRDEYPVPKNALLCGR 536
+DN GMWN+RS+ E+ YLGQ+ Y+RV + +RDE P+P N + CG+
Sbjct: 489 MDNQGMWNVRSQKAEQWYLGQELYMRVKGEGEEDPSTIPVRDENPIPGNVIRCGK 543
>At4g25240 Pollen-specific protein SKU5 homologue (SKS1)
Length = 589
Score = 500 bits (1287), Expect = e-142
Identities = 258/544 (47%), Positives = 351/544 (64%), Gaps = 23/544 (4%)
Query: 12 FLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENL 71
FL+ LL S DP+ + ++++Y PLGV QQ I +NGQFPGP ++A TN N+
Sbjct: 11 FLLCFALLSAVSFAA-DPFVSYDFRVSYLTASPLGVPQQVIAVNGQFPGPLLNATTNYNV 69
Query: 72 IISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFP 131
+++V+N+L EP L++W GIQ RRNSWQDGV GTNCPIPP NFTY QVKDQIG++FY P
Sbjct: 70 VVNVFNHLDEPLLLTWPGIQMRRNSWQDGVLGTNCPIPPRWNFTYQFQVKDQIGSFFYSP 129
Query: 132 SLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPF 191
SL +A+G FG I I +R IP+PFP P G+ + GDW H+ LRR L++G L
Sbjct: 130 SLNFQRASGGFGPIVINNRDIIPIPFPQPDGELIFIIGDWYTQDHKALRRALDSGKELGM 189
Query: 192 PDGLLINGRG---WNGN--------TFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLV 240
PDG+LING+G +N + TF V+ GKTYR R+ NVG++TS+NFRIQ HSL LV
Sbjct: 190 PDGVLINGKGPYKYNSSVPDGIDYLTFHVEPGKTYRIRVHNVGISTSLNFRIQNHSLLLV 249
Query: 241 EVEGSHTLQNTYSSLDIHLGQSYSVLVMANQ-PVKDYYIVVSTRFTRRV----LTTTSIL 295
E EG +T Q ++ D+H+GQSYS LV +Q DYYIV S RF +T +IL
Sbjct: 250 ETEGHYTSQANFTDFDVHVGQSYSFLVTMDQDATSDYYIVASARFVNETVWQRVTGVAIL 309
Query: 296 HYSYSRIGVSGPVPPGPTLDVV---SSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTR 352
HYS S+ VSGP+P P DV S++ Q +TIR N +ASG RPNPQGS+HYG I T
Sbjct: 310 HYSNSKGPVSGPLPV-PKTDVSSPWSAMSQPKTIRQNTSASGARPNPQGSFHYGQINITN 368
Query: 353 TIMLANSAP-YINGKQRYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYL 411
T +L + P ING R +N +S++ P TP++LAD + G + + P L
Sbjct: 369 TYILRSLPPTIINGALRATLNGISFVNPSTPVRLADRNKVKGAYKL-DFPDRPFNRPLRL 427
Query: 412 QTAVMGANFHEYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVAR 471
+++ A + ++++VFQN + +QS+H+DGYSFFVVG G W+ + +G YN D ++R
Sbjct: 428 DRSMINATYKGFIQVVFQNNDTKIQSFHVDGYSFFVVGMDFGIWSEDKKGSYNNWDAISR 487
Query: 472 CTTQVYPRSWTAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNA 531
T +VYP WTA+ ++LDNVG+WNIR EN +R YLG++ Y+R+ P + + E P N
Sbjct: 488 STIEVYPGGWTAVLISLDNVGVWNIRVENLDRWYLGEETYMRITNPEEDGKTEMDPPDNV 547
Query: 532 LLCG 535
L CG
Sbjct: 548 LYCG 551
>At5g51480 predicted GPI-anchored protein
Length = 592
Score = 484 bits (1246), Expect = e-137
Identities = 249/544 (45%), Positives = 344/544 (62%), Gaps = 22/544 (4%)
Query: 12 FLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENL 71
F+ + L+F S DPY + + ++Y PLGV QQ I +NG+FPGP I+A TN N+
Sbjct: 10 FVFSFALIFGFSF-AGDPYVSYDFTLSYITASPLGVPQQVIAVNGKFPGPVINATTNYNV 68
Query: 72 IISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFP 131
++V N+L EP L++W G+Q RRNSWQDGV GTNCPIPP NFTY Q+KDQIG+YFY P
Sbjct: 69 HVNVLNHLDEPLLLTWPGVQMRRNSWQDGVLGTNCPIPPNWNFTYDFQLKDQIGSYFYSP 128
Query: 132 SLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPF 191
SL +A+G FG + I +R +P+PF P G+ + GDW H LRR+L++G L
Sbjct: 129 SLNFQRASGGFGALIINNRDLVPIPFTEPDGEIIFIIGDWYTQNHTALRRILDSGKELGM 188
Query: 192 PDGLLINGRG---WNGN--------TFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLV 240
PDG+LING+G +N + T VD GKTYR R+ NVG++TS+NFRIQ H L L+
Sbjct: 189 PDGVLINGKGPFKYNSSVPDGIEHETVNVDPGKTYRIRVHNVGISTSLNFRIQNHKLLLI 248
Query: 241 EVEGSHTLQNTYSSLDIHLGQSYSVLVMANQ-PVKDYYIVVSTRFTRRV----LTTTSIL 295
E EG +T Q ++ D+H+GQSYS LV +Q DYYIV S RF +T IL
Sbjct: 249 ETEGRYTSQMNFTDFDVHVGQSYSFLVTMDQNATSDYYIVASARFVNETVWQRVTGVGIL 308
Query: 296 HYSYSRIGVSGPVPPGPTLDV---VSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTR 352
HYS S+ SGP+P T DV S++ Q R I+ N +ASG RPNPQGS+HYG I TR
Sbjct: 309 HYSNSKGPASGPLPVSAT-DVNHPWSAMNQPRAIKQNTSASGARPNPQGSFHYGQINITR 367
Query: 353 TIMLANSAP-YINGKQRYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSPTGGNAYL 411
T +L + P INGK R +N +S++ P TP++LAD + G + + L
Sbjct: 368 TYILRSLPPTKINGKLRATLNGISFVNPSTPMRLADDHKVKGDYMLDFPDRPLDEKLPRL 427
Query: 412 QTAVMGANFHEYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVAR 471
++++ A + +++++FQN + +QS+HIDGY+F+VV G W+ + YN D VAR
Sbjct: 428 SSSIINATYKGFIQVIFQNNDTKIQSFHIDGYAFYVVAMDFGIWSEDRNSSYNNWDAVAR 487
Query: 472 CTTQVYPRSWTAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNA 531
T +VYP +WTA+ ++LDNVG+WNIR EN +R YLGQ+ Y+R+ P ++ E P+N
Sbjct: 488 STVEVYPGAWTAVLISLDNVGVWNIRVENLDRWYLGQETYMRIINPEENGSTEMDPPENV 547
Query: 532 LLCG 535
+ CG
Sbjct: 548 MYCG 551
>At4g39830 putative L-ascorbate oxidase
Length = 582
Score = 216 bits (551), Expect = 2e-56
Identities = 161/553 (29%), Positives = 257/553 (46%), Gaps = 63/553 (11%)
Query: 8 VSSLFLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVT 67
V +L ++ + LFF+SV C+ R F W++ Y P ++ I ING+FPGP I A
Sbjct: 13 VFNLMVLCFIALFFSSVLCQGKIRRFKWEVKYEFKSPDCFEKLVITINGKFPGPTIKAQQ 72
Query: 68 NENLIISVYN-YLTEPFLISWNGIQHRRNSWQDGVSG-TNCPIPPGKNFTYTLQVKDQIG 125
+ +++ + N ++TE + W+GI+ W DGV G T CPI PG+ F Y V D+ G
Sbjct: 73 GDTIVVELKNSFMTENVAVHWHGIRQIGTPWFDGVEGVTQCPILPGEVFIYQF-VVDRPG 131
Query: 126 TYFYFPSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLEN 185
TY Y GM + +G G I++ P PF D L DW H+ +
Sbjct: 132 TYMYHSHYGMQRESGLIGMIQVSPPATEPEPFTYDY-DRNFLLTDWY---HKSMSEKATG 187
Query: 186 GHNLPF-----PDGLLINGRG-WNGNT------------------------FTVDQGKTY 215
++PF P L+I GRG +N + TV GKTY
Sbjct: 188 LASIPFKWVGEPQSLMIQGRGRFNCSNNLTTPPSLVSGVCNVSNADCSRFILTVIPGKTY 247
Query: 216 RFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYSSLDIHLGQSYSVLVMANQPVKD 275
R RI ++ ++++F+I+GH+L +VE +G + T +L ++ G++YSVL+ A+Q +
Sbjct: 248 RLRIGSLTALSALSFQIEGHNLTVVEADGHYVEPFTVKNLFVYSGETYSVLLKADQNPRR 307
Query: 276 YYIVVSTRFTRRVLT--TTSILHYSYSRIGVSGPVPPGPTLDVVSSVFQARTIRWNLTAS 333
Y + S+ +R T T++L+Y + P PT + + V + R L S
Sbjct: 308 NYWITSSIVSRPATTPPATAVLNYYPNH-----PRRRPPTSESSNIVPEWNDTRSRLAQS 362
Query: 334 GPRPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAVNSVSYIAPDTPLKLADYFNIPG 393
+G H + I+L N+ +NG +R++VN+VSY P TP +A N+
Sbjct: 363 LAIKARRGFIHALPENSDKVIVLLNTQNEVNGYRRWSVNNVSYHHPKTPYLIALKQNLTN 422
Query: 394 VF---------YVG---GISTSPTGGNAYLQTAVMGANFHEYVEIVFQNW------ENSV 435
F Y I P NA + F+ V+++ QN +
Sbjct: 423 AFDWRFTAPENYDSRNYDIFAKPLNANATTSDGIYRLRFNSTVDVILQNANTMNANNSET 482
Query: 436 QSWHIDGYSFFVVGFGSGQWTPN-SRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGMW 494
WH+ G+ F+V+G+G G++ + RYN D + + T V P WTA+ DN G+W
Sbjct: 483 HPWHLHGHDFWVLGYGEGKFNESEDPKRYNRVDPIKKNTVAVQPFGWTALRFRADNPGVW 542
Query: 495 NIRSENWERQYLG 507
+ ++G
Sbjct: 543 SFHCHIESHFFMG 555
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,619,398
Number of Sequences: 26719
Number of extensions: 642128
Number of successful extensions: 1369
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1169
Number of HSP's gapped (non-prelim): 61
length of query: 545
length of database: 11,318,596
effective HSP length: 104
effective length of query: 441
effective length of database: 8,539,820
effective search space: 3766060620
effective search space used: 3766060620
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC134967.11


