

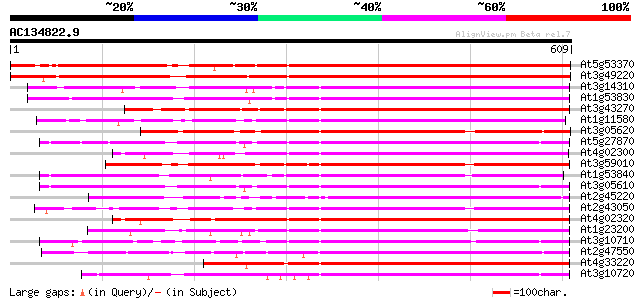
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.9 + phase: 0
(609 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g53370 pectinesterase 766 0.0
At3g49220 pectinesterase - like protein 754 0.0
At3g14310 pectin methylesterase like protein 479 e-135
At1g53830 unknown protein 476 e-134
At3g43270 pectinesterase like protein 445 e-125
At1g11580 unknown protein 432 e-121
At3g05620 putative pectinesterase 414 e-116
At5g27870 pectin methyl-esterase - like protein 412 e-115
At4g02300 putative pectinesterase 409 e-114
At3g59010 pectinesterase precursor-like protein 403 e-112
At1g53840 unknown protein 402 e-112
At3g05610 putative pectinesterase 402 e-112
At2g45220 pectinesterase like protein 402 e-112
At2g43050 putative pectinesterase 400 e-112
At4g02320 pectinesterase - like protein 400 e-111
At1g23200 putative pectinesterase 397 e-111
At3g10710 putative pectinesterase 393 e-109
At2g47550 unknown protein 389 e-108
At4g33220 pectinesterase like protein 387 e-107
At3g10720 pectinesterase like protein 384 e-107
>At5g53370 pectinesterase
Length = 587
Score = 766 bits (1977), Expect = 0.0
Identities = 385/611 (63%), Positives = 467/611 (76%), Gaps = 28/611 (4%)
Query: 1 MDFDRLGPSDPGGSSRRSTAPLTPSTTSNSSPKSNKKLIMLSILAAVLIIASAISAALIT 60
M +DRLGPS P +++ A P K+ KLI+ ++ AVL++ + +
Sbjct: 1 MGYDRLGPSGPSNPNQKDPATSLPELQK----KTKTKLILFTL--AVLVVG-VVCFGIFA 53
Query: 61 VVRSRASSNNSNLLHSKPTQAISRTCSKTRYPSLCINSLLDFPGSTSASEQELVHISFNM 120
+R+ S L KPTQAISRTCSK+ YP+LCI++LLDFPGS +A E EL+HISFN
Sbjct: 54 GIRAVDSGKTEPKLTRKPTQAISRTCSKSLYPNLCIDTLLDFPGSLTADENELIHISFNA 113
Query: 121 THRHISKALFASSGLSYTVANPRVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDD 180
T + SKAL+ SS ++YT PRVR+AY+ CLEL+D+S+DA+ ++ S++ +S D+
Sbjct: 114 TLQKFSKALYTSSTITYTQMPPRVRSAYDSCLELLDDSVDALTRALSSVVV----VSGDE 169
Query: 181 GESRQFSNVAGSTEDVMTWLSAALTNQDTCLEGFEDTSGT---VKDQMVGNLKDLSELVS 237
S DVMTWLS+A+TN DTC +GF++ G VKDQ++G +KDLSE+VS
Sbjct: 170 SHS-----------DVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQVIGAVKDLSEMVS 218
Query: 238 NSLAIFSASGDNDFTGVPIQNKRRLMGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIV 297
N LAIF A D +GVP+ N R+L+G + + E P WL++ DR LL P S IQADI V
Sbjct: 219 NCLAIF-AGKVKDLSGVPVVNNRKLLGTEE-TEELPNWLKREDRELLGTPTSAIQADITV 276
Query: 298 SKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKT 357
SK G +GT KTI EAIKKAPEHS RRF+IYV+AGRYEE NLKVG+KKTN+MFIGDG+GKT
Sbjct: 277 SKDG-SGTFKTIAEAIKKAPEHSSRRFVIYVKAGRYEEENLKVGRKKTNLMFIGDGKGKT 335
Query: 358 VITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRC 417
VITG +S+ D +TTFHTA+FAA+G GF+ RD+TFENYAGP KHQAVALRVG DHAVVYRC
Sbjct: 336 VITGGKSIADDLTTFHTATFAATGAGFIVRDMTFENYAGPAKHQAVALRVGGDHAVVYRC 395
Query: 418 NIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQN 477
NI+GYQDA YVHSNRQFFREC IYGTVDFIFGNAAV+ Q CNIYARKPMAQQK TITAQN
Sbjct: 396 NIIGYQDALYVHSNRQFFRECEIYGTVDFIFGNAAVILQSCNIYARKPMAQQKITITAQN 455
Query: 478 RKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGW 537
RKDPNQNTGISIH C++L PDL +SKGS TYLGRPWK+YSR VYM+S MGDH+ P GW
Sbjct: 456 RKDPNQNTGISIHACKLLATPDLEASKGSYPTYLGRPWKLYSRVVYMMSDMGDHIDPRGW 515
Query: 538 LEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLP 597
LEWNG FAL +LYYGEYMN G G+ IGQRVKWPGY VITST+EA+++TVAQFISGSSWLP
Sbjct: 516 LEWNGPFALDSLYYGEYMNKGLGSGIGQRVKWPGYHVITSTVEASKFTVAQFISGSSWLP 575
Query: 598 STGVAFLAGLS 608
STGV+F +GLS
Sbjct: 576 STGVSFFSGLS 586
>At3g49220 pectinesterase - like protein
Length = 598
Score = 754 bits (1948), Expect = 0.0
Identities = 382/619 (61%), Positives = 479/619 (76%), Gaps = 33/619 (5%)
Query: 1 MDFDRLGPSDPGGSSRRST--APLTPSTTSNSSPKSN-----KKLIMLSILAAVLIIASA 53
M ++RLGPS GS ST AP+ +++ P++N KKL++ SI+ A+ +I
Sbjct: 1 MGYERLGPSGATGSVTTSTTTAPILNQVSTSEQPENNNRRSKKKLVVSSIVLAISLI--- 57
Query: 54 ISAALITVVRSRASSNNSNL-LHSKPTQAISRTCSKTRYPSLCINSLLDFPGSTSASE-Q 111
++AA+ VRSR N S L KP+QAIS+ C TR+P LC++SL+DFPGS +AS +
Sbjct: 58 LAAAIFAGVRSRLKLNQSVPGLARKPSQAISKACELTRFPELCVDSLMDFPGSLAASSSK 117
Query: 112 ELVHISFNMTHRHISKALFASSGLSYTVANPRVRAAYEDCLELMDESMDAIRSSMDSLMT 171
+L+H++ NMT H S AL++S+ LS+ PR R+AY+ C+EL+D+S+DA+ ++ S+++
Sbjct: 118 DLIHVTVNMTLHHFSHALYSSASLSFVDMPPRARSAYDSCVELLDDSVDALSRALSSVVS 177
Query: 172 TSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQDTCLEGFEDTS-GTVKDQMVGNLK 230
+S+ +DV TWLSAALTN DTC EGF+ G VKD M L+
Sbjct: 178 SSA-----------------KPQDVTTWLSAALTNHDTCTEGFDGVDDGGVKDHMTAALQ 220
Query: 231 DLSELVSNSLAIFSASGD-NDFTGVPIQNKRRLMGMSDISREFPKWLEKRDRRLLSLPVS 289
+LSELVSN LAIFSAS D +DF GVPIQN RRL+G+ + +FP+W+ ++R +L +PVS
Sbjct: 221 NLSELVSNCLAIFSASHDGDDFAGVPIQN-RRLLGVEEREEKFPRWMRPKEREILEMPVS 279
Query: 290 EIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMF 349
+IQADIIVSK G NGT KTI+EAIKKAP++S RR IIYV+AGRYEENNLKVG+KK N+MF
Sbjct: 280 QIQADIIVSKDG-NGTCKTISEAIKKAPQNSTRRIIIYVKAGRYEENNLKVGRKKINLMF 338
Query: 350 IGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGS 409
+GDG+GKTVI+G +S+ D +TTFHTASFAA+G GF+ARDITFEN+AGP KHQAVALR+G+
Sbjct: 339 VGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARDITFENWAGPAKHQAVALRIGA 398
Query: 410 DHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQ 469
DHAV+YRCNI+GYQD YVHSNRQFFREC+IYGTVDFIFGNAAVV Q C+IYARKPM Q
Sbjct: 399 DHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGNAAVVLQNCSIYARKPMDFQ 458
Query: 470 KNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMG 529
KNTITAQNRKDPNQNTGISIH R+L A DL ++ GS +TYLGRPWK++SRTVYM+SY+G
Sbjct: 459 KNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTYLGRPWKLFSRTVYMMSYIG 518
Query: 530 DHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQF 589
HVH GWLEWN FAL TLYYGEY+N GPG+ +GQRV WPGYRVI ST EANR+TVA+F
Sbjct: 519 GHVHTRGWLEWNTTFALDTLYYGEYLNSGPGSGLGQRVSWPGYRVINSTAEANRFTVAEF 578
Query: 590 ISGSSWLPSTGVAFLAGLS 608
I GSSWLPSTGV+FLAGLS
Sbjct: 579 IYGSSWLPSTGVSFLAGLS 597
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 479 bits (1233), Expect = e-135
Identities = 274/619 (44%), Positives = 367/619 (59%), Gaps = 59/619 (9%)
Query: 20 APLTPSTTSNSSPKSNKKLIMLSILAAVLIIASAISAALITVVRSRASSNNSNLLHSKPT 79
AP S + K NKKL++LS A+L +A+ + + + AS N S +
Sbjct: 2 APSMKEIFSKDNFKKNKKLVLLSAAVALLFVAA------VAGISAGASKANEKRTLSPSS 55
Query: 80 QAISRT-CSKTRYPSLCINSLLDFPGSTSASEQELVHISFNMT-----HRH--ISKALFA 131
A+ R+ CS TRYP LCI++++ G S+++++ S N+T H + + K +
Sbjct: 56 HAVLRSSCSSTRYPELCISAVVTAGGVELTSQKDVIEASVNLTITAVEHNYFTVKKLIKK 115
Query: 132 SSGLSYTVANPRVRAAYEDCLELMDESMDAIRSSMDSL--MTTSSTLSNDDGESRQFSNV 189
GL+ PR + A DCLE +DE++D + +++ L T TL G
Sbjct: 116 RKGLT-----PREKTALHDCLETIDETLDELHETVEDLHLYPTKKTLREHAG-------- 162
Query: 190 AGSTEDVMTWLSAALTNQDTCLEGF--EDTSGTVKDQMVGNLKDLSELVSNSLAIFSASG 247
D+ T +S+A+TNQ+TCL+GF +D V+ ++ + + SN+LA+
Sbjct: 163 -----DLKTLISSAITNQETCLDGFSHDDADKQVRKALLKGQIHVEHMCSNALAMIKNMT 217
Query: 248 DNDFTGVP-----IQNKRRLM-------------GMSDISRE-FPKWLEKRDRRLLSLPV 288
D D N R+L G ++ E +P WL DRRLL
Sbjct: 218 DTDIANFEQKAKITSNNRKLKEENQETTVAVDIAGAGELDSEGWPTWLSAGDRRLLQ--G 275
Query: 289 SEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIM 348
S ++AD V+ + G+GT KT+ A+ APE+S +R++I+++AG Y EN ++V KKK NIM
Sbjct: 276 SGVKADATVA-ADGSGTFKTVAAAVAAAPENSNKRYVIHIKAGVYREN-VEVAKKKKNIM 333
Query: 349 FIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVG 408
F+GDGR +T+ITG R+V DG TTFH+A+ AA G F+ARDITF+N AGP KHQAVALRVG
Sbjct: 334 FMGDGRTRTIITGSRNVVDGSTTFHSATVAAVGERFLARDITFQNTAGPSKHQAVALRVG 393
Query: 409 SDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQ 468
SD + Y C+++ YQD YVHSNRQFF +C I GTVDFIFGNAAVV Q C+I+AR+P +
Sbjct: 394 SDFSAFYNCDMLAYQDTLYVHSNRQFFVKCLIAGTVDFIFGNAAVVLQDCDIHARRPNSG 453
Query: 469 QKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYM 528
QKN +TAQ R DPNQNTGI I CRI DL S KGS TYLGRPWK YS+TV M S +
Sbjct: 454 QKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLQSVKGSFPTYLGRPWKEYSQTVIMQSAI 513
Query: 529 GDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQ 588
D + P GW EW G FAL TL Y EY N G GA RVKW G++VIT+ EA +YT Q
Sbjct: 514 SDVIRPEGWSEWTGTFALNTLTYREYSNTGAGAGTANRVKWRGFKVITAAAEAQKYTAGQ 573
Query: 589 FISGSSWLPSTGVAFLAGL 607
FI G WL STG F GL
Sbjct: 574 FIGGGGWLSSTGFPFSLGL 592
>At1g53830 unknown protein
Length = 587
Score = 476 bits (1225), Expect = e-134
Identities = 268/603 (44%), Positives = 361/603 (59%), Gaps = 32/603 (5%)
Query: 20 APLTPSTTSNSSPKSNKKLIMLSILAAVLIIASAISAALITVVRSRASSNNSNLLHSKPT 79
AP+ + S K+NKKLI+ S A+L++AS + A T +++ + L S
Sbjct: 2 APIKEFISKFSDFKNNKKLILSSAAIALLLLASIVGIAATTTNQNK--NQKITTLSSTSH 59
Query: 80 QAISRTCSKTRYPSLCINSLLDFPGSTSASEQELVHISFNMTHRHISKALFASSGL--SY 137
+ CS T YP LC +++ G S++E++ S N+T + + FA L
Sbjct: 60 AILKSVCSSTLYPELCFSAVAATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKR 119
Query: 138 TVANPRVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVM 197
PR A DCLE +DE++D + +++ L S + +D+
Sbjct: 120 KGLTPREVTALHDCLETIDETLDELHVAVEDLHQYPKQKS-----------LRKHADDLK 168
Query: 198 TWLSAALTNQDTCLEGF--EDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVP 255
T +S+A+TNQ TCL+GF +D V+ ++ + + SN+LA+ + D
Sbjct: 169 TLISSAITNQGTCLDGFSYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFE 228
Query: 256 IQNK---------RRLMGMS-DISRE-FPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNG 304
+++K R+L ++ D+ + +PKWL DRRLL S I+AD V+ G +G
Sbjct: 229 LRDKSSTFTNNNNRKLKEVTGDLDSDGWPKWLSVGDRRLLQ--GSTIKADATVADDG-SG 285
Query: 305 TVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRS 364
T+ A+ APE S +RF+I+++AG Y EN ++V KKKTNIMF+GDGRGKT+ITG R+
Sbjct: 286 DFTTVAAAVAAAPEKSNKRFVIHIKAGVYREN-VEVTKKKTNIMFLGDGRGKTIITGSRN 344
Query: 365 VGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQD 424
V DG TTFH+A+ AA G F+ARDITF+N AGP KHQAVALRVGSD + Y+C++ YQD
Sbjct: 345 VVDGSTTFHSATVAAVGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQD 404
Query: 425 ACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQN 484
YVHSNRQFF +C+I GTVDFIFGNAA V Q C+I AR+P + QKN +TAQ R DPNQN
Sbjct: 405 TLYVHSNRQFFVKCHITGTVDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQN 464
Query: 485 TGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDF 544
TGI I +CRI DL + KG+ TYLGRPWK YSRTV M S + D + P GW EW+G F
Sbjct: 465 TGIVIQNCRIGGTSDLLAVKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSF 524
Query: 545 ALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFL 604
AL TL Y EY+N G GA RVKW GY+VITS EA +T QFI G WL STG F
Sbjct: 525 ALDTLTYREYLNRGGGAGTANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFS 584
Query: 605 AGL 607
L
Sbjct: 585 LSL 587
>At3g43270 pectinesterase like protein
Length = 527
Score = 445 bits (1145), Expect = e-125
Identities = 235/483 (48%), Positives = 313/483 (64%), Gaps = 15/483 (3%)
Query: 125 ISKALFASSGLSYTVANPRVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESR 184
I+KA+ S RV A DC++L+D S+ + L S + +G+
Sbjct: 58 ITKAVAIVSKFDKKAGKSRVSNAIVDCVDLLD-------SAAEELSWIISASQSPNGKDN 110
Query: 185 QFSNVAGSTEDVMTWLSAALTNQDTCLEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIFS 244
+V D+ TW+SAAL+NQDTCL+GFE T+G +K + G L + V N L +
Sbjct: 111 STGDVGS---DLRTWISAALSNQDTCLDGFEGTNGIIKKIVAGGLSKVGTTVRNLLTMVH 167
Query: 245 ASGDNDFTGVPIQNKRRLMGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNG 304
+ PI+ + S S+ FP W++ DR+LL + AD +V+ + G G
Sbjct: 168 SPPSKP-KPKPIKAQTMTKAHSGFSK-FPSWVKPGDRKLLQTDNITV-ADAVVA-ADGTG 223
Query: 305 TVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRS 364
TI++A+ AP++S +R++I+V+ G Y EN +++ KKK NIM +GDG TVITG RS
Sbjct: 224 NFTTISDAVLAAPDYSTKRYVIHVKRGVYVEN-VEIKKKKWNIMMVGDGIDATVITGNRS 282
Query: 365 VGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQD 424
DG TTF +A+FA SG GF+ARDITF+N AGPEKHQAVA+R +D V YRC + GYQD
Sbjct: 283 FIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVAIRSDTDLGVFYRCAMRGYQD 342
Query: 425 ACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQN 484
Y HS RQFFREC I GTVDFIFG+A VFQ C I A++ + QKN+ITAQ RKDPN+
Sbjct: 343 TLYAHSMRQFFRECIITGTVDFIFGDATAVFQSCQIKAKQGLPNQKNSITAQGRKDPNEP 402
Query: 485 TGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDF 544
TG +I I DL + + TYLGRPWK+YSRTV+M +YM D ++P GWLEWNG+F
Sbjct: 403 TGFTIQFSNIAADTDLLLNLNTTATYLGRPWKLYSRTVFMQNYMSDAINPVGWLEWNGNF 462
Query: 545 ALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFL 604
AL TLYYGEYMN GPGA++ +RVKWPGY V+ ++ EAN +TV+Q I G+ WLPSTG+ F+
Sbjct: 463 ALDTLYYGEYMNSGPGASLDRRVKWPGYHVLNTSAEANNFTVSQLIQGNLWLPSTGITFI 522
Query: 605 AGL 607
AGL
Sbjct: 523 AGL 525
>At1g11580 unknown protein
Length = 557
Score = 432 bits (1111), Expect = e-121
Identities = 253/587 (43%), Positives = 348/587 (59%), Gaps = 56/587 (9%)
Query: 30 SSPKSNKKLIMLSILAAVLIIASA--ISAALITVVRSRASSNNSNLLHSKPTQAISRTCS 87
S PKS K + +L+ V I+ S +A LI+V ++N+ +LL + S+ C
Sbjct: 10 SKPKSLKHKNLCLVLSFVAILGSVAFFTAQLISV---NTNNNDDSLLTT------SQICH 60
Query: 88 KTRYPSLCINSLLDFP--GSTSASEQELVHI-------SFNMTHRHISKALFASSGLSYT 138
C L +F + + +L+H+ T +S+A S+G+
Sbjct: 61 GAHDQDSCQALLSEFTTLSLSKLNRLDLLHVFLKNSVWRLESTMTMVSEARIRSNGV--- 117
Query: 139 VANPRVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMT 198
R +A + DC E+MD S D + SSM+ L + L + +SNV T
Sbjct: 118 ----RDKAGFADCEEMMDVSKDRMMSSMEELRGGNYNLES-------YSNVH-------T 159
Query: 199 WLSAALTNQDTCLEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVPIQN 258
WLS+ LTN TCLE D S K + L+DL +LAIF + +P ++
Sbjct: 160 WLSSVLTNYMTCLESISDVSVNSKQIVKPQLEDLVSRARVALAIFVSV-------LPARD 212
Query: 259 KRRLMGMSDISREFPKWLEKRDRRLL--SLPVSEIQADIIVSKSGGNGTVKTITEAIKKA 316
+++ IS FP WL DR+LL S ++ A+++V+K G G KT+ EA+ A
Sbjct: 213 DLKMI----ISNRFPSWLTALDRKLLESSPKTLKVTANVVVAKDG-TGKFKTVNEAVAAA 267
Query: 317 PEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTAS 376
PE+S R++IYV+ G Y+E + +GKKK N+M +GDG+ T+ITG +V DG TTF +A+
Sbjct: 268 PENSNTRYVIYVKKGVYKET-IDIGKKKKNLMLVGDGKDATIITGSLNVIDGSTTFRSAT 326
Query: 377 FAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFR 436
AA+G GFMA+DI F+N AGP KHQAVALRV +D V+ RC I YQD Y H+ RQF+R
Sbjct: 327 VAANGDGFMAQDIWFQNTAGPAKHQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYR 386
Query: 437 ECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILP 496
+ I GTVDFIFGN+AVVFQ C+I AR P A QKN +TAQ R+D NQNT ISI C+I
Sbjct: 387 DSYITGTVDFIFGNSAVVFQNCDIVARNPGAGQKNMLTAQGREDQNQNTAISIQKCKITA 446
Query: 497 APDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMN 556
+ DLA KGS++T+LGRPWK+YSRTV M S++ +H+ P GW W+G+FAL TLYYGEY N
Sbjct: 447 SSDLAPVKGSVKTFLGRPWKLYSRTVIMQSFIDNHIDPAGWFPWDGEFALSTLYYGEYAN 506
Query: 557 FGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAF 603
GPGA +RV W G++VI + EA ++TVA+ I G WL TGV F
Sbjct: 507 TGPGADTSKRVNWKGFKVIKDSKEAEQFTVAKLIQGGLWLKPTGVTF 553
>At3g05620 putative pectinesterase
Length = 543
Score = 414 bits (1065), Expect = e-116
Identities = 221/468 (47%), Positives = 290/468 (61%), Gaps = 27/468 (5%)
Query: 143 RVRAAYEDCLELMDESMDAIRSSMDSL--MTTSSTLSNDDGESRQFSNVAGSTEDVMTWL 200
R + A EDC EL+ S+ + SM + + + DDG + A + ++ TWL
Sbjct: 100 REQVAIEDCKELVGFSVTELAWSMLEMNKLHGGGGIDLDDGSH----DAAAAGGNLKTWL 155
Query: 201 SAALTNQDTCLEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVPIQNKR 260
SAA++NQDTCLEGFE T ++ + G+L+ +++LVSN L +++ +P + R
Sbjct: 156 SAAMSNQDTCLEGFEGTERKYEELIKGSLRQVTQLVSNVLDMYT-----QLNALPFKASR 210
Query: 261 RLMGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHS 320
++ P+WL + D L+ + V G G +TI EAI +AP HS
Sbjct: 211 NESVIAS-----PEWLTETDESLMMRHDPSVMHPNTVVAIDGKGKYRTINEAINEAPNHS 265
Query: 321 RRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAAS 380
+R++IYV+ G Y+EN + + KKKTNIM +GDG G+T+ITG R+ G+TTF TA+ A S
Sbjct: 266 TKRYVIYVKKGVYKEN-IDLKKKKTNIMLVGDGIGQTIITGDRNFMQGLTTFRTATVAVS 324
Query: 381 GPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNI 440
G GF+A+DITF N AGP+ QAVALRV SD + YRC++ GYQD Y HS RQF+R+C I
Sbjct: 325 GRGFIAKDITFRNTAGPQNRQAVALRVDSDQSAFYRCSVEGYQDTLYAHSLRQFYRDCEI 384
Query: 441 YGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDL 500
YGT+DFIFGN A V Q C IY R P+ QK TITAQ RK PNQNTG I + +L
Sbjct: 385 YGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPNQNTGFVIQNSYVL----- 439
Query: 501 ASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPG 560
+ TYLGRPWK+YSRTVYM +YM V P GWLEW G+FAL TL+YGEY N GPG
Sbjct: 440 ----ATQPTYLGRPWKLYSRTVYMNTYMSQLVQPRGWLEWFGNFALDTLWYGEYNNIGPG 495
Query: 561 AAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGLS 608
RVKWPGY ++ A +TV FI G WLP+TGV F AGL+
Sbjct: 496 WRSSGRVKWPGYHIMDKR-TALSFTVGSFIDGRRWLPATGVTFTAGLA 542
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 412 bits (1059), Expect = e-115
Identities = 224/578 (38%), Positives = 339/578 (57%), Gaps = 29/578 (5%)
Query: 33 KSNKKLIMLSILAAVLIIASAISAALITVVRSRASSNNSNLLHSKPTQAISRTCSKTRYP 92
K K+ +++SI ++VL+I+ ++ + V ++ + + S +AI C+ T Y
Sbjct: 11 KRKKRYVIISI-SSVLLISMVVAVTIGVSVNKSDNAGDEEITTS--VKAIKDVCAPTDYK 67
Query: 93 SLCINSLLDFPGSTSASEQELVHISFNMTHRHISKALFASSGLSYTVANPRVRAAYEDCL 152
C ++L TS ELV +FN T + IS S + +PR + A + C
Sbjct: 68 ETCEDTLRKDAKDTS-DPLELVKTAFNATMKQISDVAKKSQTMIELQKDPRAKMALDQCK 126
Query: 153 ELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQDTCLE 212
ELMD ++ + S + L +F V + + WLSA ++++ TCL+
Sbjct: 127 ELMDYAIGELSKSFEEL------------GKFEFHKVDEALVKLRIWLSATISHEQTCLD 174
Query: 213 GFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTG---VPIQNKRRLMGMSDIS 269
GF+ T G + + LK +L N LA+ + +++ G +P N RRL+ S
Sbjct: 175 GFQGTQGNAGETIKKALKTAVQLTHNGLAMVTEM--SNYLGQMQIPEMNSRRLL-----S 227
Query: 270 REFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVR 329
+EFP W++ R RRLL+ P+SE++ DI+V++ G +G KTI EA+ P+ F+++++
Sbjct: 228 QEFPSWMDARARRLLNAPMSEVKPDIVVAQDG-SGQYKTINEALNFVPKKKNTTFVVHIK 286
Query: 330 AGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDI 389
G Y+E ++V + T+++FIGDG KTVI+G +S DG+TT+ TA+ A G F+A++I
Sbjct: 287 EGIYKEY-VQVNRSMTHLVFIGDGPDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNI 345
Query: 390 TFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFG 449
FEN AG KHQAVA+RV +D ++ Y C GYQD Y HS+RQF+R+C I GT+DF+FG
Sbjct: 346 AFENTAGAIKHQAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFG 405
Query: 450 NAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIET 509
+AA VFQ C + RKP+ Q ITA RKDP ++TG + C I+ PD + K +T
Sbjct: 406 DAAAVFQNCTLLVRKPLLNQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKT 465
Query: 510 YLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKW 569
YLGRPWK YSRT+ M +++ D V P GW W G+F L TL+Y E N GPGAAI +RV W
Sbjct: 466 YLGRPWKEYSRTIIMNTFIPDFVPPEGWQPWLGEFGLNTLFYSEVQNTGPGAAITKRVTW 525
Query: 570 PGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
PG + + S E ++T AQ+I G +W+P GV ++ GL
Sbjct: 526 PGIKKL-SDEEILKFTPAQYIQGDAWIPGKGVPYILGL 562
>At4g02300 putative pectinesterase
Length = 532
Score = 409 bits (1052), Expect = e-114
Identities = 232/509 (45%), Positives = 304/509 (59%), Gaps = 46/509 (9%)
Query: 112 ELVHISFNMTHRHISKALFASSGLS--YTVANPRV----RAAYEDCLELMDESMDAIRSS 165
EL+ N+T I K ASS S T P + R A+EDCL L+D+++ + ++
Sbjct: 55 ELIIADLNLT---ILKVNLASSNFSDLQTRLFPNLTHYERCAFEDCLGLLDDTISDLETA 111
Query: 166 MDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQDTCLEGFEDTSGTVKDQM 225
+ L ++S + D+ L+ +T QDTCL+GF + + M
Sbjct: 112 VSDLRSSSLEFN-----------------DISMLLTNVMTYQDTCLDGFSTSDNENNNDM 154
Query: 226 V----GNLK----DLSELVSNSLAIFSASGDNDFTGVPIQNKRRLMGMSDISREFPKWLE 277
NLK D+S +SNSL + I K+ S++ E+P WL
Sbjct: 155 TYELPENLKEIILDISNNLSNSLHMLQV----------ISRKKPSPKSSEVDVEYPSWLS 204
Query: 278 KRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENN 337
+ D+RLL PV E ++ V+ G G TI +A+ AP S RFIIY++ G Y EN
Sbjct: 205 ENDQRLLEAPVQETNYNLSVAIDG-TGNFTTINDAVFAAPNMSETRFIIYIKGGEYFEN- 262
Query: 338 LKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGP 397
+++ KKKT IMFIGDG GKTVI RS DG +TF T + G G++A+DI+F N AGP
Sbjct: 263 VELPKKKTMIMFIGDGIGKTVIKANRSRIDGWSTFQTPTVGVKGKGYIAKDISFVNSAGP 322
Query: 398 EKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQK 457
K QAVA R GSDH+ YRC GYQD YVHS +QF+REC+IYGT+DFIFGNAAVVFQ
Sbjct: 323 AKAQAVAFRSGSDHSAFYRCEFDGYQDTLYVHSAKQFYRECDIYGTIDFIFGNAAVVFQN 382
Query: 458 CNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKM 517
++YARKP K TAQ+R +Q TGISI +CRIL APDL K + + YLGRPW+
Sbjct: 383 SSLYARKPNPGHKIAFTAQSRNQSDQPTGISILNCRILAAPDLIPVKENFKAYLGRPWRK 442
Query: 518 YSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITS 577
YSRTV + S++ D +HP GWLE DFAL+TLYYGEYMN GPGA + +RV WPG+R I +
Sbjct: 443 YSRTVIIKSFIDDLIHPAGWLEGKKDFALETLYYGEYMNEGPGANMAKRVTWPGFRRIEN 502
Query: 578 TLEANRYTVAQFISGSSWLPSTGVAFLAG 606
EA ++TV FI GS+WL STG+ F G
Sbjct: 503 QTEATQFTVGPFIDGSTWLNSTGIPFSLG 531
>At3g59010 pectinesterase precursor-like protein
Length = 529
Score = 403 bits (1036), Expect = e-112
Identities = 222/505 (43%), Positives = 312/505 (60%), Gaps = 37/505 (7%)
Query: 105 STSASEQELVHISFNMTHRHISKALFASSGLSYTVANPRVRAAYEDCLELMDESMDAIRS 164
STS+++ L + ++ H +++L + LS+ + + DCLEL+D+++D +
Sbjct: 59 STSSNKGFLSSVQLSLDHALFARSLAFNLTLSHRTSQTLMLDPVNDCLELLDDTLDMLYR 118
Query: 165 SMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQDTCLEGFEDTSGTVKDQ 224
+ + D + +DV TWLSAALTNQ+TC + + S K+
Sbjct: 119 IV--------VIKRKDHVN----------DDVHTWLSAALTNQETCKQSLSEKSSFNKEG 160
Query: 225 MVGN--LKDLSELVSNSLAIFSASGDNDFTGVPIQNKRRLMGMSDISREFPKWLEKRDRR 282
+ + ++L+ L++NSL +F + + + R+L+ D FP W+ DR+
Sbjct: 161 IAIDSFARNLTGLLTNSLDMFVSDKQKSSSSSNLTGGRKLLSDHD----FPTWVSSSDRK 216
Query: 283 LLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGK 342
LL V E++ +V+ + G+GT ++ EA+ + S R +I++ AG Y+EN L +
Sbjct: 217 LLEASVEELRPHAVVA-ADGSGTHMSVAEALASLEKGSGRS-VIHLTAGTYKEN-LNIPS 273
Query: 343 KKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQA 402
K+ N+M +GDG+GKTVI G RS G T+ +A+ AA G GF+ARDITF N AGP QA
Sbjct: 274 KQKNVMLVGDGKGKTVIVGSRSNRGGWNTYQSATVAAMGDGFIARDITFVNSAGPNSEQA 333
Query: 403 VALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYA 462
VALRVGSD +VVYRC+I GYQD+ Y S RQF+RE +I GTVDFIFGN+AVVFQ CN+ +
Sbjct: 334 VALRVGSDRSVVYRCSIDGYQDSLYTLSKRQFYRETDITGTVDFIFGNSAVVFQSCNLVS 393
Query: 463 RKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTV 522
RK + Q N +TAQ R DPNQNTGISIH+CRI GS +TYLGRPWK YSRTV
Sbjct: 394 RKGSSDQ-NYVTAQGRSDPNQNTGISIHNCRI---------TGSTKTYLGRPWKQYSRTV 443
Query: 523 YMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEAN 582
M S++ +HP GW W+ +FALKTLYYGE+ N GPG+++ RV W GY + EA
Sbjct: 444 VMQSFIDGSIHPSGWSPWSSNFALKTLYYGEFGNSGPGSSVSGRVSWAGYHPALTLTEAQ 503
Query: 583 RYTVAQFISGSSWLPSTGVAFLAGL 607
+TV+ FI G+SWLPSTGV F +GL
Sbjct: 504 GFTVSGFIDGNSWLPSTGVVFDSGL 528
>At1g53840 unknown protein
Length = 586
Score = 402 bits (1034), Expect = e-112
Identities = 227/584 (38%), Positives = 345/584 (58%), Gaps = 41/584 (7%)
Query: 33 KSNKKLIMLSILAAVLIIASAISAALITVV-RSRASSNNSNLLHSKPTQAISRTCSKTRY 91
K+ K+L++LSI + V++IA I+A + TVV +++ S S P+ ++ CS TR+
Sbjct: 24 KTRKRLLLLSI-SVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAICSVTRF 82
Query: 92 PSLCINSLLDFPGSTSASEQELVHISFNMTHRHISKALFASSGLSYTVANPRVRAAYEDC 151
P CI+S+ P S + + L +S + + LS + R+++A C
Sbjct: 83 PESCISSISKLPSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDERIKSALRVC 142
Query: 152 LELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQDTCL 211
+L+++++D + + T+S D E ++ + + ED+ TWLSA +T+ +TC
Sbjct: 143 GDLIEDALDRL----------NDTVSAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCF 192
Query: 212 EGFED--------TSGTVKDQMVGNLKDLSELVSNSLAIFSA--SGDNDFTGVPIQNKRR 261
+ ++ + T+ + + +E SNSLAI S S +D G+PI +RR
Sbjct: 193 DSLDELKQNKTEYANSTITQNLKSAMSRSTEFTSNSLAIVSKILSALSDL-GIPIHRRRR 251
Query: 262 LMGMS-DISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHS 320
LM S +F KW +R L + ++ D+ V+ G G V T+ EA+ K P+ S
Sbjct: 252 LMSHHHQQSVDFEKWARRR-----LLQTAGLKPDVTVA-GDGTGDVLTVNEAVAKVPKKS 305
Query: 321 RRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAAS 380
+ F+IYV++G Y EN + + K K N+M GDG+GKT+I+G ++ DG T+ TA+FA
Sbjct: 306 LKMFVIYVKSGTYVEN-VVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFAIQ 364
Query: 381 GPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNI 440
G GF+ +DI N AG KHQAVA R GSD +V Y+C+ G+QD Y HSNRQF+R+C++
Sbjct: 365 GKGFIMKDIGIINTAGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDV 424
Query: 441 YGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDL 500
GT+DFIFG+AAVVFQ C I R+P++ Q NTITAQ +KDPNQ++G+SI C I
Sbjct: 425 TGTIDFIFGSAAVVFQGCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTI------ 478
Query: 501 ASSKGSI--ETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEW-NGDFALKTLYYGEYMNF 557
S+ G++ TYLGRPWK +S TV M + +G V P GW+ W +G ++ YGEY N
Sbjct: 479 -SANGNVIAPTYLGRPWKEFSTTVIMETVIGAVVRPSGWMSWVSGVDPPASIVYGEYKNT 537
Query: 558 GPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGV 601
GPG+ + QRVKW GY+ + S EA ++TVA + G+ W+P+TGV
Sbjct: 538 GPGSDVTQRVKWAGYKPVMSDAEAAKFTVATLLHGADWIPATGV 581
>At3g05610 putative pectinesterase
Length = 669
Score = 402 bits (1033), Expect = e-112
Identities = 218/580 (37%), Positives = 336/580 (57%), Gaps = 29/580 (5%)
Query: 33 KSNKKLIMLSILAAVLIIASAISAALITVVRSRASSNNSNLLHSKPTQAISRTCSKTRYP 92
K ++ I+++I ++VL+I+ ++ + + + + +A+ C+ T Y
Sbjct: 10 KRKRRYIVITI-SSVLLISMVVAVTVGVSLNKHDGDSKGKAEVNASVKAVKDVCAPTDYR 68
Query: 93 SLCINSLLDFPGSTSASEQELVHISFNMTHRHISKALFASSGLSYTVANPRVRAAYEDCL 152
C ++L+ G + ELV +FN+T + I+ A S + + R R A + C
Sbjct: 69 KTCEDTLIK-NGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIMELQKDSRTRMALDQCK 127
Query: 153 ELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQDTCLE 212
ELMD ++D + +S + L +F + + ++ WLSAA+++++TCLE
Sbjct: 128 ELMDYALDELSNSFEEL------------GKFEFHLLDEALINLRIWLSAAISHEETCLE 175
Query: 213 GFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTG---VPIQNKRRLMGMSDIS 269
GF+ T G + M LK EL N LAI S ++F G +P N RRL+ +
Sbjct: 176 GFQGTQGNAGETMKKALKTAIELTHNGLAIISEM--SNFVGQMQIPGLNSRRLL-----A 228
Query: 270 REFPKWLEKRDRRLLSLPV--SEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIY 327
FP W+++R R+LL S+++ DI+V++ G +G KTI EA++ P+ F+++
Sbjct: 229 EGFPSWVDQRGRKLLQAAAAYSDVKPDIVVAQDG-SGQYKTINEALQFVPKKRNTTFVVH 287
Query: 328 VRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMAR 387
++AG Y+E ++V K ++++FIGDG KT+I+G ++ DG+TT+ TA+ A G F+A+
Sbjct: 288 IKAGLYKEY-VQVNKTMSHLVFIGDGPDKTIISGNKNYKDGITTYRTATVAIVGNYFIAK 346
Query: 388 DITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFI 447
+I FEN AG KHQAVA+RV SD ++ + C GYQD Y HS+RQFFR+C I GT+DF+
Sbjct: 347 NIGFENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFL 406
Query: 448 FGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSI 507
FG+AA VFQ C + RKP+ Q ITA RKDP ++TG C I PD + K +
Sbjct: 407 FGDAAAVFQNCTLLVRKPLPNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVKETS 466
Query: 508 ETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRV 567
+ YLGRPWK YSRT+ M +++ D V P GW W GDF LKTL+Y E N GPG+A+ RV
Sbjct: 467 KAYLGRPWKEYSRTIIMNTFIPDFVQPQGWQPWLGDFGLKTLFYSEVQNTGPGSALANRV 526
Query: 568 KWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
W G + + S + ++T AQ+I G W+P GV + GL
Sbjct: 527 TWAGIKTL-SEEDILKFTPAQYIQGDDWIPGKGVPYTTGL 565
>At2g45220 pectinesterase like protein
Length = 511
Score = 402 bits (1033), Expect = e-112
Identities = 239/525 (45%), Positives = 306/525 (57%), Gaps = 50/525 (9%)
Query: 86 CSKTRYPSLCINSLLDFPGSTSA-SEQELVHISFNMTHRHISKALFASSGLSYTVANPRV 144
CS+T P C L + SE E + IS + A + L + R
Sbjct: 34 CSQTPNPKPCEYFLTHNSNNEPIKSESEFLKISMKLVLDRAILAKTHAFTLGPKCRDTRE 93
Query: 145 RAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAAL 204
+AA+EDC++L D ++ I +MD NV S D TWLS AL
Sbjct: 94 KAAWEDCIKLYDLTVSKINETMDP-------------------NVKCSKLDAQTWLSTAL 134
Query: 205 TNQDTCLEGFEDTSGT--VKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVPIQNKRRL 262
TN DTC GF + T V M N+ S L+ N+LAI F P +
Sbjct: 135 TNLDTCRAGFLELGVTDIVLPLMSNNV---SNLLCNTLAINKVP----FNYTPPEKDG-- 185
Query: 263 MGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRR 322
FP W++ DR+LL S D V G+G KTI EAI A R
Sbjct: 186 ---------FPSWVKPGDRKLLQ---SSTPKDNAVVAKDGSGNFKTIKEAIDAASGSGR- 232
Query: 323 RFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGP 382
F+IYV+ G Y EN L++ KK N+M GDG GKT+ITG +SVG G TTF++A+ AA G
Sbjct: 233 -FVIYVKQGVYSEN-LEIRKK--NVMLRGDGIGKTIITGSKSVGGGTTTFNSATVAAVGD 288
Query: 383 GFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYG 442
GF+AR ITF N AG QAVALR GSD +V Y+C+ YQD YVHSNRQF+R+C++YG
Sbjct: 289 GFIARGITFRNTAGASNEQAVALRSGSDLSVFYQCSFEAYQDTLYVHSNRQFYRDCDVYG 348
Query: 443 TVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLAS 502
TVDFIFGNAA V Q CNI+AR+P ++ NTITAQ R DPNQNTGI IH+ R+ A DL
Sbjct: 349 TVDFIFGNAAAVLQNCNIFARRPRSKT-NTITAQGRSDPNQNTGIIIHNSRVTAASDLRP 407
Query: 503 SKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAA 562
GS +TYLGRPW+ YSRTV+M + + + P GWLEW+G+FALKTL+Y E+ N GPGA+
Sbjct: 408 VLGSTKTYLGRPWRQYSRTVFMKTSLDSLIDPRGWLEWDGNFALKTLFYAEFQNTGPGAS 467
Query: 563 IGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
RV WPG+RV+ S EA+++TV F++G SW+PS+ V F +GL
Sbjct: 468 TSGRVTWPGFRVLGSASEASKFTVGTFLAGGSWIPSS-VPFTSGL 511
>At2g43050 putative pectinesterase
Length = 518
Score = 400 bits (1029), Expect = e-112
Identities = 246/589 (41%), Positives = 343/589 (57%), Gaps = 83/589 (14%)
Query: 28 SNSSPKSNKKL---IMLSILAAVLIIASAISAALITVVRSRASSNNSNLLHSKPTQAISR 84
++SS SN K+ +M ++ L + SA I S+NSN S+ ++ S
Sbjct: 2 ASSSSISNHKIPNTLMFLVIVNFLYLIQTNSAVSI--------SSNSNSHFSRFSRHRSS 53
Query: 85 TCSKTRYPSLCINSLLDFPGSTSASEQELVHISFNMTHRHISKALFASSGLSYTVANPRV 144
SKT+ L A+ QE +M H ++++L + LS+
Sbjct: 54 PSSKTKQGFL-------------ATVQE------SMNHALLARSLAFNLTLSHRTVQTHT 94
Query: 145 RAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAAL 204
DCLEL+D+++D + S + D+ E EDV TWLSAAL
Sbjct: 95 FDPIHDCLELLDDTLDML-----------SRIHADNDE-----------EDVHTWLSAAL 132
Query: 205 TNQDTCLEGFEDTSGTVKDQMVGNL--KDLSELVSNSLAIFSASGDNDFTGVPIQNKRRL 262
TNQDTC + ++ S + K + + ++L+ L+++SL +F V +++K R
Sbjct: 133 TNQDTCEQSLQEKSESYKHGLAMDFVARNLTGLLTSSLDLF----------VSVKSKHRK 182
Query: 263 MGMSDISRE--FPKWLEKRD-RRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAP-E 318
+ +S++ FP ++ + RRLL PV E+ D +V+ G +GT KTI EA+
Sbjct: 183 L----LSKQEYFPTFVPSSEQRRLLEAPVEELNVDAVVAPDG-SGTHKTIGEALLSTSLA 237
Query: 319 HSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFA 378
S R IY++AG Y EN + + K+ N+M +GDG+GKTVI G RS G TT+ TA+ A
Sbjct: 238 SSGGRTKIYLKAGTYHEN-INIPTKQKNVMLVGDGKGKTVIVGSRSNRGGWTTYKTATVA 296
Query: 379 ASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFREC 438
A G GF+ARD+TF N AGP+ QAVALRVG+D +VV+RC++ GYQD+ Y HS RQF+RE
Sbjct: 297 AMGEGFIARDMTFVNNAGPKSEQAVALRVGADKSVVHRCSVEGYQDSLYTHSKRQFYRET 356
Query: 439 NIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAP 498
+I GTVDFIFGN+AVVFQ CNI ARKP+ Q+N +TAQ R +P QNTGI+I +CRI
Sbjct: 357 DITGTVDFIFGNSAVVFQSCNIAARKPLPGQRNFVTAQGRSNPGQNTGIAIQNCRI---- 412
Query: 499 DLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFG 558
TYLGRPWK YSRTV M S++G +HP GW W+G F LK+L+YGEY N G
Sbjct: 413 -----TAESMTYLGRPWKEYSRTVVMQSFIGGSIHPSGWSPWSGGFGLKSLFYGEYGNSG 467
Query: 559 PGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
PG+++ RVKW G + EA ++TVA FI G+ WLPSTGV+F GL
Sbjct: 468 PGSSVSGRVKWSGCHPSLTVTEAEKFTVASFIDGNIWLPSTGVSFDPGL 516
>At4g02320 pectinesterase - like protein
Length = 518
Score = 400 bits (1028), Expect = e-111
Identities = 222/505 (43%), Positives = 310/505 (60%), Gaps = 34/505 (6%)
Query: 112 ELVHISFNMTHRHISKALFASSGLSYTVA------NPRVRAAYEDCLELMDESMDAIRSS 165
ELV + N T IS +SS S + + R A++DCLEL+D+++ + ++
Sbjct: 39 ELVVTALNQT---ISNVNLSSSNFSDLLQRLGSNLSHRDLCAFDDCLELLDDTVFDLTTA 95
Query: 166 MDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQDTCLEGFEDTSGTVKDQM 225
+ L + S L N V LSAA+TN TCL+GF + D+
Sbjct: 96 ISKLRSHSPELHN-----------------VKMLLSAAMTNTRTCLDGFASSDN---DEN 135
Query: 226 VGNLKDLSELVSNSL--AIFSASGDNDFTGVPIQN-KRRLMGMSDISREFPKWLEKRDRR 282
+ N + + V+ SL ++F+ S + ++N + G FP W+ DR
Sbjct: 136 LNNNDNKTYGVAESLKESLFNISSHVSDSLAMLENIPGHIPGKVKEDVGFPMWVSGSDRN 195
Query: 283 LLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGK 342
LL PV E + +++V+++G G TI EAI AP S RF+IY++ G Y EN +++ +
Sbjct: 196 LLQDPVDETKVNLVVAQNG-TGNYTTIGEAISAAPNSSETRFVIYIKCGEYFEN-IEIPR 253
Query: 343 KKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQA 402
+KT IMFIGDG G+TVI RS DG T FH+A+ G GF+A+D++F NYAGPEKHQA
Sbjct: 254 EKTMIMFIGDGIGRTVIKANRSYADGWTAFHSATVGVRGSGFIAKDLSFVNYAGPEKHQA 313
Query: 403 VALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYA 462
VALR SD + YRC+ YQD YVHS++QF+REC+IYGTVDFIFG+A+VVFQ C++YA
Sbjct: 314 VALRSSSDLSAYYRCSFESYQDTIYVHSHKQFYRECDIYGTVDFIFGDASVVFQNCSLYA 373
Query: 463 RKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTV 522
R+P QK TAQ R++ + TGISI RIL APDL + + + YLGRPW++YSRTV
Sbjct: 374 RRPNPNQKIIYTAQGRENSREPTGISIISSRILAAPDLIPVQANFKAYLGRPWQLYSRTV 433
Query: 523 YMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEAN 582
M S++ D V P GWL+W DFAL+TLYYGEYMN GPG+ + RV+WPG++ I + EA+
Sbjct: 434 IMKSFIDDLVDPAGWLKWKDDFALETLYYGEYMNEGPGSNMTNRVQWPGFKRIETVEEAS 493
Query: 583 RYTVAQFISGSSWLPSTGVAFLAGL 607
+++V FI G+ WL ST + F L
Sbjct: 494 QFSVGPFIDGNKWLNSTRIPFTLDL 518
>At1g23200 putative pectinesterase
Length = 554
Score = 397 bits (1021), Expect = e-111
Identities = 235/549 (42%), Positives = 317/549 (56%), Gaps = 57/549 (10%)
Query: 85 TCSKTRYPSLCINSLLDFPGSTSASEQELVHISFNMTHRHISKAL----FASSGLSYTVA 140
+C +T YPS+C + + + P T + + + + +A+ SS +
Sbjct: 37 SCKQTPYPSVCDHHMSNSPLKTLDDQTDGFTFHDLVVSSTMDQAVQLHRLVSSLKQHHSL 96
Query: 141 NPRVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWL 200
+ +A DCLEL ++++D + S S Q+S S D T L
Sbjct: 97 HKHATSALFDCLELYEDTIDQLNHSRRSY--------------GQYS----SPHDRQTSL 138
Query: 201 SAALTNQDTCLEGFED---TSGTVKDQMVGNLKDLSELVSNSLAIFSASGDND------- 250
SAA+ NQDTC GF D TS K V ++L++ +SNSLA+ A+ + +
Sbjct: 139 SAAIANQDTCRNGFRDFKLTSSYSKYFPVQFHRNLTKSISNSLAVTKAAAEAEAVAEKYP 198
Query: 251 ---FTGVPIQNK-------RRLMGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVSKS 300
FT Q RRL+ SD +FP W DR+LL + +AD++V+K
Sbjct: 199 STGFTKFSKQRSSAGGGSHRRLLLFSD--EKFPSWFPLSDRKLLEDSKTTAKADLVVAKD 256
Query: 301 GGNGTVKTITEAIKKAPEHSRR--RFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTV 358
G +G +I +A+ A + RR R +IYV+AG Y EN + + K N+M IGDG T+
Sbjct: 257 G-SGHYTSIQQAVNAAAKLPRRNQRLVIYVKAGVYREN-VVIKKSIKNVMVIGDGIDSTI 314
Query: 359 ITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCN 418
+TG R+V DG TTF +A+FA SG GF+A+ ITFEN AGPEKHQAVALR SD +V Y C+
Sbjct: 315 VTGNRNVQDGTTTFRSATFAVSGNGFIAQGITFENTAGPEKHQAVALRSSSDFSVFYACS 374
Query: 419 IVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNR 478
GYQD Y+HS+RQF R CNIYGTVDFIFG+A + Q CNIYARKPM+ QKNTITAQ+R
Sbjct: 375 FKGYQDTLYLHSSRQFLRNCNIYGTVDFIFGDATAILQNCNIYARKPMSGQKNTITAQSR 434
Query: 479 KDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWL 538
K+P++ TG I + A ETYLGRPW+ +SRTV+M +G V P GWL
Sbjct: 435 KEPDETTGFVIQSSTVATAS---------ETYLGRPWRSHSRTVFMKCNLGALVSPAGWL 485
Query: 539 EWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPS 598
W+G FAL TLYYGEY N G GA++ RVKWPGY VI + EA ++TV F+ G+ W+ +
Sbjct: 486 PWSGSFALSTLYYGEYGNTGAGASVSGRVKWPGYHVIKTVTEAEKFTVENFLDGNYWITA 545
Query: 599 TGVAFLAGL 607
TGV GL
Sbjct: 546 TGVPVNDGL 554
>At3g10710 putative pectinesterase
Length = 561
Score = 393 bits (1010), Expect = e-109
Identities = 236/580 (40%), Positives = 328/580 (55%), Gaps = 43/580 (7%)
Query: 33 KSNKKLIMLSILAAVLIIASAISAALITVVRSRA-----SSNNSNLLHSKPTQAISRTCS 87
+ +K I + ++ V++ I A T+ ++ ++NN + + + ++ C
Sbjct: 20 RKTRKNIAIIAVSLVILAGIVIGAVFGTMAHKKSPETVETNNNGDSI----SVSVKAVCD 75
Query: 88 KTRYPSLCINSLLDFPGSTSASEQELVHISFNMTHRHISKALFASSGLSYTVANPRVRAA 147
T + C +L P ++S + +EL + +T +SKA+ A S ++ + +
Sbjct: 76 VTLHKEKCFETLGSAPNASSLNPEELFRYAVKITIAEVSKAINAFSS---SLGDEKNNIT 132
Query: 148 YEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQ 207
C EL+D ++D+L T ++ SN D V +D+ TWLS+A T Q
Sbjct: 133 MNACAELLD-------LTIDNLNNTLTSSSNGD------VTVPELVDDLRTWLSSAGTYQ 179
Query: 208 DTCLEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVPIQNKRRLMGMSD 267
TC+E + +LK+ +EL SN+LAI + G + +RRL+ +D
Sbjct: 180 RTCVETLAPDMRPFGES---HLKNSTELTSNALAIITWLGK---IADSFKLRRRLLTTAD 233
Query: 268 ISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIY 327
+ +F RRLL ADI+V+K G +G +TI A++ PE S +R IIY
Sbjct: 234 VEVDF-----HAGRRLLQSTDLRKVADIVVAKDG-SGKYRTIKRALQDVPEKSEKRTIIY 287
Query: 328 VRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMAR 387
V+ G Y EN +KV KK N++ +GDG K++++G+ +V DG TF TA+FA G GFMAR
Sbjct: 288 VKKGVYFEN-VKVEKKMWNVIVVGDGESKSIVSGRLNVIDGTPTFKTATFAVFGKGFMAR 346
Query: 388 DITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFI 447
D+ F N AGP KHQAVAL V +D YRC + YQD YVH+ RQF+REC I GTVDFI
Sbjct: 347 DMGFINTAGPSKHQAVALMVSADLTAFYRCTMNAYQDTLYVHAQRQFYRECTIIGTVDFI 406
Query: 448 FGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSI 507
FGN+A V Q C I R+PM Q+NTITAQ R DPN NTGISIH C I P DL +
Sbjct: 407 FGNSASVLQSCRILPRRPMKGQQNTITAQGRTDPNMNTGISIHRCNISPLGDLT----DV 462
Query: 508 ETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRV 567
T+LGRPWK +S TV M SY+ + GWL W GD A T++YGEY N GPGA+ RV
Sbjct: 463 MTFLGRPWKNFSTTVIMDSYLHGFIDRKGWLPWTGDSAPDTIFYGEYKNTGPGASTKNRV 522
Query: 568 KWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
KW G R + ST EANR+TV FI G WLP+T V F +GL
Sbjct: 523 KWKGLRFL-STKEANRFTVKPFIDGGRWLPATKVPFRSGL 561
>At2g47550 unknown protein
Length = 560
Score = 389 bits (1000), Expect = e-108
Identities = 245/593 (41%), Positives = 327/593 (54%), Gaps = 56/593 (9%)
Query: 35 NKKLIMLSILAAVLIIASAISAALITVVRSRASSNNSNLLHSKPTQAISRTCSKTRYPSL 94
++KL+ L LA + + S +A I + + S +N C PS
Sbjct: 2 SQKLMFLFTLACLSSLPSPFISAQIPAIGNATSPSN--------------ICRFAPDPSY 47
Query: 95 CINSLLDFPGSTSASEQELVHISFNMTHRHISKALFASSGLSYTVANPRVRAAYEDCLEL 154
C + L + PG + + + S + R IS + A VA A EDC L
Sbjct: 48 CRSVLPNQPGDIYSYGRLSLRRSLSRARRFISM-IDAELDRKGKVAAKSTVGALEDCKFL 106
Query: 155 MDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSAALTNQDTCLEGF 214
+MD + SS + +T TLS EDV T+LSAA+TN+ TCLEG
Sbjct: 107 ASLTMDYLLSSSQTADSTK-TLS------------LSRAEDVHTFLSAAITNEQTCLEGL 153
Query: 215 EDTSGTVKDQMVGNLKDLSELVSNSLAIFS----------------ASGDNDFTGVPIQN 258
+ T+ ++ + G+L + ++L SLA+FS + F G +N
Sbjct: 154 KSTAS--ENGLSGDLFNDTKLYGVSLALFSKGWVPRRQRSRPIWQPQARFKKFFG--FRN 209
Query: 259 KRRLMGMSDISREFPKWLEKRDRRLLSLPVSEIQ-ADIIVSKSGGNGTVKTITEAIKKAP 317
+ + M++ +R + + R+LL +Q +DI+ G G TI AI AP
Sbjct: 210 GKLPLKMTERARAVYNTVTR--RKLLQSDADAVQVSDIVTVIQNGTGNFTTINAAIAAAP 267
Query: 318 ---EHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHT 374
+ S F+IYV AG YEE ++V K K +M IGDG +TVITG RSV DG TTF++
Sbjct: 268 NKTDGSNGYFLIYVTAGLYEE-YVEVPKNKRYVMMIGDGINQTVITGNRSVVDGWTTFNS 326
Query: 375 ASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQF 434
A+F SGP F+ +IT N AGP K QAVALR G D +V Y C+ YQD Y HS RQF
Sbjct: 327 ATFILSGPNFIGVNITIRNTAGPTKGQAVALRSGGDLSVFYSCSFEAYQDTLYTHSLRQF 386
Query: 435 FRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRI 494
+REC++YGTVDFIFGNAAVV Q CN+Y R+P Q N +TAQ R DPNQNTG +IH C I
Sbjct: 387 YRECDVYGTVDFIFGNAAVVLQNCNLYPRQPRKGQSNEVTAQGRTDPNQNTGTAIHGCTI 446
Query: 495 LPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEY 554
PA DLA+S +++TYLGRPWK YSRTV M +Y+ + P GW W+GDFAL TLYY EY
Sbjct: 447 RPADDLATSNYTVKTYLGRPWKEYSRTVVMQTYIDGFLEPSGWNAWSGDFALSTLYYAEY 506
Query: 555 MNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
N GPG+ RV WPGY VI +T +A+ +TV F+ G W+ TGV F+ GL
Sbjct: 507 NNTGPGSDTTNRVTWPGYHVINAT-DASNFTVTNFLVGEGWIGQTGVPFVGGL 558
>At4g33220 pectinesterase like protein
Length = 404
Score = 387 bits (994), Expect = e-107
Identities = 202/406 (49%), Positives = 257/406 (62%), Gaps = 13/406 (3%)
Query: 211 LEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVP--------IQNKRRL 262
+EGF+ TSG VK + G+L L ++ L + P R+L
Sbjct: 1 MEGFDGTSGLVKSLVAGSLDQLYSMLRELLPLVQPEQKPKAVSKPGPIAKGPKAPPGRKL 60
Query: 263 MGMS-DISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSR 321
D S +FP W+ DR+LL + + G G I +AIKKAP++S
Sbjct: 61 RDTDEDESLQFPDWVRPDDRKLLESNGRTYDVSVALD---GTGNFTKIMDAIKKAPDYSS 117
Query: 322 RRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASG 381
RF+IY++ G Y EN +++ KKK NI+ +GDG TVI+G RS DG TTF +A+FA SG
Sbjct: 118 TRFVIYIKKGLYLEN-VEIKKKKWNIVMLGDGIDVTVISGNRSFIDGWTTFRSATFAVSG 176
Query: 382 PGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIY 441
GF+ARDITF+N AGPEKHQAVALR SD +V +RC + GYQD Y H+ RQF+REC I
Sbjct: 177 RGFLARDITFQNTAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYTHTMRQFYRECTIT 236
Query: 442 GTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLA 501
GTVDFIFG+ VVFQ C I A++ + QKNTITAQ RKD NQ +G SI I DL
Sbjct: 237 GTVDFIFGDGTVVFQNCQILAKRGLPNQKNTITAQGRKDVNQPSGFSIQFSNISADADLV 296
Query: 502 SSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGA 561
+ TYLGRPWK+YSRTV++ + M D V P GWLEWN DFAL TL+YGE+MN+GPG+
Sbjct: 297 PYLNTTRTYLGRPWKLYSRTVFIRNNMSDVVRPEGWLEWNADFALDTLFYGEFMNYGPGS 356
Query: 562 AIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
+ RVKWPGY V ++ +AN +TV+QFI G+ WLPSTGV F GL
Sbjct: 357 GLSSRVKWPGYHVFNNSDQANNFTVSQFIKGNLWLPSTGVTFSDGL 402
>At3g10720 pectinesterase like protein
Length = 619
Score = 384 bits (987), Expect = e-107
Identities = 235/556 (42%), Positives = 307/556 (54%), Gaps = 43/556 (7%)
Query: 79 TQAISRTCSKTRYPSLCINSLLDFPGSTSASEQELVHISFNMTHRHISKALFASSGLSYT 138
+Q+ S C T YP LC ++L+ S+ + + + S+ + +
Sbjct: 76 SQSPSLACKSTPYPKLC-RTILNAVKSSPSDPYRYGKFTIKQCLKQASRLSKVITSYARR 134
Query: 139 VANPRVRAAYE------DCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGS 192
V + A E DC EL + S++ + + L T A
Sbjct: 135 VESKPGSATAEEIGAVADCGELSELSVNYLETVTTELKTAQVM-------------TAAL 181
Query: 193 TEDVMTWLSAALTNQDTCLEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFT 252
E V + LS +TNQ TCL+G + + + +L+ L S SL + S + + +
Sbjct: 182 VEHVNSLLSGVVTNQQTCLDGLVEAKSGFAAAIGSPMGNLTRLYSISLGLVSHALNRNLK 241
Query: 253 GVPIQNKRRLMGMSDISREFPKWLEK---------RDRRLLSLPVSEIQ----ADIIVSK 299
+ L G + RE + L K +D R S + E+ I+VSK
Sbjct: 242 RFKASKGKILGGGNSTYREPLETLIKGLRKTCDNDKDCRKTSRNLGELGETSGGSILVSK 301
Query: 300 SGGNGTVK-----TITEAIKKAPEHSRRR---FIIYVRAGRYEENNLKVGKKKTNIMFIG 351
+ G K TIT+AI AP ++R F+IY R G YEE + V K N+M +G
Sbjct: 302 AVIVGPFKSDNFTTITDAIAAAPNNTRPEDGYFVIYAREGVYEEY-IVVPINKKNLMLMG 360
Query: 352 DGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDH 411
DG KT+ITG +V DG TT++ +SFA G FMA D+TF N AGPEKHQAVALR ++
Sbjct: 361 DGINKTIITGNHNVVDGWTTYNCSSFAVVGERFMAVDVTFRNTAGPEKHQAVALRNNAEG 420
Query: 412 AVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKN 471
+ YRC+ GYQD YVHS RQF+REC+IYGTVDFIFGNAA +FQ CNIYARKPMA+QKN
Sbjct: 421 SSFYRCSFEGYQDTLYVHSLRQFYRECDIYGTVDFIFGNAAAIFQNCNIYARKPMAKQKN 480
Query: 472 TITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDH 531
ITA R DPNQNTGISI +C I APDLA+ S T+LGRPWK YSRTV+M SY+ D
Sbjct: 481 AITAHGRLDPNQNTGISIINCTIKAAPDLAAEPKSAMTFLGRPWKPYSRTVFMQSYISDI 540
Query: 532 VHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFIS 591
V P GWLEWNG L T+YYGEY NFGPGA QRV+W GY ++ + EA +TV F
Sbjct: 541 VQPVGWLEWNGTIGLDTIYYGEYSNFGPGANTNQRVQWLGYNLL-NLAEAMNFTVYNFTM 599
Query: 592 GSSWLPSTGVAFLAGL 607
G +WLP T + F GL
Sbjct: 600 GDTWLPQTDIPFYGGL 615
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,395,879
Number of Sequences: 26719
Number of extensions: 564892
Number of successful extensions: 2042
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 92
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 1629
Number of HSP's gapped (non-prelim): 148
length of query: 609
length of database: 11,318,596
effective HSP length: 105
effective length of query: 504
effective length of database: 8,513,101
effective search space: 4290602904
effective search space used: 4290602904
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC134822.9


