

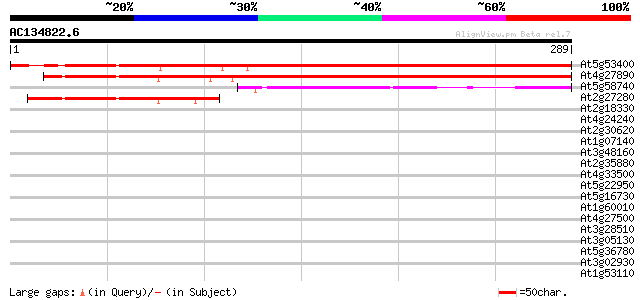
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.6 + phase: 0
(289 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g53400 unknown protein 328 2e-90
At4g27890 putative protein 311 3e-85
At5g58740 unknown protein 69 2e-12
At2g27280 unknown protein 62 3e-10
At2g18330 AAA-type like ATPase 39 0.003
At4g24240 putative DNA-binding protein 36 0.028
At2g30620 putative histone H1 protein 35 0.047
At1g07140 Unknown protein (F10K1.15) 35 0.047
At3g48160 putative protein 34 0.080
At2g35880 unknown protein 34 0.080
At4g33500 unknown protein 34 0.10
At5g22950 unknown protein (At5g22950) 33 0.18
At5g16730 putative protein 33 0.18
At1g60010 unknown protein 33 0.18
At4g27500 unknown protein 32 0.31
At3g28510 hypothetical protein 32 0.31
At3g05130 hypothetical protein 32 0.31
At5g36780 putative protein 32 0.40
At3g02930 unknown protein 32 0.40
At1g53110 proton pump interactor, putative 32 0.52
>At5g53400 unknown protein
Length = 304
Score = 328 bits (841), Expect = 2e-90
Identities = 177/314 (56%), Positives = 217/314 (68%), Gaps = 36/314 (11%)
Query: 1 MAIISDYQDDNQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFD 60
MAIIS+ +++ SSSS+P PF +T +NP FLEKVFDF+ ++S +F
Sbjct: 1 MAIISEVEEE-------SSSSRPMI---FPFRATLSSANPLGFLEKVFDFLGEQS-DFLK 49
Query: 61 KDSAEKVVLSAVRAAR--VKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEK--SEAVTEKI 116
K SAE ++ AVRAA+ +KKA+ AEK + EK KL ++K E+V I
Sbjct: 50 KPSAEDEIVVAVRAAKEKLKKAEKKKAEKESVKPVEKKAEKEIVKLVEKKVEKESVKPTI 109
Query: 117 DEKAA---------------------PNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSG 155
+A PN+GNG DLE YSW Q LQE+ VN+PVP GTK+
Sbjct: 110 AASSAEPIEVEKPKEEEEKKESGPIVPNKGNGTDLENYSWIQNLQEVTVNIPVPTGTKAR 169
Query: 156 FVICEIKKNHLKVGLKGQPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKC 215
V+CEIKKN LKVGLKGQ PI+D ELY+S+KPD+CYW+IEDQ +SILLTK DQM+WWKC
Sbjct: 170 TVVCEIKKNRLKVGLKGQDPIVDGELYRSVKPDDCYWNIEDQKVISILLTKSDQMEWWKC 229
Query: 216 LVKGDPEINTQKVEPASSKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKF 275
VKG+PEI+TQKVEP +SKLGDLD ETR TVEKMMFDQRQK MGLPTSEELQK+E++KKF
Sbjct: 230 CVKGEPEIDTQKVEPETSKLGDLDPETRSTVEKMMFDQRQKQMGLPTSEELQKQEILKKF 289
Query: 276 MSQHPNMDFSGAKF 289
MS+HP MDFS AKF
Sbjct: 290 MSEHPEMDFSNAKF 303
>At4g27890 putative protein
Length = 293
Score = 311 bits (797), Expect = 3e-85
Identities = 158/290 (54%), Positives = 205/290 (70%), Gaps = 20/290 (6%)
Query: 18 SSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSAVRAA-- 75
S + +PS +PF+++FDPSNP FLEKV D I KES NF KD+AEK +++AV AA
Sbjct: 5 SEMEEARPSM-VPFTASFDPSNPIAFLEKVLDVIGKES-NFLKKDTAEKEIVAAVMAAKQ 62
Query: 76 RVKKAKSVAAEKAKIAAKEKAKAAAEK----KLNDEKSEAVT------------EKIDEK 119
R+++A+ EK + + E K + +L K E++ EK
Sbjct: 63 RLREAEKKKLEKESVKSMEVEKPKKDSLKPTELEKPKEESLMATDPMEIEKPKEEKESGP 122
Query: 120 AAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQPPIIDR 179
PN+GNG+D EKYSW Q LQE+ +N+P+P GTKS V CEIKKN LKVGLKGQ I+D
Sbjct: 123 IVPNKGNGLDFEKYSWGQNLQEVTINIPMPEGTKSRSVTCEIKKNRLKVGLKGQDLIVDG 182
Query: 180 ELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPASSKLGDLD 239
E + S+KPD+C+W+IEDQ +S+LLTK DQM+WWK VKG+PEI+TQKVEP +SKLGDLD
Sbjct: 183 EFFNSVKPDDCFWNIEDQKMISVLLTKQDQMEWWKYCVKGEPEIDTQKVEPETSKLGDLD 242
Query: 240 SETRMTVEKMMFDQRQKSMGLPTSEELQKEEMMKKFMSQHPNMDFSGAKF 289
ETR +VEKMMFDQRQK MGLP S+E++K++M+KKFM+Q+P MDFS AKF
Sbjct: 243 PETRASVEKMMFDQRQKQMGLPRSDEIEKKDMLKKFMAQNPGMDFSNAKF 292
>At5g58740 unknown protein
Length = 158
Score = 69.3 bits (168), Expect = 2e-12
Identities = 48/178 (26%), Positives = 81/178 (44%), Gaps = 45/178 (25%)
Query: 118 EKAAPNQG-----NGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKG 172
EK AP + NG + + W QTL+E+N+ + +P C+I+ H++VG+KG
Sbjct: 3 EKLAPEKRHDFIHNGQKV--FEWDQTLEEVNMYITLPPNVHPKSFHCKIQSKHIEVGIKG 60
Query: 173 QPPIIDRELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPEINTQKVEPAS 232
PP ++ +L +K D +W++ED + + I L K ++ W + G
Sbjct: 61 NPPYLNHDLSAPVKTDCSFWTLED-DIMHITLQKREKGQTWASPILGQ------------ 107
Query: 233 SKLGDLDSETRMTVEKMMFDQRQKSMGLPTSEEL-QKEEMMKKFMSQHPNMDFSGAKF 289
G LD P + +L QK M+++F ++P DFS A+F
Sbjct: 108 ---GQLD---------------------PYATDLEQKRLMLQRFQEENPGFDFSQAQF 141
>At2g27280 unknown protein
Length = 764
Score = 62.4 bits (150), Expect = 3e-10
Identities = 36/104 (34%), Positives = 65/104 (61%), Gaps = 7/104 (6%)
Query: 10 DNQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVL 69
+ + KT S + +PS +PF+++FDPS+P FLEKVF+F+ K+S NF KD A ++
Sbjct: 397 ETEKKTESMDVEKVRPST-LPFNASFDPSDPLGFLEKVFEFVGKKS-NFLVKDKAVNAII 454
Query: 70 SAVRAA--RVKKAKSVAAEKAKIAAKE---KAKAAAEKKLNDEK 108
+AV A R+K+ + + ++A + K+ + +A ++KK + +
Sbjct: 455 TAVTDAKERLKEEEKESVKQATVKIKKYGLQIRAPSQKKQSSSR 498
Score = 40.4 bits (93), Expect = 0.001
Identities = 25/53 (47%), Positives = 31/53 (58%), Gaps = 7/53 (13%)
Query: 43 FLEKVFDFIAKESTNFFDKDSAEKVVLSAVRAAR------VKKAKSVAAEKAK 89
FLE V DFI KES NF KD+AEK + AV A+ KK +S+ EK +
Sbjct: 360 FLENVLDFIGKES-NFLRKDTAEKEITDAVTTAKERLRETEKKTESMDVEKVR 411
>At2g18330 AAA-type like ATPase
Length = 636
Score = 38.9 bits (89), Expect = 0.003
Identities = 29/119 (24%), Positives = 56/119 (46%), Gaps = 12/119 (10%)
Query: 14 KTSSSSSSQPKPSKPIPFSSTFDPS------------NPTTFLEKVFDFIAKESTNFFDK 61
K+SS+S S+ KP P S FDP N + ++VFD + K+ +
Sbjct: 51 KSSSNSKSETKPDSDEPKGSGFDPESLERGAKALREINSSPHSKQVFDLMRKQEKTRLAE 110
Query: 62 DSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
+AEK A++A++ + + AE + +++A+A A+ +++ + D +A
Sbjct: 111 LAAEKEHNEAIQASKDIERQRKLAEDQRNLVQQQAQAKAQNLRYEDELARKRMQTDNEA 169
>At4g24240 putative DNA-binding protein
Length = 353
Score = 35.8 bits (81), Expect = 0.028
Identities = 33/143 (23%), Positives = 56/143 (39%), Gaps = 4/143 (2%)
Query: 9 DDNQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVV 68
+ NQTK SSSSS P + P + F PS P + ++ + E
Sbjct: 164 NQNQTKNGSSSSSPPMLANGAPSTINFAPSPPVSATNSFMSSHRCDTDSTHMSSGFEFTN 223
Query: 69 LSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGM 128
S + +R K S A+ K + + ++ KK + V I A ++ +
Sbjct: 224 PSQLSGSRGKPPLSSASLKRRCNSSPSSRCHCSKK----RKSRVKRVIRVPAVSSKMADI 279
Query: 129 DLEKYSWTQTLQELNVNVPVPNG 151
+++SW + Q+ P P G
Sbjct: 280 PSDEFSWRKYGQKPIKGSPHPRG 302
>At2g30620 putative histone H1 protein
Length = 273
Score = 35.0 bits (79), Expect = 0.047
Identities = 36/135 (26%), Positives = 57/135 (41%), Gaps = 3/135 (2%)
Query: 16 SSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSAVRAA 75
S+ S++ PKP+ P+ +T V AK + K +A KVV A A
Sbjct: 130 SARSAATPKPAAPVKKKATVVAKPKGKVAAAVAPAKAKAAAKGTKKPAA-KVVAKAKVTA 188
Query: 76 RVKKAKSVAAEKAK-IAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPNQGNGMDLEKYS 134
+ K + A K+K +AA K KA A K E+ A + + +P + +K +
Sbjct: 189 KPKAKVTAAKPKSKSVAAVSKTKAVAAKPKAKER-PAKASRTSTRTSPGKKVAAPAKKVA 247
Query: 135 WTQTLQELNVNVPVP 149
T+ +V V P
Sbjct: 248 VTKKAPAKSVKVKSP 262
>At1g07140 Unknown protein (F10K1.15)
Length = 228
Score = 35.0 bits (79), Expect = 0.047
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 6/78 (7%)
Query: 39 NPTTFLEKVFDFIAKESTNFFDKDSA------EKVVLSAVRAARVKKAKSVAAEKAKIAA 92
N TF++K + E KD+A EK+ + + +AK+V K ++ A
Sbjct: 145 NCKTFMQKFKEVAESEEEKEESKDAADTAGLLEKLTVEETKTEEKTEAKAVETAKTEVKA 204
Query: 93 KEKAKAAAEKKLNDEKSE 110
+EK ++ AEK +K+E
Sbjct: 205 EEKKESEAEKSGEAKKTE 222
>At3g48160 putative protein
Length = 323
Score = 34.3 bits (77), Expect = 0.080
Identities = 24/86 (27%), Positives = 34/86 (38%), Gaps = 9/86 (10%)
Query: 5 SDYQDDNQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFD---------FIAKES 55
SD +DD++ + SSSQ SKP + DPS EK FI E+
Sbjct: 75 SDDEDDDEESSQPHSSSQTDSSKPGSLPQSSDPSKIDNRREKSLGLLTQNFIKLFICSEA 134
Query: 56 TNFFDKDSAEKVVLSAVRAARVKKAK 81
D A K++L + + K
Sbjct: 135 IRIISLDDAAKLLLGDAHNTSIMRTK 160
>At2g35880 unknown protein
Length = 432
Score = 34.3 bits (77), Expect = 0.080
Identities = 22/66 (33%), Positives = 33/66 (49%), Gaps = 1/66 (1%)
Query: 61 KDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKA 120
KDS+ V + ++ K + KAK K+ K AEK+ +EK+ AV K +E+
Sbjct: 352 KDSSSSTVKKPITKSQPKLETQEKSVKAKEKKKKVKKEEAEKRGEEEKATAVAAKPEEQ- 410
Query: 121 APNQGN 126
PN N
Sbjct: 411 KPNSNN 416
>At4g33500 unknown protein
Length = 724
Score = 33.9 bits (76), Expect = 0.10
Identities = 35/111 (31%), Positives = 51/111 (45%), Gaps = 15/111 (13%)
Query: 13 TKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSAV 72
+K+S+SSSS P+ S P FD + T + F F D EK + +
Sbjct: 53 SKSSASSSSPPENSAP----EKFDLVSSTQLKDGSHVF------RFGDASEIEKYLEAEE 102
Query: 73 RAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEKAAPN 123
+A V+ V + AKIA + + +KKL E TEK +E AAP+
Sbjct: 103 KARCVE----VETQNAKIAEEASEVSRKQKKLVSSIIETSTEK-EETAAPS 148
>At5g22950 unknown protein (At5g22950)
Length = 229
Score = 33.1 bits (74), Expect = 0.18
Identities = 19/71 (26%), Positives = 31/71 (42%)
Query: 49 DFIAKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEK 108
+F+ + N D + E+ + V A AAE KE+ K A+K +
Sbjct: 144 EFVNEAIDNALDSEDMEEEIDEEVDKVLTAIAGETAAELPVAVRKERIKVPAQKASTSRE 203
Query: 109 SEAVTEKIDEK 119
EAV E +D++
Sbjct: 204 EEAVAEGVDDE 214
>At5g16730 putative protein
Length = 853
Score = 33.1 bits (74), Expect = 0.18
Identities = 20/66 (30%), Positives = 36/66 (54%), Gaps = 2/66 (3%)
Query: 70 SAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDE--KAAPNQGNG 127
+ R +++K+ A E+ K+KAKA E K +++E VT K+D+ KA +
Sbjct: 96 TTTRLSQIKEDLKKANERISSLEKDKAKALDELKQAKKEAEQVTLKLDDALKAQKHVEEN 155
Query: 128 MDLEKY 133
++EK+
Sbjct: 156 SEIEKF 161
>At1g60010 unknown protein
Length = 173
Score = 33.1 bits (74), Expect = 0.18
Identities = 22/77 (28%), Positives = 37/77 (47%), Gaps = 5/77 (6%)
Query: 68 VLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVT-EKIDEKAAPNQGN 126
V+ +RA + K K +E +KEK K ++EKK+++E + E DEK N
Sbjct: 97 VMKVLRAKKYAKTKKHQSE----TSKEKKKPSSEKKIDEESDKNQNLETKDEKQRSVLTN 152
Query: 127 GMDLEKYSWTQTLQELN 143
+W +LQ ++
Sbjct: 153 SASSRSKTWRPSLQSIS 169
>At4g27500 unknown protein
Length = 612
Score = 32.3 bits (72), Expect = 0.31
Identities = 48/208 (23%), Positives = 76/208 (36%), Gaps = 20/208 (9%)
Query: 11 NQTKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSA------ 64
++T+ + ++P+P K P S+ + EK D + ++ D D
Sbjct: 401 SETEVVPKAKAKPQP-KEEPVSAPKPDATVAQNTEKAKDAVKVKNVADDDDDEVYGLGKP 459
Query: 65 ---EKVVLSAV-----RAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKI 116
EK V +A + + KAK K K+A K AKAA + EK E +K
Sbjct: 460 QKEEKPVDAATAKEMRKQEEIAKAKQAMERKKKLAEKAAAKAAIRAQKEAEKKE---KKE 516
Query: 117 DEKAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQPPI 176
EK A + G + E + PV + + K + +G P
Sbjct: 517 QEKKAKKKTGGNTETETEEVPEASEEEIEAPVQEEKPQKEKVFKEKPIRNRTRGRG-PET 575
Query: 177 IDRELYKSIKPDECYWSIEDQNTVSILL 204
I R + K K YW + +LL
Sbjct: 576 IPRAILKR-KKSTNYWVYAAPAALVVLL 602
>At3g28510 hypothetical protein
Length = 530
Score = 32.3 bits (72), Expect = 0.31
Identities = 29/84 (34%), Positives = 37/84 (43%), Gaps = 9/84 (10%)
Query: 60 DKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKE-----KAKAAAEKKLN---DEKSEA 111
D D K ++ + + K K E+ K A KE KA+ A EKK DEK E
Sbjct: 445 DADICIKRLVKTLEEEKEKARKLAEEEEKKKAEKEAKKMKKAEEAEEKKKKTEEDEKKEK 504
Query: 112 VTEKIDEKAAPNQ-GNGMDLEKYS 134
V K + Q GN +DL K S
Sbjct: 505 VKAKEENGNVSQQNGNSIDLNKKS 528
>At3g05130 hypothetical protein
Length = 634
Score = 32.3 bits (72), Expect = 0.31
Identities = 41/203 (20%), Positives = 91/203 (44%), Gaps = 10/203 (4%)
Query: 60 DKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDEK 119
+K+S ++V+ + + + K + +I +K K EK + ++K+E K + K
Sbjct: 201 EKESNLEIVIGKLESENERLVKERKVREEEIEGVKKEKIGLEKIMEEKKNEIDGLKREIK 260
Query: 120 AAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGL-KGQPPIID 178
++ N M++ K ++EL + N T E L +GL K ++
Sbjct: 261 VLLSEKNEMEIVKIEQKGVIEELERKLDKLNETVRSLTKEEKVLRDLVIGLEKNLDESME 320
Query: 179 RELYKSIKPDECYWSIEDQNTVSILLTKHDQMDWWKCLVKGDPE-INTQKVEPASSKLGD 237
+E ++ D ++ + T+ ++ +++ K L++ E +N Q + KL D
Sbjct: 321 KESGMMVEID----ALGKERTIK--ESEVERLIGEKNLIEKQMEMLNVQSSD--KGKLID 372
Query: 238 LDSETRMTVEKMMFDQRQKSMGL 260
S ++ +E+ +F + +K + L
Sbjct: 373 QLSREKVELEERIFSRERKLVEL 395
>At5g36780 putative protein
Length = 565
Score = 32.0 bits (71), Expect = 0.40
Identities = 30/122 (24%), Positives = 53/122 (42%), Gaps = 9/122 (7%)
Query: 14 KTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKV---FDFIAKESTNFFDKDSAEKVVLS 70
KT S K ++ S++ + N T K D I E +++ E+ +
Sbjct: 365 KTIKRSEKVHKMNREDSSSNSSEDGNVITDKRKKETKSDVIVYEKPKKKEEEIDEEALKE 424
Query: 71 AVRAARVKKAKSVAAEKAKIAAKEKAKAA------AEKKLNDEKSEAVTEKIDEKAAPNQ 124
R +++KA+ V K K+ K AKAA AEKKL + + +A + +P++
Sbjct: 425 RKREEQLEKARLVMERKRKLQEKAAAKAAIRAQKEAEKKLKECEKKAKKKAAANSTSPSE 484
Query: 125 GN 126
+
Sbjct: 485 SD 486
>At3g02930 unknown protein
Length = 806
Score = 32.0 bits (71), Expect = 0.40
Identities = 31/123 (25%), Positives = 51/123 (41%), Gaps = 6/123 (4%)
Query: 13 TKTSSSSSSQPKPSKPIPFSSTFDPSNPTTFLEKVFDFIAKESTNFFDKDSAEKVVLSAV 72
TK S+S S + + F S P+T E T A +V S
Sbjct: 32 TKPDSNSPSPTQQQSRLSFERPSSNSKPSTDKRSPKAPTPPEKTQI----RAVRVSESQP 87
Query: 73 RAARVKKAKSVAAEKAKIAAKEKAKAAAEKKLNDEKSEAVTEKIDE--KAAPNQGNGMDL 130
++ ++K+ A E EKAKA + K +++E +EK+DE +A ++
Sbjct: 88 QSVQIKEDLKKANELIASLENEKAKALDQLKEARKEAEEASEKLDEALEAQKKSLENFEI 147
Query: 131 EKY 133
EK+
Sbjct: 148 EKF 150
>At1g53110 proton pump interactor, putative
Length = 439
Score = 31.6 bits (70), Expect = 0.52
Identities = 23/95 (24%), Positives = 47/95 (49%), Gaps = 3/95 (3%)
Query: 48 FDFIAKESTNFFDKDSAEKVVLSAVRAARVKKAKSVAAEKAKIAAKEKAKAAAE-KKLND 106
FD E +K+ E+ + R +++KA+ K K+ K AKA KK +
Sbjct: 276 FDIPVYEKLGKEEKEIDEETLKEKKREEQLEKARLAMERKRKLHEKAAAKAVIRVKKEAE 335
Query: 107 EKSEAVTEKIDEKAAPNQGNGMDLEKYSWTQTLQE 141
+K + + ++ +K A + + +D+++ T+T+ E
Sbjct: 336 KKRKELDKRAKKKKAVCKSSSVDVDR--TTETVSE 368
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.126 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,613,352
Number of Sequences: 26719
Number of extensions: 290176
Number of successful extensions: 1652
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 77
Number of HSP's that attempted gapping in prelim test: 1493
Number of HSP's gapped (non-prelim): 201
length of query: 289
length of database: 11,318,596
effective HSP length: 98
effective length of query: 191
effective length of database: 8,700,134
effective search space: 1661725594
effective search space used: 1661725594
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC134822.6


