

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.5 - phase: 0
(195 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
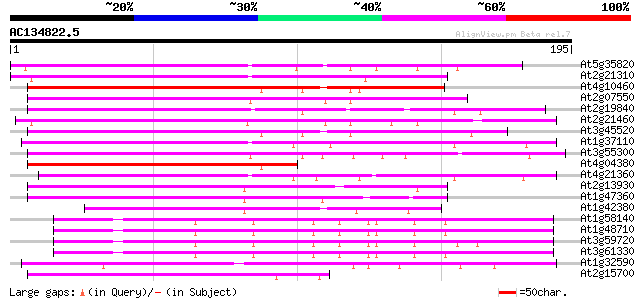
Score E
Sequences producing significant alignments: (bits) Value
At5g35820 copia-like retrotransposable element 110 6e-25
At2g21310 putative retroelement pol polyprotein 110 6e-25
At4g10460 putative retrotransposon 101 2e-22
At2g07550 putative retroelement pol polyprotein 99 1e-21
At2g19840 copia-like retroelement pol polyprotein 97 4e-21
At2g21460 putative retroelement pol polyprotein 97 5e-21
At3g45520 copia-like polyprotein 94 5e-20
At1g37110 92 2e-19
At3g55300 putative protein 87 4e-18
At4g04380 putative polyprotein 81 4e-16
At4g21360 putative transposable element 80 5e-16
At2g13930 putative retroelement pol polyprotein 80 7e-16
At1g47360 polyprotein, putative 79 2e-15
At1g42380 hypothetical protein 60 6e-10
At1g58140 hypothetical protein 53 1e-07
At1g48710 hypothetical protein 53 1e-07
At3g59720 copia-type reverse transcriptase-like protein 52 2e-07
At3g61330 copia-type polyprotein 51 3e-07
At1g32590 hypothetical protein, 5' partial 51 4e-07
At2g15700 copia-like retroelement pol polyprotein 49 1e-06
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 110 bits (274), Expect = 6e-25
Identities = 81/196 (41%), Positives = 113/196 (57%), Gaps = 20/196 (10%)
Query: 1 MKVL-QKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDK 59
+KVL Q + KSL +R LKQ+LY +KM E+ ++ E + +F K++ DL N++V + DED+
Sbjct: 105 IKVLDQLFMAKSLPNRIYLKQRLYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQ 164
Query: 60 ALLLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTK--KLTKSKDLRANENSEGLCV 117
A++LL SLP+ F+ K+T+ Y K T LEEI SA+R+K +L S L N NS+GL V
Sbjct: 165 AIVLLMSLPRQFDQLKETLKYCKT-TLHLEEITSAIRSKILELGASGKLLKN-NSDGLFV 222
Query: 118 -SRGNGGGRG---NRGSSKSGNKERYK-CFKCHKFGHFKRDF----------SEDNENFA 162
RG RG N+ S+S +K K C+ C K GHFK+ S A
Sbjct: 223 QDRGRSETRGKGPNKNKSRSKSKGAGKTCWICGKEGHFKKQCYVWKERNKQGSTSERGEA 282
Query: 163 QVVSEEYEDAGALVVS 178
V+ DA ALVVS
Sbjct: 283 STVTARVTDAAALVVS 298
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 110 bits (274), Expect = 6e-25
Identities = 62/160 (38%), Positives = 96/160 (59%), Gaps = 9/160 (5%)
Query: 1 MKVLQK-YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDK 59
++VL K + KSL +R LKQ+LY +KM +S +I E + +F K++ DL N++V++ DED+
Sbjct: 97 IRVLDKLFMAKSLPNRIYLKQRLYGYKMSDSMTIEENVNDFFKLISDLENVKVSVPDEDQ 156
Query: 60 ALLLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRANENSEGLCVSR 119
A++LL SLPK F+ KDT+ YGK T L+EI A+R+K L + +NS +
Sbjct: 157 AIVLLMSLPKQFDQLKDTLKYGKT-TLALDEITGAIRSKVLELGASGKMLKNSSDALFVQ 215
Query: 120 GNG-------GGRGNRGSSKSGNKERYKCFKCHKFGHFKR 152
G N+ S+S ++E+ C+ C K GHFK+
Sbjct: 216 DRGRSEKRDKSSERNKSQSRSKSREKKVCWVCGKEGHFKK 255
>At4g10460 putative retrotransposon
Length = 1230
Score = 101 bits (252), Expect = 2e-22
Identities = 63/156 (40%), Positives = 97/156 (61%), Gaps = 13/156 (8%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y +K+L +R LKQ+LYS+KM E+ S+ + EF +++ DL N V + DED+A+LLL S
Sbjct: 105 YMSKALPNRIYLKQKLYSYKMQENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMS 164
Query: 67 LPKSFEHFKDTILYGKEGTT-TLEEIQSALRTKKL---TKSKDLRANENSEGLCV----- 117
LPK F+ KDT+ YG TT +++E+ +A+ +K+L + K +R +EGL V
Sbjct: 165 LPKQFDQLKDTLKYGSGRTTLSVDEVVAAIYSKELELGSNKKSIRG--QAEGLYVKDKPE 222
Query: 118 SRG--NGGGRGNRGSSKSGNKERYKCFKCHKFGHFK 151
+RG +GN+G S+S +K C+ C + GHFK
Sbjct: 223 TRGMSEQKEKGNKGRSRSRSKGWKGCWICGEEGHFK 258
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 99.0 bits (245), Expect = 1e-21
Identities = 57/160 (35%), Positives = 94/160 (58%), Gaps = 7/160 (4%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y +K+L +R KQ+LYSFKM E+ S+ + EF +I+ DL N+ V + DED+A+LLL +
Sbjct: 107 YMSKALPNRIYPKQKLYSFKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTA 166
Query: 67 LPKSFEHFKDTILYGK-EGTTTLEEIQSALRTKKLTKSKDLRA-NENSEGLCV-----SR 119
LPK+F+ KDT+ Y + TL+E+ +A+ +K+L ++ +EGL V ++
Sbjct: 167 LPKAFDQLKDTLKYSSGKSILTLDEVAAAIYSKELELGSVKKSIKVQAEGLYVKDKNENK 226
Query: 120 GNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRDFSEDNE 159
G G +G K +K++ C+ C + GHF+ N+
Sbjct: 227 GKGEQKGKGKGKKGKSKKKPGCWTCGEEGHFRSSCPNQNK 266
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 97.4 bits (241), Expect = 4e-21
Identities = 63/188 (33%), Positives = 108/188 (56%), Gaps = 12/188 (6%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y KSL +R L+ ++Y+++M +SK++ E + EF K++ DL N+++ + DE +A+L+L +
Sbjct: 77 YLVKSLPNRVYLQLKVYNYRMQDSKTLEENVDEFQKMISDLNNLQIQVPDEVQAILILSA 136
Query: 67 LPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKL-TKSKDLRANENSEGLCVSRGNGGGR 125
LP S++ K+T+ YG+EG L+++ SA ++K+L + + EGL V RG R
Sbjct: 137 LPDSYDMLKETLKYGREG-IKLDDVISAAKSKELELRDSSGGSRPVGEGLYV-RGKSQAR 194
Query: 126 GNRGSSKSGNKERYKCFKCHKFGHFKRD----FSEDNENFA---QVVSEEYEDAGALVVS 178
G+ G + K+ C+ C K GHFKR ++ N A +V ++ +D LV S
Sbjct: 195 GSDGPKSTEGKK--VCWICGKEGHFKRQCYKWLEKNKANGAGETALVKDDAQDLVGLVAS 252
Query: 179 CWEDDEGE 186
EG+
Sbjct: 253 EVNMSEGK 260
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 97.1 bits (240), Expect = 5e-21
Identities = 73/199 (36%), Positives = 110/199 (54%), Gaps = 14/199 (7%)
Query: 3 VLQK-YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKAL 61
VL K Y +K+L +R KQ+LYSFKM E+ SI + EF +I+ DL N V + DED+A+
Sbjct: 102 VLDKLYMSKALPNRIYQKQKLYSFKMSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAI 161
Query: 62 LLLCSLPKSFEHFKDTILYG-KEGTTTLEEIQSALRTKKLTKSKDLRA-NENSEGLCV-- 117
LLL SLPK F+ +DT+ YG T +L+E+ +A+ +K+L + ++ +EGL V
Sbjct: 162 LLLMSLPKPFDQLRDTLKYGLGRVTLSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKE 221
Query: 118 ---SRGNGGGRGNRGSS-KSGNKERYK--CFKCHKFGHFKRDFSEDNENFAQVVSEEYED 171
+RG RGN ++ KS +K R K C+ C + + ++SE N + VSE
Sbjct: 222 KTETRGRTEQRGNNNNNKKSRSKSRSKKGCWICGESSNGSSNYSEANGLY---VSEALSS 278
Query: 172 AGALVVSCWEDDEGEVSHL 190
+ W D G H+
Sbjct: 279 TDIHLEDEWVMDTGCSYHM 297
>At3g45520 copia-like polyprotein
Length = 1363
Score = 94.0 bits (232), Expect = 5e-20
Identities = 63/177 (35%), Positives = 98/177 (54%), Gaps = 12/177 (6%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y +K+L +R LKQ+LYSFKM E+ SI + EF I+ DL N+ V + DED+A+LLL S
Sbjct: 107 YMSKALPNRIYLKQKLYSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMS 166
Query: 67 LPKSFEHFKDTILYGKEGTT-TLEEIQSALRTKKL---TKSKDLRANENSEGLCV----- 117
LPK F+ KDT+ Y T +L+E+ +A+ +++L + K ++ +EGL V
Sbjct: 167 LPKPFDQLKDTLKYSSGKTVLSLDEVAAAIYSRELEFGSVKKSIKG--QAEGLYVKDKAE 224
Query: 118 SRGNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRDFSEDNE-NFAQVVSEEYEDAG 173
+RG + +S +K + C+ C + GH K N+ F S + E +G
Sbjct: 225 NRGRSEQKDKGKGKRSKSKSKRGCWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSG 281
>At1g37110
Length = 1356
Score = 92.0 bits (227), Expect = 2e-19
Identities = 61/207 (29%), Positives = 111/207 (53%), Gaps = 22/207 (10%)
Query: 5 QKYKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLL 64
+KY SL +R + +LYSFKM+ + +I + + EF +I+ +L ++E+ +++E +A+L+L
Sbjct: 113 KKYMETSLPNRIYTQLKLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILIL 172
Query: 65 CSLPKSFEHFKDTILYGKEGTTTLEEIQSALRT--KKLTKSKDLRANE------NSEGLC 116
SLP S K T+ YG + T T++++ S+ ++ ++L ++ DL + G
Sbjct: 173 NSLPASHIQLKHTLKYGNK-TLTVQDVTSSAKSLERELAEAVDLDKGQAAVLYTTERGRP 231
Query: 117 VSRGNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKRD-------FSEDNENFAQVVSEEY 169
+ R N G +G S+S +K + C+ C K GH K+D + + A V++E+
Sbjct: 232 LVRNNQKGGQGKGRSRSNSKTKVPCWYCKKEGHVKKDCYSRKKKMESEGQGEAGVITEKL 291
Query: 170 EDAGALVVS------CWEDDEGEVSHL 190
+ AL V+ W D G SH+
Sbjct: 292 VFSEALSVNEQMVKDLWILDSGCTSHM 318
>At3g55300 putative protein
Length = 262
Score = 87.4 bits (215), Expect = 4e-18
Identities = 67/210 (31%), Positives = 104/210 (48%), Gaps = 24/210 (11%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y+TK+LA+ LK + SFKM+E+KSI E L K++ DLA++++N DED+AL +
Sbjct: 40 YQTKTLANMIYLKHKFTSFKMVENKSIEENLDTSLKLVADLASLDINTSDEDQALQFMAG 99
Query: 67 LPKSFEHFKDTILYGK-EGTTTLEEIQSALRTKKL-TKSKDLR--ANENSEGLCVS-RGN 121
LP ++ DT+ YG + T TL + S+ +K++ K K L + +SE L +G
Sbjct: 100 LPPQYDSLVDTLKYGSGKDTLTLNAVISSAYSKEIELKEKGLLNVSKPDSEALLGDFKGR 159
Query: 122 GGGRGNR----GSSKSGNK------ERYKCFKCHKFGHFKRDFSEDNENFAQVVSEEYED 171
+GN+ GSS K CF C K H+KR+ + + A++ + E
Sbjct: 160 SNNKGNKRPCTGSSSGNGKFQSRPSNNTSCFICGKEDHWKRECPQ-RKKHAELANIATEP 218
Query: 172 AGALVVSC--------WEDDEGEVSHLDID 193
LV++ W D G H D
Sbjct: 219 PQPLVLTVSTQSTCKEWVMDSGRTFHSTSD 248
>At4g04380 putative polyprotein
Length = 778
Score = 80.9 bits (198), Expect = 4e-16
Identities = 43/95 (45%), Positives = 64/95 (67%), Gaps = 1/95 (1%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y +K+L ++ LKQ+LY FKM E+ S+ + EF I+ DL N+ V + DED+ +LLL S
Sbjct: 76 YMSKALPNQIYLKQKLYRFKMSENLSMEGNIDEFLHIVADLENLNVLVSDEDQTILLLMS 135
Query: 67 LPKSFEHFKDTILYGKEGTT-TLEEIQSALRTKKL 100
LPKSF+ KDT+ Y T TL+E+ +A+ +K+L
Sbjct: 136 LPKSFDQLKDTLQYSSGKTVLTLDEVTAAIYSKEL 170
>At4g21360 putative transposable element
Length = 1308
Score = 80.5 bits (197), Expect = 5e-16
Identities = 58/201 (28%), Positives = 110/201 (53%), Gaps = 23/201 (11%)
Query: 11 SLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCSLPKS 70
SL +R + +LYSFKM+++ SI + EF +I+ +L ++++ + +E +A+L+L SLP S
Sbjct: 121 SLPNRIYTQLRLYSFKMVDNLSIDQNTDEFLRIVAELGSLQIQVGEEVQAILILNSLPPS 180
Query: 71 FEHFKDTILYGKEGTTTLEEIQSALRT--KKLTKSKD-LRANENSEGLCVSRG-----NG 122
+ K T+ YG + T +++++ S+ ++ ++L++ K+ +RA ++ RG N
Sbjct: 181 YIQLKHTLKYGNK-TLSVQDVVSSAKSLERELSEQKETIRAPASTALYTAERGRPQTKNT 239
Query: 123 GGRGNRGSSKSGNKERYKCFKCHKFGHFKRD-------FSEDNENFAQVVSEEYEDAGAL 175
G+G +G +S +K R C+ C K GH K+D + + A V++E+ + AL
Sbjct: 240 QGQG-KGRGRSNSKSRLTCWFCKKEGHVKKDCYAGKRKLENEGQGKAGVITEKLVYSEAL 298
Query: 176 VV------SCWEDDEGEVSHL 190
+ W D G H+
Sbjct: 299 SMYDQEAKDKWVIDSGCTYHM 319
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 80.1 bits (196), Expect = 7e-16
Identities = 49/154 (31%), Positives = 81/154 (51%), Gaps = 11/154 (7%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
+ +SL HR + Y+FKM E+K I E + +F KI+ DL ++++++ DE +A+LLL S
Sbjct: 82 FMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLKIVADLNHLQIDVTDEVQAILLLSS 141
Query: 67 LPKSFEHFKDTILY-GKEGTTTLEEIQSALRTKKLTKSKDLRANENSEGLCVSRGNGGGR 125
LP ++ +T+ Y L+++ A R K+ S++ R +RG G+
Sbjct: 142 LPARYDGLVETMKYSNSREKLRLDDVMVAARDKERELSQNNRPVVEGH---FARGRPDGK 198
Query: 126 GNRGSSKSGNKERYK-------CFKCHKFGHFKR 152
N +K N+ R K C+ C K GHFK+
Sbjct: 199 NNNQGNKGKNRSRSKSADGKRVCWICGKEGHFKK 232
>At1g47360 polyprotein, putative
Length = 1182
Score = 79.0 bits (193), Expect = 2e-15
Identities = 52/154 (33%), Positives = 83/154 (53%), Gaps = 11/154 (7%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
+ +SL HR + Y+FKM E+K I E + +F KI+ DL ++++ + DE +A+LLL S
Sbjct: 72 FMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLKIVADLNHLQIEVTDEVQAILLLSS 131
Query: 67 LPKSFEHFKDTILY-GKEGTTTLEEIQSALRTKKLTKSKDLR-------ANENSEGLCVS 118
LP ++ +T+ Y L+++ R K+ S++ R A EG +
Sbjct: 132 LPARYDGLVETMKYSNSREKLRLDDVMVGARDKERELSQNNRPVAEGHYARGRPEGKNNN 191
Query: 119 RGNGGGRGNRGSSKSGNKERYKCFKCHKFGHFKR 152
+GN G NR SKS + +R C+ C K GHFK+
Sbjct: 192 QGNKG--KNRSRSKSADGKRV-CWICGKEGHFKK 222
>At1g42380 hypothetical protein
Length = 143
Score = 60.5 bits (145), Expect = 6e-10
Identities = 42/131 (32%), Positives = 69/131 (52%), Gaps = 9/131 (6%)
Query: 27 MLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCSLPKSFEHFKDTILY-GKEGT 85
M E+K I E + +F KI+ DL +I++ + DE +A+LLL SLP ++ +T+ Y
Sbjct: 1 MQENKKIDENIDDFLKIVVDLNHIQIEVRDEVQAILLLSSLPSRYDGLVETMKYSNSREK 60
Query: 86 TTLEEIQSALRTKKLTKSKDLRANENSEGLCVSR---GNGGGRGNRGSSKSGNKE---RY 139
L ++ A R K+ S++ R +EG R G +GN+G ++S +K +
Sbjct: 61 LRLNDVMVAARDKEREMSQNNRL--IAEGHYARRRPVGKNNNQGNKGKNRSWSKSADGKR 118
Query: 140 KCFKCHKFGHF 150
C+ C K HF
Sbjct: 119 VCWICGKEKHF 129
>At1g58140 hypothetical protein
Length = 1320
Score = 52.8 bits (125), Expect = 1e-07
Identities = 57/223 (25%), Positives = 94/223 (41%), Gaps = 52/223 (23%)
Query: 16 QLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLL---LCSLPKSFE 72
Q L+ + + +M E + +S+ F+++L N++ N E D ++ L SL FE
Sbjct: 114 QTLRGEFEALQMKEGELVSDY---FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFE 170
Query: 73 HFKDTILYGKE-GTTTLEEIQSALRTKKLTKSK-----------DLRANENSE------- 113
H I K+ T+E++ +L+ + K K + EN +
Sbjct: 171 HIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGG 230
Query: 114 GLCVSRGNGG---GRG---------------NRGSSKSGNKERY-----KCFKCHKFGHF 150
G RG GG GRG +RG K K RY KC+ C KFGH+
Sbjct: 231 GQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHY 290
Query: 151 ----KRDFSEDNENFAQVVSEEYEDAGALVVSCWEDDEGEVSH 189
K ++ E A V E+ ++ L+++ ++ DE E +H
Sbjct: 291 ASECKAPSNKKFEEKANYVEEKIQEEDMLLMASYKKDEQEENH 333
>At1g48710 hypothetical protein
Length = 1352
Score = 52.8 bits (125), Expect = 1e-07
Identities = 57/223 (25%), Positives = 94/223 (41%), Gaps = 52/223 (23%)
Query: 16 QLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLL---LCSLPKSFE 72
Q L+ + + +M E + +S+ F+++L N++ N E D ++ L SL FE
Sbjct: 114 QTLRGEFEALQMKEGELVSDY---FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFE 170
Query: 73 HFKDTILYGKE-GTTTLEEIQSALRTKKLTKSK-----------DLRANENSE------- 113
H I K+ T+E++ +L+ + K K + EN +
Sbjct: 171 HIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDIIEQVLNMQITKEENGQSYQRRGG 230
Query: 114 GLCVSRGNGG---GRG---------------NRGSSKSGNKERY-----KCFKCHKFGHF 150
G RG GG GRG +RG K K RY KC+ C KFGH+
Sbjct: 231 GQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHY 290
Query: 151 ----KRDFSEDNENFAQVVSEEYEDAGALVVSCWEDDEGEVSH 189
K ++ E A V E+ ++ L+++ ++ DE E +H
Sbjct: 291 ASECKAPSNKKFEEKANYVEEKIQEEDMLLMASYKKDEQEENH 333
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 52.4 bits (124), Expect = 2e-07
Identities = 57/223 (25%), Positives = 95/223 (42%), Gaps = 52/223 (23%)
Query: 16 QLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLL---LCSLPKSFE 72
Q L+ + + +M E + +S+ F+++L N++ N E D ++ L SL FE
Sbjct: 114 QTLRGEFEALQMKEGELVSDY---FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFE 170
Query: 73 HFKDTILYGKE-GTTTLEEIQSALRTKKLTKSK-----------DLRANENSE------- 113
H I K+ T+E++ +L+ + K K + EN +
Sbjct: 171 HIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGG 230
Query: 114 GLCVSRGNGG---GRG---------------NRGSSKSGNKERY-----KCFKCHKFGHF 150
G RG GG GRG +RG K K RY KC+ C KFGH+
Sbjct: 231 GQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHY 290
Query: 151 KRDF-SEDNENF---AQVVSEEYEDAGALVVSCWEDDEGEVSH 189
+ + N+ F A V E+ ++ L+++ ++ DE E +H
Sbjct: 291 ASECKAPSNKKFKEKANYVEEKIQEEDMLLMASYKKDEQEENH 333
>At3g61330 copia-type polyprotein
Length = 1352
Score = 51.2 bits (121), Expect = 3e-07
Identities = 56/223 (25%), Positives = 94/223 (42%), Gaps = 52/223 (23%)
Query: 16 QLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLL---LCSLPKSFE 72
Q L+ + + +M E + +S+ F+++L N++ N E D ++ L SL FE
Sbjct: 114 QTLRGEFEALQMKEGELVSDY---FSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFE 170
Query: 73 HFKDTILYGKE-GTTTLEEIQSALRTKKLTKSK-----------DLRANENSE------- 113
H I K+ T+E++ +L+ + K K + EN +
Sbjct: 171 HIVTVIEETKDLEAMTIEQLLGSLQAYEEKKKKKEDIAEQVLNMQITKEENGQSYQRRGG 230
Query: 114 GLCVSRGNGG---GRG---------------NRGSSKSGNKERY-----KCFKCHKFGHF 150
G RG GG GRG +RG K K RY KC+ C KFGH+
Sbjct: 231 GQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHY 290
Query: 151 ----KRDFSEDNENFAQVVSEEYEDAGALVVSCWEDDEGEVSH 189
K ++ E A V E+ ++ L+++ ++ DE + +H
Sbjct: 291 ASECKAPSNKKFEEKAHYVEEKIQEEDMLLMASYKKDEQKENH 333
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 50.8 bits (120), Expect = 4e-07
Identities = 50/213 (23%), Positives = 89/213 (41%), Gaps = 30/213 (14%)
Query: 5 QKYKTKSLAHRQLLKQQLYSFKMLESK---SISEQLAEFNKILDDLANIEVNMEDEDKAL 61
+KY+ L++ SF++LE K +I+ + +I +D+ N+ +M D
Sbjct: 59 RKYQGNDRVQSAQLQRLRRSFEVLEMKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVE 118
Query: 62 LLLCSLPKSFEHFKDTILYGKEGTTTLEEIQSALRTKKLTKSKDLRANENSEGLCVS--- 118
+L +L + F + I +E E L++ + ++L ++ E + +
Sbjct: 119 KILRTLVEKFTYVVCAI---EESNNIKELTVDGLQSSLMVHEQNLSRHDVEERVLKAETQ 175
Query: 119 ------RGNGG----GRGNRGSSKSG----NKERYKCFKCHKFGHFKRDFS--EDNENFA 162
RG GG GRG G G N++ +CFKCHK GH+K + E N+
Sbjct: 176 WRPDGGRGRGGSPSRGRGRGGYQGRGRGYVNRDTVECFKCHKMGHYKAECPSWEKEANYV 235
Query: 163 QVVSE-----EYEDAGALVVSCWEDDEGEVSHL 190
++ + E G W D G +H+
Sbjct: 236 EMEEDLLLMAHVEQIGDEEKQIWFLDSGCSNHM 268
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 49.3 bits (116), Expect = 1e-06
Identities = 34/110 (30%), Positives = 61/110 (54%), Gaps = 5/110 (4%)
Query: 7 YKTKSLAHRQLLKQQLYSFKMLESKSISEQLAEFNKILDDLANIEVNMEDEDKALLLLCS 66
Y SL +R L+ + Y++KM +S+SI + +F K++ DL NI V++ +E +A+LLL S
Sbjct: 116 YMETSLPNRIYLQLKFYTYKMTDSRSIDGNVDDFLKLVTDLNNIGVDVTEEVQAILLLSS 175
Query: 67 LPKSFEHFKDTILYGKEGTTTLEEI--QSALRTKKLTKSKDL---RANEN 111
L E+ + EG +E + L+ +K +D+ +A++N
Sbjct: 176 LYDRKENSQKVESSQSEGLYVEQEADQRRGLKKEKANHGEDVLRAKADQN 225
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.132 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,590,691
Number of Sequences: 26719
Number of extensions: 208497
Number of successful extensions: 1077
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 955
Number of HSP's gapped (non-prelim): 132
length of query: 195
length of database: 11,318,596
effective HSP length: 94
effective length of query: 101
effective length of database: 8,807,010
effective search space: 889508010
effective search space used: 889508010
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC134822.5


