

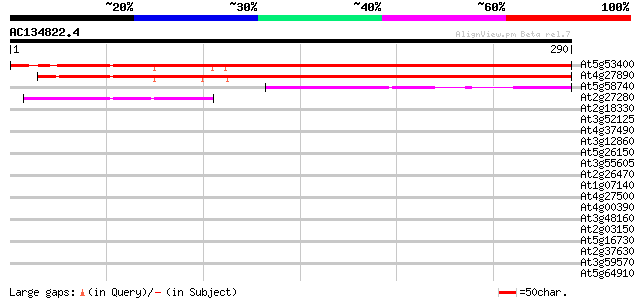
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.4 - phase: 0
(290 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g53400 unknown protein 330 5e-91
At4g27890 putative protein 309 1e-84
At5g58740 unknown protein 67 1e-11
At2g27280 unknown protein 59 3e-09
At2g18330 AAA-type like ATPase 37 0.016
At3g52125 unknown protein 35 0.062
At4g37490 cyclin cyc1 33 0.23
At3g12860 nucleolar protein, putative 32 0.40
At5g26150 putative protein 31 0.68
At3g55605 unknown protein 31 0.89
At2g26470 unknown protein 31 0.89
At1g07140 Unknown protein (F10K1.15) 31 0.89
At4g27500 unknown protein 30 1.2
At4g00390 unknown protein 30 1.5
At3g48160 putative protein 30 1.5
At2g03150 Unknown protein (T18E12.18; At2g03150) 30 1.5
At5g16730 putative protein 30 2.0
At2g37630 putative MYB family transcription factor 30 2.0
At3g59570 putative protein 29 2.6
At5g64910 unknown protein 29 3.4
>At5g53400 unknown protein
Length = 304
Score = 330 bits (846), Expect = 5e-91
Identities = 177/312 (56%), Positives = 217/312 (68%), Gaps = 30/312 (9%)
Query: 1 MAIISDYQDETQTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDS 60
MAIIS+ ++E SSSS+P PF +T + P FLEKVFDF+ ++S DF K S
Sbjct: 1 MAIISEVEEE----SSSSRPM---IFPFRATLSSANPLGFLEKVFDFLGEQS-DFLKKPS 52
Query: 61 AEKMVLSAVRAAK--VKKAKAVAAEKAKIASQEKAKAAAGIKVND---EKSGVGT----- 110
AE ++ AVRAAK +KKA+ AEK + EK +K+ + EK V
Sbjct: 53 AEDEIVVAVRAAKEKLKKAEKKKAEKESVKPVEKKAEKEIVKLVEKKVEKESVKPTIAAS 112
Query: 111 ------------EKKDGESGLAAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVI 158
E++ ESG PN+GNG DLE YSW Q LQE+ VN+PVP GTK+ V+
Sbjct: 113 SAEPIEVEKPKEEEEKKESGPIVPNKGNGTDLENYSWIQNLQEVTVNIPVPTGTKARTVV 172
Query: 159 CEIKKNHLKVGLKGQPPIIDGELYKFIKADECYWSIEDQSTVSILLTKHDQMDWWKCLVK 218
CEIKKN LKVGLKGQ PI+DGELY+ +K D+CYW+IEDQ +SILLTK DQM+WWKC VK
Sbjct: 173 CEIKKNRLKVGLKGQDPIVDGELYRSVKPDDCYWNIEDQKVISILLTKSDQMEWWKCCVK 232
Query: 219 GDPLINTQKVEPESSKLGELDSETRMTVEKMMFDQRQKSMGLPTSEELEKQEMMKKFMSQ 278
G+P I+TQKVEPE+SKLG+LD ETR TVEKMMFDQRQK MGLPTSEEL+KQE++KKFMS+
Sbjct: 233 GEPEIDTQKVEPETSKLGDLDPETRSTVEKMMFDQRQKQMGLPTSEELQKQEILKKFMSE 292
Query: 279 HPNMDFSGAKLS 290
HP MDFS AK +
Sbjct: 293 HPEMDFSNAKFN 304
>At4g27890 putative protein
Length = 293
Score = 309 bits (791), Expect = 1e-84
Identities = 158/291 (54%), Positives = 209/291 (71%), Gaps = 17/291 (5%)
Query: 15 SSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLSAVRAAK- 73
S + +PS +PF+++FDPS P AFLEKV D I KES +F KD+AEK +++AV AAK
Sbjct: 5 SEMEEARPSM-VPFTASFDPSNPIAFLEKVLDVIGKES-NFLKKDTAEKEIVAAVMAAKQ 62
Query: 74 -VKKAKAVAAEKAKIASQEKAKAAAG------IKVNDEKSGVGTE-------KKDGESGL 119
+++A+ EK + S E K ++ E+S + T+ K++ ESG
Sbjct: 63 RLREAEKKKLEKESVKSMEVEKPKKDSLKPTELEKPKEESLMATDPMEIEKPKEEKESGP 122
Query: 120 AAPNQGNGMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQPPIIDG 179
PN+GNG+D EKYSW Q LQE+ +N+P+P GTKS V CEIKKN LKVGLKGQ I+DG
Sbjct: 123 IVPNKGNGLDFEKYSWGQNLQEVTINIPMPEGTKSRSVTCEIKKNRLKVGLKGQDLIVDG 182
Query: 180 ELYKFIKADECYWSIEDQSTVSILLTKHDQMDWWKCLVKGDPLINTQKVEPESSKLGELD 239
E + +K D+C+W+IEDQ +S+LLTK DQM+WWK VKG+P I+TQKVEPE+SKLG+LD
Sbjct: 183 EFFNSVKPDDCFWNIEDQKMISVLLTKQDQMEWWKYCVKGEPEIDTQKVEPETSKLGDLD 242
Query: 240 SETRMTVEKMMFDQRQKSMGLPTSEELEKQEMMKKFMSQHPNMDFSGAKLS 290
ETR +VEKMMFDQRQK MGLP S+E+EK++M+KKFM+Q+P MDFS AK +
Sbjct: 243 PETRASVEKMMFDQRQKQMGLPRSDEIEKKDMLKKFMAQNPGMDFSNAKFN 293
>At5g58740 unknown protein
Length = 158
Score = 67.0 bits (162), Expect = 1e-11
Identities = 42/159 (26%), Positives = 74/159 (46%), Gaps = 38/159 (23%)
Query: 133 YSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQPPIIDGELYKFIKADECYW 192
+ W QTL+E+N+ + +P C+I+ H++VG+KG PP ++ +L +K D +W
Sbjct: 21 FEWDQTLEEVNMYITLPPNVHPKSFHCKIQSKHIEVGIKGNPPYLNHDLSAPVKTDCSFW 80
Query: 193 SIEDQSTVSILLTKHDQMDWWKCLVKGDPLINTQKVEPESSKLGELDSETRMTVEKMMFD 252
++ED + I L K ++ W + G G+LD
Sbjct: 81 TLED-DIMHITLQKREKGQTWASPILGQ---------------GQLD------------- 111
Query: 253 QRQKSMGLPTSEELE-KQEMMKKFMSQHPNMDFSGAKLS 290
P + +LE K+ M+++F ++P DFS A+ S
Sbjct: 112 --------PYATDLEQKRLMLQRFQEENPGFDFSQAQFS 142
>At2g27280 unknown protein
Length = 764
Score = 58.9 bits (141), Expect = 3e-09
Identities = 35/98 (35%), Positives = 55/98 (55%), Gaps = 2/98 (2%)
Query: 8 QDETQTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLS 67
+ E +T S + T+PF+++FDPS P FLEKVF+F+ K+S +F KD A +++
Sbjct: 397 ETEKKTESMDVEKVRPSTLPFNASFDPSDPLGFLEKVFEFVGKKS-NFLVKDKAVNAIIT 455
Query: 68 AVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEK 105
AV AK ++ K E K A+ + K I+ +K
Sbjct: 456 AVTDAK-ERLKEEEKESVKQATVKIKKYGLQIRAPSQK 492
Score = 38.5 bits (88), Expect = 0.004
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 7/53 (13%)
Query: 40 FLEKVFDFIAKESTDFFDKDSAEKMVLSAVRAAK------VKKAKAVAAEKAK 86
FLE V DFI KES +F KD+AEK + AV AK KK +++ EK +
Sbjct: 360 FLENVLDFIGKES-NFLRKDTAEKEITDAVTTAKERLRETEKKTESMDVEKVR 411
>At2g18330 AAA-type like ATPase
Length = 636
Score = 36.6 bits (83), Expect = 0.016
Identities = 25/109 (22%), Positives = 49/109 (44%), Gaps = 13/109 (11%)
Query: 9 DETQTPSSSSQPKPSKTIPFSSTFDPSK------------PTAFLEKVFDFIAKESTDFF 56
D + +S S+ KP P S FDP + ++VFD + K+
Sbjct: 49 DHKSSSNSKSETKPDSDEPKGSGFDPESLERGAKALREINSSPHSKQVFDLMRKQEKTRL 108
Query: 57 DKDSAEKMVLSAVRAAK-VKKAKAVAAEKAKIASQEKAKAAAGIKVNDE 104
+ +AEK A++A+K +++ + +A ++ + Q+ A ++ DE
Sbjct: 109 AELAAEKEHNEAIQASKDIERQRKLAEDQRNLVQQQAQAKAQNLRYEDE 157
>At3g52125 unknown protein
Length = 443
Score = 34.7 bits (78), Expect = 0.062
Identities = 44/167 (26%), Positives = 61/167 (36%), Gaps = 29/167 (17%)
Query: 8 QDETQTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLS 67
Q + + P +S PS+T S+ DP K + D+I E D F LS
Sbjct: 295 QSKDEMPPPASLQGPSET----SSTDPGKRGHHMG---DYIPLEELDKF---------LS 338
Query: 68 AVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKKDGESGLAAPNQGNG 127
A +KA AAEKAKI + E G+G+ +K G+A P
Sbjct: 339 KCNDAAAQKATKEAAEKAKIQADNVGHKLLSKMGWKEGEGIGSSRK----GMADPIMAGD 394
Query: 128 MDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQP 174
+ L V P K I E K + +G K +P
Sbjct: 395 V---------KTNNLGVGASAPGEVKPEDDIYEQYKKRMMLGYKHRP 432
>At4g37490 cyclin cyc1
Length = 428
Score = 32.7 bits (73), Expect = 0.23
Identities = 31/111 (27%), Positives = 46/111 (40%), Gaps = 9/111 (8%)
Query: 11 TQTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLSAVR 70
+Q P+ + K + + K A KV D I S DS E++ L A R
Sbjct: 61 SQNPTLLVEDNLKKPVVKRNAVPKPKKVAGKPKVVDVIEISS------DSDEELGLVAAR 114
Query: 71 AAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKKDGESGLAA 121
K K KA A ++KAA G++ ++ V + D E+ LAA
Sbjct: 115 EKKATKKKATTYTSVLTA---RSKAACGLEKKQKEKIVDIDSADVENDLAA 162
>At3g12860 nucleolar protein, putative
Length = 477
Score = 32.0 bits (71), Expect = 0.40
Identities = 23/87 (26%), Positives = 44/87 (50%), Gaps = 2/87 (2%)
Query: 32 FDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLSAVRAAKVKKAKAVAAEKAKI-ASQ 90
F + TAF EK+ + + +E DF+DK A + + ++ + + K E+ K +
Sbjct: 388 FSDNSTTAFGEKLREQV-EERLDFYDKGVAPRKNVDVMKEKRKTEEKEEEKEEEKSKKKK 446
Query: 91 EKAKAAAGIKVNDEKSGVGTEKKDGES 117
+K+KA G ++ +G +KK +S
Sbjct: 447 KKSKAVEGEELTATDNGHSKKKKKTKS 473
>At5g26150 putative protein
Length = 703
Score = 31.2 bits (69), Expect = 0.68
Identities = 37/151 (24%), Positives = 64/151 (41%), Gaps = 26/151 (17%)
Query: 67 SAVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKKDGESGLAAPNQGN 126
+A+ A+++KAK A +A +Q A+ + E V +E+KD + ++A N
Sbjct: 346 AALAVAEIEKAKCRTAVEAAEKAQRMAELEGQRRKQAEMKAV-SEEKDKDRAVSALAH-N 403
Query: 127 GMDLEKYSWTQTLQELNVNVPVPNGTKSGFVICEIKKNHLKVGLKGQPPIIDGEL----- 181
+ KYS + ++E E NH K+G G P+ +GEL
Sbjct: 404 DVRYRKYS-IEEIEEAT----------------ERFANHRKIGEGGYGPVYNGELDHTPV 446
Query: 182 -YKFIKADECYWSIEDQSTVSILLT-KHDQM 210
K ++ D + Q V +L + +H M
Sbjct: 447 AIKVLRPDAAQGKKQFQQEVEVLCSIRHPHM 477
>At3g55605 unknown protein
Length = 258
Score = 30.8 bits (68), Expect = 0.89
Identities = 33/137 (24%), Positives = 59/137 (42%), Gaps = 21/137 (15%)
Query: 16 SSSQPKPS-------KTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLSA 68
SS PKPS + P+S+ +P K L +V D K+S + D D+ E+ + S+
Sbjct: 34 SSLVPKPSLLRPFVSRGFPYSTATEPLKSDQTLIQVIDSEIKDSFEADDHDADEETIDSS 93
Query: 69 VRAAKVK-----KAKAVAAE------KAKIASQEKAKAAAGIKVNDEKSGVGTEKKDGES 117
K++ + + E K +++ A V+D++ G G +K ES
Sbjct: 94 DFPFKIEDNPGHRTVTLTREYNGEQIKVEVSMPGLAMDENEDDVDDDEDGDGRHEKSNES 153
Query: 118 GL---AAPNQGNGMDLE 131
+ + +G+ LE
Sbjct: 154 SIPLVVTVTKKSGLSLE 170
>At2g26470 unknown protein
Length = 487
Score = 30.8 bits (68), Expect = 0.89
Identities = 31/128 (24%), Positives = 53/128 (41%), Gaps = 6/128 (4%)
Query: 11 TQTPSSSSQPKPSKTIPFSSTFDPSK-PTAFLEKVFDFIAKESTDFFDKDSAEKMVLSAV 69
T+ P + K +K+ + D K PTA + D I K +KD + V +
Sbjct: 249 TKQPKTDEGDKETKSTDANIIVDLKKEPTAEKDTFSDSIKKIEELDGEKDMSN--VAKNL 306
Query: 70 RAAKVKKAKAVAAEKAKIASQE---KAKAAAGIKVNDEKSGVGTEKKDGESGLAAPNQGN 126
++ KA+ + + +AS K G K + G K+ GES + P +
Sbjct: 307 EFQEIVKAEPFVEDNSAVASLPEPVKNDVQEGTKEEGKSLKTGLTKETGESSASIPAESY 366
Query: 127 GMDLEKYS 134
+DL++ S
Sbjct: 367 ILDLQRIS 374
>At1g07140 Unknown protein (F10K1.15)
Length = 228
Score = 30.8 bits (68), Expect = 0.89
Identities = 25/85 (29%), Positives = 40/85 (46%), Gaps = 14/85 (16%)
Query: 40 FLEKVFDFIAKESTDFFDKDSA------EKMVLSAVRAAKVKKAKAVAAEKAKIASQEKA 93
F++K + E KD+A EK+ + + + +AKAV K ++ ++EK
Sbjct: 149 FMQKFKEVAESEEEKEESKDAADTAGLLEKLTVEETKTEEKTEAKAVETAKTEVKAEEKK 208
Query: 94 KAAAGIKVNDEKSGVGTEKKDGESG 118
++ A EKS G KK ESG
Sbjct: 209 ESEA------EKS--GEAKKTEESG 225
>At4g27500 unknown protein
Length = 612
Score = 30.4 bits (67), Expect = 1.2
Identities = 30/114 (26%), Positives = 45/114 (39%), Gaps = 11/114 (9%)
Query: 18 SQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLSAV-----RAA 72
S PKP T+ + + +K ++ V D E EK V +A +
Sbjct: 421 SAPKPDATV--AQNTEKAKDAVKVKNVADDDDDEVYGLGKPQKEEKPVDAATAKEMRKQE 478
Query: 73 KVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKKDGESGLAAPNQGN 126
++ KAK K K+A + AKAA + +K EKK+ E GN
Sbjct: 479 EIAKAKQAMERKKKLAEKAAAKAA----IRAQKEAEKKEKKEQEKKAKKKTGGN 528
>At4g00390 unknown protein
Length = 364
Score = 30.0 bits (66), Expect = 1.5
Identities = 29/120 (24%), Positives = 47/120 (39%), Gaps = 22/120 (18%)
Query: 6 DYQDETQTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMV 65
D + SSS + +P K++P ++ P+K TA +A
Sbjct: 17 DVETSEDDSSSSEEDEPIKSLPATTAAAPAKSTAV------------------SAATPAK 58
Query: 66 LSAVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVG----TEKKDGESGLAA 121
++V AA K+ AV+A + E + + KSG G ++KKD S A
Sbjct: 59 STSVSAAAPSKSTAVSAAADSDSGSESETDSDSESTDPPKSGSGKTIASKKKDDPSSSTA 118
>At3g48160 putative protein
Length = 323
Score = 30.0 bits (66), Expect = 1.5
Identities = 23/82 (28%), Positives = 31/82 (37%), Gaps = 9/82 (10%)
Query: 6 DYQDETQTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFD---------FIAKESTDFF 56
D +E+ P SSSQ SK + DPSK EK FI E+
Sbjct: 79 DDDEESSQPHSSSQTDSSKPGSLPQSSDPSKIDNRREKSLGLLTQNFIKLFICSEAIRII 138
Query: 57 DKDSAEKMVLSAVRAAKVKKAK 78
D A K++L + + K
Sbjct: 139 SLDDAAKLLLGDAHNTSIMRTK 160
>At2g03150 Unknown protein (T18E12.18; At2g03150)
Length = 1336
Score = 30.0 bits (66), Expect = 1.5
Identities = 33/152 (21%), Positives = 56/152 (36%), Gaps = 20/152 (13%)
Query: 10 ETQTPSSSSQPKPSKTIPFSSTFDPSKP--------TAFLEKVFDFIAKESTDFFDK--- 58
E T +SS++ K T DPS T +K+ +AK D
Sbjct: 787 EESTGTSSNKKIVKKVAETGDTSDPSAKANEQTPAKTIVKKKIIKRVAKRKVAEIDNKMD 846
Query: 59 -------DSAEKMVLSAVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTE 111
DS EK V+ + + + + + + A+ + V E T+
Sbjct: 847 GDSKKDGDSDEKKVMEVGKKSSDSGSVEMKPTAESLEDVKDENASKTVDVKQETGSPDTK 906
Query: 112 KKDGESGLAAPNQGNGMD--LEKYSWTQTLQE 141
KK+G S + + G D EK + ++T+ E
Sbjct: 907 KKEGASSSSKKDTKTGEDKKAEKKNNSETMSE 938
>At5g16730 putative protein
Length = 853
Score = 29.6 bits (65), Expect = 2.0
Identities = 19/71 (26%), Positives = 39/71 (54%), Gaps = 4/71 (5%)
Query: 50 KESTDFFDKDSAEKMVLSAVRAAKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVG 109
K+ + DKD + +++ + +++K +A +K+++ SQE+ + ++ I DE
Sbjct: 751 KDESQDDDKDDSVEVIFKMWESCQIEKKEAFPDKKSELESQEEEEDSSKI---DESDKTS 807
Query: 110 TEKKDGESGLA 120
TE D E+G A
Sbjct: 808 TENID-ETGNA 817
>At2g37630 putative MYB family transcription factor
Length = 367
Score = 29.6 bits (65), Expect = 2.0
Identities = 14/54 (25%), Positives = 31/54 (56%)
Query: 226 QKVEPESSKLGELDSETRMTVEKMMFDQRQKSMGLPTSEELEKQEMMKKFMSQH 279
+K+E +K+ L E + +EK+ + R++ +GL E + Q++ ++ S+H
Sbjct: 296 EKMEEIEAKMKALREEQKNAMEKIEGEYREQLVGLRRDAEAKDQKLADQWTSRH 349
>At3g59570 putative protein
Length = 549
Score = 29.3 bits (64), Expect = 2.6
Identities = 16/45 (35%), Positives = 22/45 (48%)
Query: 23 SKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLS 67
S +P SS + P F + +FDF + TD F + S K LS
Sbjct: 238 SSFVPPSSPYGFPSPGPFADDIFDFPSLPVTDLFGRSSLGKKGLS 282
>At5g64910 unknown protein
Length = 487
Score = 28.9 bits (63), Expect = 3.4
Identities = 25/113 (22%), Positives = 45/113 (39%), Gaps = 9/113 (7%)
Query: 12 QTPSSSSQPKPSKTIPFSSTFDPSKPTAFLEKVFDFIAKESTDFFDKDSAEKMVLSAVRA 71
+T +SSS K + ++T ++PT E+ D + + + K+ E A
Sbjct: 20 KTGASSSASKNDDAVVEATTTQETQPTQETEETEDKVESPAPEEEGKNEEEANENQEEEA 79
Query: 72 AKVKKAKAVAAEKAKIASQEKAKAAAGIKVNDEKSGVGTEKKDGESGLAAPNQ 124
AKV+ A + A ++K +EK E K+ E P++
Sbjct: 80 AKVESKAAEEGGNEEEAKEDK---------EEEKEEAAREDKEEEEEAVKPDE 123
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.127 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,601,940
Number of Sequences: 26719
Number of extensions: 287036
Number of successful extensions: 1040
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 1013
Number of HSP's gapped (non-prelim): 51
length of query: 290
length of database: 11,318,596
effective HSP length: 98
effective length of query: 192
effective length of database: 8,700,134
effective search space: 1670425728
effective search space used: 1670425728
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC134822.4


