

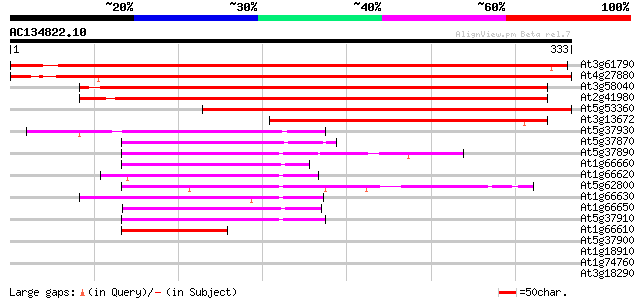
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.10 - phase: 0
(333 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g61790 seven in absentia-like protein 569 e-163
At4g27880 putative zinc finger protein 554 e-158
At3g58040 unknown protein 469 e-133
At2g41980 putative RING zinc finger protein 457 e-129
At5g53360 ring finger E3 ligase (SINAT5) 429 e-120
At3g13672 seven in absentia-like protein 219 2e-57
At5g37930 unknown protein (At5g37930) 113 1e-25
At5g37870 putative protein 112 3e-25
At5g37890 unknown protein 100 1e-21
At1g66660 hypothetical protein 99 3e-21
At1g66620 hypothetical protein 97 2e-20
At5g62800 SIAH1 - like protein 96 2e-20
At1g66630 hypothetical protein 96 2e-20
At1g66650 hypothetical protein 96 3e-20
At5g37910 putative protein 95 5e-20
At1g66610 hypothetical protein 69 5e-12
At5g37900 putative protein 40 0.002
At1g18910 hypothetical protein 35 0.075
At1g74760 34 0.097
At3g18290 zinc finger protein, putative 34 0.13
>At3g61790 seven in absentia-like protein
Length = 326
Score = 569 bits (1467), Expect = e-163
Identities = 268/333 (80%), Positives = 282/333 (84%), Gaps = 10/333 (3%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
MDLDS++C S+ D D++EI HQ H ++ S + N S +
Sbjct: 1 MDLDSMDCTSTMDVTDDEEI--------HQDRHSYASVSKHHHTNNNTTNVNAAASGLLP 52
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEK 120
TSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCK RVHNRCPTCRQELGDIRCLALEK
Sbjct: 53 TTTSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKARVHNRCPTCRQELGDIRCLALEK 112
Query: 121 VAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLR 180
VAESLELPCK+ SLGCPEIFPYYSKLKHET+CNFRPY+CPYAGSECS GDI FLVAHLR
Sbjct: 113 VAESLELPCKHMSLGCPEIFPYYSKLKHETVCNFRPYSCPYAGSECSVTGDIPFLVAHLR 172
Query: 181 DDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 240
DDHKVDMH+GCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF
Sbjct: 173 DDHKVDMHSGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAF 232
Query: 241 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
LRFMGDE EARNY YSLEVG GRKLIWEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFS
Sbjct: 233 LRFMGDETEARNYNYSLEVGGYGRKLIWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFS 292
Query: 301 GGDRKELKLRVTGRIWKEQQ--NPDGGVCIPNL 331
GGDRKELKLRVTGRIWKEQQ GG CIPNL
Sbjct: 293 GGDRKELKLRVTGRIWKEQQQSGEGGGACIPNL 325
>At4g27880 putative zinc finger protein
Length = 327
Score = 554 bits (1428), Expect = e-158
Identities = 266/337 (78%), Positives = 288/337 (84%), Gaps = 15/337 (4%)
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHS-EFSSLKPRSGGNNNH---GVIGS 56
M+ DS+ECVSS+ +EI H + H S +FSS K G ++G
Sbjct: 1 METDSMECVSSTG----NEI-------HQNGNGHQSYQFSSTKTHGGAAAAAVVTNIVGP 49
Query: 57 TAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCL 116
TA APATSV+ELLECPVCT SMYPPIHQCHNGHTLCSTCK RVHNRCPTCRQELGDIRCL
Sbjct: 50 TATAPATSVYELLECPVCTYSMYPPIHQCHNGHTLCSTCKVRVHNRCPTCRQELGDIRCL 109
Query: 117 ALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLV 176
ALEKVAESLELPCK+Y+LGCPEIFPYYSKLKHE++CNFRPY+CPYAGSEC VGDI FLV
Sbjct: 110 ALEKVAESLELPCKFYNLGCPEIFPYYSKLKHESLCNFRPYSCPYAGSECGIVGDIPFLV 169
Query: 177 AHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPV 236
AHLRDDHKVDMH G TFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGM PV
Sbjct: 170 AHLRDDHKVDMHAGSTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMGPV 229
Query: 237 YMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMA 296
YMAFLRFMGDE +AR+Y+YSLEVG +GRKL WEGTPRSIRDSHRKVRDS+DGLIIQRNMA
Sbjct: 230 YMAFLRFMGDEEDARSYSYSLEVGGSGRKLTWEGTPRSIRDSHRKVRDSNDGLIIQRNMA 289
Query: 297 LFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
LFFSGGDRKELKLRVTG+IWKEQ +PD G+ IPNL S
Sbjct: 290 LFFSGGDRKELKLRVTGKIWKEQHSPDSGLSIPNLSS 326
>At3g58040 unknown protein
Length = 308
Score = 469 bits (1207), Expect = e-133
Identities = 219/278 (78%), Positives = 237/278 (84%), Gaps = 6/278 (2%)
Query: 42 KPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHN 101
KP S G IG I V+ELLECPVCTN MYPPIHQC NGHTLCS CK RV N
Sbjct: 37 KPGSAG------IGKYGIHSNNGVYELLECPVCTNLMYPPIHQCPNGHTLCSNCKLRVQN 90
Query: 102 RCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPY 161
CPTCR ELG+IRCLALEKVAESLE+PC+Y +LGC +IFPYYSKLKHE C FRPYTCPY
Sbjct: 91 TCPTCRYELGNIRCLALEKVAESLEVPCRYQNLGCHDIFPYYSKLKHEQHCRFRPYTCPY 150
Query: 162 AGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQY 221
AGSECS GDI LV HL+DDHKVDMH GCTFNHRYVKSNP EVENATWMLTVF+CFG+
Sbjct: 151 AGSECSVTGDIPTLVVHLKDDHKVDMHDGCTFNHRYVKSNPHEVENATWMLTVFNCFGRQ 210
Query: 222 FCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRK 281
FCLHFEAFQLGMAPVYMAFLRFMGDENEA+ ++YSLEVGA+GRKL W+G PRSIRDSHRK
Sbjct: 211 FCLHFEAFQLGMAPVYMAFLRFMGDENEAKKFSYSLEVGAHGRKLTWQGIPRSIRDSHRK 270
Query: 282 VRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
VRDS DGLII RN+AL+FSGGDR+ELKLRVTGRIWKE+
Sbjct: 271 VRDSQDGLIIPRNLALYFSGGDRQELKLRVTGRIWKEE 308
>At2g41980 putative RING zinc finger protein
Length = 305
Score = 457 bits (1177), Expect = e-129
Identities = 212/278 (76%), Positives = 236/278 (84%), Gaps = 5/278 (1%)
Query: 42 KPRSGGNNNHGVIGSTAIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHN 101
KP G+ + G S+ V+ELLECPVCTN MYPPIHQC NGHTLCS+CK RV N
Sbjct: 33 KPTKSGSGSIGKFHSS-----NGVYELLECPVCTNLMYPPIHQCPNGHTLCSSCKLRVQN 87
Query: 102 RCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPY 161
CPTCR ELG+IRCLALEKVAESLE+PC+Y +LGC +IFPYYSKLKHE C FR Y+CPY
Sbjct: 88 TCPTCRYELGNIRCLALEKVAESLEVPCRYQNLGCQDIFPYYSKLKHEQHCRFRSYSCPY 147
Query: 162 AGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQY 221
AGSECS GDI LV HL+DDHKVDMH GCTFNHRYVKSNP EVENATWMLTVF+CFG+
Sbjct: 148 AGSECSVTGDIPTLVDHLKDDHKVDMHDGCTFNHRYVKSNPHEVENATWMLTVFNCFGRQ 207
Query: 222 FCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRK 281
FCLHFEAFQLGMAPVYMAFLRFMGDENEA+ ++YSLEVGA+ RKL W+G PRSIRDSHRK
Sbjct: 208 FCLHFEAFQLGMAPVYMAFLRFMGDENEAKKFSYSLEVGAHSRKLTWQGIPRSIRDSHRK 267
Query: 282 VRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
VRDS DGLII RN+AL+FSG D++ELKLRVTGRIWKE+
Sbjct: 268 VRDSQDGLIIPRNLALYFSGSDKEELKLRVTGRIWKEE 305
>At5g53360 ring finger E3 ligase (SINAT5)
Length = 231
Score = 429 bits (1103), Expect = e-120
Identities = 197/219 (89%), Positives = 208/219 (94%)
Query: 115 CLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINF 174
C ALEKVAESLELPCKYY+LGC IFPYYSKLKHE+ CNFRPY+CPYAGSEC+AVGDI F
Sbjct: 12 CFALEKVAESLELPCKYYNLGCLGIFPYYSKLKHESQCNFRPYSCPYAGSECAAVGDITF 71
Query: 175 LVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMA 234
LVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVF CFGQYFCLHFEAFQLGMA
Sbjct: 72 LVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFQCFGQYFCLHFEAFQLGMA 131
Query: 235 PVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRN 294
PVYMAFLRFMGDE++ARNYTYSLEVG +GRK WEGTPRS+RDSHRKVRDSHDGLIIQRN
Sbjct: 132 PVYMAFLRFMGDEDDARNYTYSLEVGGSGRKQTWEGTPRSVRDSHRKVRDSHDGLIIQRN 191
Query: 295 MALFFSGGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
MALFFSGGD+KELKLRVTGRIWKEQQNPD GVCI ++CS
Sbjct: 192 MALFFSGGDKKELKLRVTGRIWKEQQNPDSGVCITSMCS 230
>At3g13672 seven in absentia-like protein
Length = 216
Score = 219 bits (557), Expect = 2e-57
Identities = 98/168 (58%), Positives = 130/168 (77%), Gaps = 3/168 (1%)
Query: 155 RPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTV 214
+PY CP++G++C GDI L+ HLR+DH V+M G +F+HRYV +P+ + +ATWMLT+
Sbjct: 45 KPYNCPHSGAKCDVTGDIQRLLLHLRNDHNVEMSDGRSFSHRYVHHDPKHLHHATWMLTL 104
Query: 215 FHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRS 274
C G+ FCL+FEAF L P+YMAF++FMGDE EA +++YSL+VG NGRKL W+G PRS
Sbjct: 105 LDCCGRKFCLYFEAFHLRKTPMYMAFMQFMGDEEEAMSFSYSLQVGGNGRKLTWQGVPRS 164
Query: 275 IRDSHRKVRDSHDGLIIQRNMALFFSGGDR---KELKLRVTGRIWKEQ 319
IRDSH+ VRDS DGLII R +ALFFS + KELKL+V+GR+W+EQ
Sbjct: 165 IRDSHKTVRDSQDGLIITRKLALFFSTDNNTTDKELKLKVSGRVWREQ 212
Score = 31.2 bits (69), Expect = 0.82
Identities = 13/25 (52%), Positives = 17/25 (68%)
Query: 65 VHELLECPVCTNSMYPPIHQCHNGH 89
VHELL+ PV TN + I++C N H
Sbjct: 14 VHELLDFPVHTNQISSAIYECPNDH 38
>At5g37930 unknown protein (At5g37930)
Length = 349
Score = 113 bits (283), Expect = 1e-25
Identities = 61/190 (32%), Positives = 93/190 (48%), Gaps = 20/190 (10%)
Query: 11 SSDGMDEDEIQHRILHPHHQQHHHHSEFSS-------------LKPRSGGNNNHGVIGST 57
SS DEDE Q++ + P + S+ S + +S ++ + T
Sbjct: 45 SSGEEDEDETQNQGMRPESEDRGSTSDDSDREVVIEERRFGKFVNSQSSSSSKDSPLSVT 104
Query: 58 AIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLA 117
+ P ++L+CP+C + PI QC NGH C+ C T+V NRCP+C +G +RC A
Sbjct: 105 LLDP-----DVLDCPICCEPLKIPIFQCDNGHLACTLCCTKVRNRCPSCTLPIGYVRCRA 159
Query: 118 LEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVA 177
+EKV E+ + C GC E Y ++ HE +C F P +CP +C G L
Sbjct: 160 MEKVIEASRVSCLNAKYGCKESTSYGNRFSHEQVCVFTPCSCPIL--DCHYTGYYKDLNN 217
Query: 178 HLRDDHKVDM 187
H+R +HK D+
Sbjct: 218 HVRAEHKDDL 227
>At5g37870 putative protein
Length = 281
Score = 112 bits (280), Expect = 3e-25
Identities = 51/128 (39%), Positives = 73/128 (56%), Gaps = 3/128 (2%)
Query: 67 ELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLE 126
++L+CP+C ++ P+ QC NGH CS+C ++ N+CP C +G IRC A+E+V ES+
Sbjct: 41 DILDCPICYQALKIPVFQCGNGHLACSSCCPKLRNKCPACALPVGHIRCRAMERVLESVL 100
Query: 127 LPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVD 186
+PC+Y LGC + Y + HE ICNF P +CP G C+ G L H H
Sbjct: 101 VPCRYADLGCTKTIYYGRESTHEKICNFSPCSCPVQG--CNYTGSYKDLYEHYDLTHSTG 158
Query: 187 MHTGCTFN 194
T +FN
Sbjct: 159 -STAYSFN 165
>At5g37890 unknown protein
Length = 286
Score = 100 bits (249), Expect = 1e-21
Identities = 61/205 (29%), Positives = 93/205 (44%), Gaps = 11/205 (5%)
Query: 67 ELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLE 126
E+L+CP+C + PI QC NGH CS+C +++N+CP C +G RC A+E V ES+
Sbjct: 47 EILDCPICYEAFTIPIFQCDNGHLACSSCCPKLNNKCPACTSPVGHNRCRAMESVLESIL 106
Query: 127 LPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVD 186
+PC LGC + Y +L HE C F CP +C+ L H R H ++
Sbjct: 107 IPCPNAKLGCKKNVSYGKELTHEKECMFSHCACP--ALDCNYTSSYKDLYTHYRITH-ME 163
Query: 187 MHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAP--VYMAFLRFM 244
++ TF S + + T ++ H A Q P VY+
Sbjct: 164 INQINTFICDIPLSVRMNISKKILIRT------EHLTNHLFAVQCFREPYGVYVTVSCIA 217
Query: 245 GDENEARNYTYSLEVGANGRKLIWE 269
E Y+Y+L +G +I++
Sbjct: 218 PSSPELSQYSYALSYTVDGHTVIYQ 242
>At1g66660 hypothetical protein
Length = 412
Score = 99.0 bits (245), Expect = 3e-21
Identities = 44/112 (39%), Positives = 65/112 (57%), Gaps = 2/112 (1%)
Query: 67 ELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLE 126
++L+CP C + PI+QC NGH CS+C +++ +C CR +GDIRC A+EKV E+
Sbjct: 166 DVLDCPTCCEPLKRPIYQCSNGHLACSSCCQKLNKKCSFCRCNIGDIRCRAMEKVIEASI 225
Query: 127 LPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAH 178
+PC GC E Y ++ HE +C F +CP S C+ V + L +H
Sbjct: 226 VPCPNAKHGCKETTTYCNQSSHEKVCKFVRCSCPV--SNCNYVSSYSNLKSH 275
>At1g66620 hypothetical protein
Length = 313
Score = 96.7 bits (239), Expect = 2e-20
Identities = 47/131 (35%), Positives = 72/131 (54%), Gaps = 4/131 (3%)
Query: 55 GSTAIAPATSVHEL--LECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGD 112
G A+A + ++ EL L+CP+C +++ PI QC NGH CS+C T++ N+CP+C +G+
Sbjct: 26 GGDAVARSGTLFELDLLDCPICCHALTSPIFQCDNGHIACSSCCTKLRNKCPSCALPIGN 85
Query: 113 IRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDI 172
R +E+V E++ + C GC E F Y +L HE C F CP C+ G
Sbjct: 86 FRSRIMERVVEAVMVTCPNVKHGCTEKFSYGKELIHEKDCRFALCYCP--APNCNYSGVY 143
Query: 173 NFLVAHLRDDH 183
L +H +H
Sbjct: 144 KDLYSHFYVNH 154
>At5g62800 SIAH1 - like protein
Length = 348
Score = 96.3 bits (238), Expect = 2e-20
Identities = 67/256 (26%), Positives = 113/256 (43%), Gaps = 30/256 (11%)
Query: 67 ELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPT--CRQELGDIRCLALEKVAES 124
++L+CPVC + P QC +GH +C+ C +V N+CP C +G+ RC A+E+V ES
Sbjct: 39 DVLDCPVCFEPLTIPTFQCDDGHIVCNFCFAKVSNKCPGPGCDLPIGNKRCFAMERVLES 98
Query: 125 LELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHK 184
+PC+ GC + Y HE CN+ +CP EC+ G N + H H
Sbjct: 99 AFVPCQNTEFGCTKSVSYEKVSSHEKECNYSQCSCP--NLECNYTGSYNIIYGHFMRRHL 156
Query: 185 VD---MHTGCTFNHRYVKSNPREVENATW-----MLTVFHCFGQYFCLHFEAFQLGMAPV 236
+ + + ++ V N +E + W +L V CF + V
Sbjct: 157 YNSTIVSSKWGYSTVDVLINIKEKVSVLWESRQKLLFVVQCFKE------------RHGV 204
Query: 237 YMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTP-RSIRDSHRKVRDSHDGLIIQRNM 295
Y+ R +E + ++Y L +G + +E + + + + ++ D D + N
Sbjct: 205 YVTVRRIAPPASEFKKFSYRLSYSIDGHNVTYESPEVKRLLEVNSQIPD--DSFMFVPNC 262
Query: 296 ALFFSGGDRKELKLRV 311
L G+ ELKL +
Sbjct: 263 LLH---GEMLELKLGI 275
>At1g66630 hypothetical protein
Length = 303
Score = 96.3 bits (238), Expect = 2e-20
Identities = 53/148 (35%), Positives = 72/148 (47%), Gaps = 5/148 (3%)
Query: 42 KPRSGGNNNHGVIGSTAIAPATSVH-ELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVH 100
+P S N GS AT + +LL+CP+C + + PI+QC NGH CS+C +V
Sbjct: 19 RPVSMENVGGTASGSEVARSATLLELDLLDCPICYHKLGAPIYQCDNGHIACSSCCKKVK 78
Query: 101 NRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPY--YSKLKHETICNFRPYT 158
+CP C +G R LEK+ E++ + C GC E PY S+ HE +C F
Sbjct: 79 YKCPYCSLRIGFFRSRILEKIVEAVVVSCPNAKYGCTEKIPYDNESESAHERVCEFTLCY 138
Query: 159 CPYAGSECSAVGDINFLVAHLRDDHKVD 186
CP EC G L H +HK D
Sbjct: 139 CP--EPECKYTGVYTDLYRHYHAEHKTD 164
>At1g66650 hypothetical protein
Length = 329
Score = 95.9 bits (237), Expect = 3e-20
Identities = 45/119 (37%), Positives = 68/119 (56%), Gaps = 3/119 (2%)
Query: 68 LLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLEL 127
+LECP C + + PI QC+NGH C C ++ RC C+ +GD+RC A+EKV ++ +
Sbjct: 83 VLECPNCFDPLKKPIFQCNNGHLACFLCCIKLKKRCSFCKLPIGDVRCRAMEKVIKAGLV 142
Query: 128 PCKYYSLGCPEIFPYYSKLK-HETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKV 185
C GC + Y ++L+ HE +C F P +CP +C+ +G L+ H R HKV
Sbjct: 143 SCSNAIYGCKQSTTYGNQLQSHEKVCVFAPCSCPI--KDCNYIGFYKDLINHFRATHKV 199
>At5g37910 putative protein
Length = 276
Score = 95.1 bits (235), Expect = 5e-20
Identities = 43/121 (35%), Positives = 63/121 (51%), Gaps = 2/121 (1%)
Query: 67 ELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLE 126
++L+CP+C ++ PI QC NGH C +C ++ N+CP C +G R A+E V ES+
Sbjct: 34 DILDCPICCEALTSPIFQCDNGHLACGSCCPKLSNKCPACTLPVGHSRSRAMESVLESIL 93
Query: 127 LPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVD 186
+PC GC + F Y + HE C F +CP S C G L AH + H +
Sbjct: 94 IPCPNVRFGCTKSFFYGKESAHEKECIFSQCSCP--SSVCDYTGSYKDLYAHYKLTHSTN 151
Query: 187 M 187
+
Sbjct: 152 I 152
>At1g66610 hypothetical protein
Length = 366
Score = 68.6 bits (166), Expect = 5e-12
Identities = 27/63 (42%), Positives = 40/63 (62%)
Query: 67 ELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLE 126
+LL+CP+C N++ PI QC GH CS+C T V N+CP C +G+ R +E+V E+
Sbjct: 52 DLLDCPICCNALTIPIFQCDKGHIACSSCCTNVSNKCPYCSLAIGNYRSRIMERVVEAFI 111
Query: 127 LPC 129
+ C
Sbjct: 112 VRC 114
>At5g37900 putative protein
Length = 241
Score = 39.7 bits (91), Expect = 0.002
Identities = 25/81 (30%), Positives = 33/81 (39%), Gaps = 2/81 (2%)
Query: 103 CPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYA 162
CP C + L +E + ES+ + C GC E F Y K HE C F +CP
Sbjct: 35 CPICCEGLTCPIFQPMENILESILVTCPNDMFGCTESFLYGKKSTHEEECIFSLCSCP-- 92
Query: 163 GSECSAVGDINFLVAHLRDDH 183
+C G L H + H
Sbjct: 93 SLDCEYSGRYEDLYDHYKLTH 113
>At1g18910 hypothetical protein
Length = 195
Score = 34.7 bits (78), Expect = 0.075
Identities = 20/69 (28%), Positives = 33/69 (46%), Gaps = 8/69 (11%)
Query: 71 CPVCTNSMYP---PIHQCHNGHTLCSTC---KTRVHNRCPTCRQELGDIRCL--ALEKVA 122
CP+C ++ P+ GH + STC T H CP C + LGD++ L+ +
Sbjct: 89 CPICHEYIFTSNSPVKALPCGHVMHSTCFQEYTCSHYTCPICSKSLGDMQVYFRMLDALL 148
Query: 123 ESLELPCKY 131
++P +Y
Sbjct: 149 AEQKMPDEY 157
>At1g74760
Length = 255
Score = 34.3 bits (77), Expect = 0.097
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 6/50 (12%)
Query: 71 CPVCTNSMYP---PIHQCHNGHTLCSTC---KTRVHNRCPTCRQELGDIR 114
CP+C ++ P+ GH + STC T H CP C + LGD++
Sbjct: 149 CPICHEYIFTSSSPVKALPCGHLMHSTCFQEYTCSHYTCPVCSKSLGDMQ 198
>At3g18290 zinc finger protein, putative
Length = 1254
Score = 33.9 bits (76), Expect = 0.13
Identities = 26/102 (25%), Positives = 41/102 (39%), Gaps = 9/102 (8%)
Query: 71 CPVCTNSMYPPIHQC------HNGHTLCSTCKTRVHNRCPTCRQELGDIRCL--ALEKVA 122
CP+C ++ H H+ C T H CP C + LGD+ L+ +
Sbjct: 1135 CPICCEFLFTSSEAVRALPCGHYMHSACFQAYTCSHYTCPICGKSLGDMAVYFGMLDALL 1194
Query: 123 ESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGS 164
+ ELP + Y C +I + K T ++ + C GS
Sbjct: 1195 AAEELP-EEYKNRCQDILCNDCERKGTTRFHWLYHKCGSCGS 1235
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,609,312
Number of Sequences: 26719
Number of extensions: 376884
Number of successful extensions: 1587
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 1445
Number of HSP's gapped (non-prelim): 148
length of query: 333
length of database: 11,318,596
effective HSP length: 100
effective length of query: 233
effective length of database: 8,646,696
effective search space: 2014680168
effective search space used: 2014680168
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC134822.10


