

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.21 - phase: 0
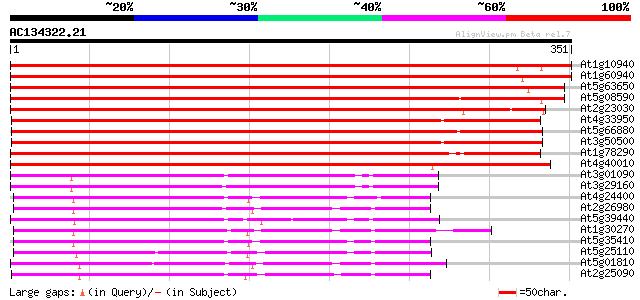
(351 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g10940 Ser/Thr kinase 602 e-172
At1g60940 putative serine/threonine-protein kinase 594 e-170
At5g63650 serine/threonine-protein kinase 579 e-166
At5g08590 serine/threonine-protein kinase 572 e-163
At2g23030 putative protein kinase 498 e-141
At4g33950 protein kinase like protein 495 e-140
At5g66880 protein kinase 481 e-136
At3g50500 protein kinase SPK-2 476 e-135
At1g78290 protein kinase 1 like protein 474 e-134
At4g40010 putative serine/threonine protein kinase 466 e-132
At3g01090 putative SNF1-related protein kinase 219 2e-57
At3g29160 SNF1-like protein kinase (AKin11) 215 3e-56
At4g24400 serine/threonine kinase-like protein 207 5e-54
At2g26980 putative protein kinase (At2g26980) 206 1e-53
At5g39440 serine/threonine-specific protein kinase -like protein 205 3e-53
At1g30270 serine/threonine kinase, putative 204 6e-53
At5g35410 serine/threonine protein kinase SOS2 (gb|AAF62923.1) 202 2e-52
At5g25110 serine/threonine protein kinase-like protein 201 6e-52
At5g01810 serine/threonine protein kinase ATPK10 200 1e-51
At2g25090 CBL-interacting protein kinase 16 (CIPK16) 200 1e-51
>At1g10940 Ser/Thr kinase
Length = 363
Score = 602 bits (1551), Expect = e-172
Identities = 293/363 (80%), Positives = 326/363 (89%), Gaps = 12/363 (3%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE+VKDIG+GNFGVARLM+ K++K+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYELVKDIGAGNFGVARLMKVKNSKELVAMKYIERGPKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTHL I MEYAAGGELF+RICSAGRFSEDEARYFFQQLISGVSYCH+MQIC
Sbjct: 61 IRFKEVVLTPTHLAIAMEYAAGGELFERICSAGRFSEDEARYFFQQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNFRKTI +IMAVQYKIPDYVHISQ+C++LLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFRKTIQKIMAVQYKIPDYVHISQDCKNLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+ +RITI EIK H WFLKNLPRELTE AQA Y++KENPT+SLQ++E+IMKIV +AK
Sbjct: 241 FVANSLKRITIAEIKKHSWFLKNLPRELTETAQAAYFKKENPTFSLQTVEEIMKIVADAK 300
Query: 301 NPPQASRSVGGFGWGG-------EEDDDEINEAEAELE-----EDEYEKRVKEAQESGEI 348
PP SRS+GGFGWGG EED +++ E E E+E EDEY+K VKE SGE+
Sbjct: 301 TPPPVSRSIGGFGWGGNGDADGKEEDAEDVEEEEEEVEEEEDDEDEYDKTVKEVHASGEV 360
Query: 349 HVN 351
++
Sbjct: 361 RIS 363
>At1g60940 putative serine/threonine-protein kinase
Length = 361
Score = 594 bits (1532), Expect = e-170
Identities = 292/361 (80%), Positives = 324/361 (88%), Gaps = 10/361 (2%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYE+VKDIG+GNFGVARLMR K++K+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYELVKDIGAGNFGVARLMRVKNSKELVAMKYIERGPKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEVVLTPTH+ I MEYAAGGELF+RICSAGRFSEDEARYFFQQLISGVSYCH+MQIC
Sbjct: 61 IRFKEVVLTPTHIAIAMEYAAGGELFERICSAGRFSEDEARYFFQQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHS PKSTVGTPAYIAPEVLSR EYDGK+
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSMPKSTVGTPAYIAPEVLSRGEYDGKM 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFEDQ+DPKNF+KTI RIMAV+YKIPDYVHISQ+C+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDQEDPKNFKKTIQRIMAVKYKIPDYVHISQDCKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + +RITI +IK HPWFLKNLPRELTE+AQA Y+RKENPT+SLQS+E+IMKIVEEAK
Sbjct: 241 FVTNSNKRITIGDIKKHPWFLKNLPRELTEIAQAAYFRKENPTFSLQSVEEIMKIVEEAK 300
Query: 301 NPPQASRSVGGFGWGGEED--------DDEINEAEAEL-EEDEYEKRVKEAQES-GEIHV 350
P + SRS+G FGWGG ED ++E+ E E E EEDEY+K VK+ S GE+ V
Sbjct: 301 TPARVSRSIGAFGWGGGEDAEGKEEDAEEEVEEVEEEEDEEDEYDKTVKQVHASMGEVRV 360
Query: 351 N 351
+
Sbjct: 361 S 361
>At5g63650 serine/threonine-protein kinase
Length = 360
Score = 579 bits (1493), Expect = e-166
Identities = 284/356 (79%), Positives = 312/356 (86%), Gaps = 9/356 (2%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KYEVVKD+G+GNFGVARL+RHK+TK+LVAMKYIERG KIDENVAREIINHRSLRHPNI
Sbjct: 1 MDKYEVVKDLGAGNFGVARLLRHKETKELVAMKYIERGRKIDENVAREIINHRSLRHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LTPTHL IVMEYA+GGELF+RIC+AGRFSE EARYFFQQLI GV YCHS+QIC
Sbjct: 61 IRFKEVILTPTHLAIVMEYASGGELFERICNAGRFSEAEARYFFQQLICGVDYCHSLQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAP LKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGK
Sbjct: 121 HRDLKLENTLLDGSPAPLLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKH 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVG YPFED DDP+NFRKTI RIMAVQYKIPDYVHISQECRHLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGGYPFEDPDDPRNFRKTIQRIMAVQYKIPDYVHISQECRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + A+RIT+KEIK HPW+LKNLP+ELTE AQA YY++E P++SLQS+EDIMKIV EA+
Sbjct: 241 FVTNSAKRITLKEIKKHPWYLKNLPKELTEPAQAAYYKRETPSFSLQSVEDIMKIVGEAR 300
Query: 301 NPPQASRSVGGFGWGGEEDDDEI---------NEAEAELEEDEYEKRVKEAQESGE 347
NP +S +V GF E+ +DE+ E E E EEDEYEK VKEA E
Sbjct: 301 NPAPSSNAVKGFDDDEEDVEDEVEEEEEEEEEEEEEEEEEEDEYEKHVKEAHSCQE 356
>At5g08590 serine/threonine-protein kinase
Length = 353
Score = 572 bits (1474), Expect = e-163
Identities = 280/350 (80%), Positives = 314/350 (89%), Gaps = 4/350 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
M+KY+VVKD+G+GNFGVARL+RHKDTK+LVAMKYIERG KIDENVAREIINHRSL+HPNI
Sbjct: 1 MDKYDVVKDLGAGNFGVARLLRHKDTKELVAMKYIERGRKIDENVAREIINHRSLKHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LTPTHL IVMEYA+GGELFDRIC+AGRFSE EARYFFQQLI GV YCHS+QIC
Sbjct: 61 IRFKEVILTPTHLAIVMEYASGGELFDRICTAGRFSEAEARYFFQQLICGVDYCHSLQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAP LKICDFGYSKSS+LHSRPKSTVGTPAYIAPEVLSRREYDGK
Sbjct: 121 HRDLKLENTLLDGSPAPLLKICDFGYSKSSILHSRPKSTVGTPAYIAPEVLSRREYDGKH 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED +DPKNFRKTI RIMAVQYKIPDYVHISQEC+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPNDPKNFRKTIQRIMAVQYKIPDYVHISQECKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FV + A+RIT+KEIK+HPW+LKNLP+EL E AQA YY+++ ++SLQS+EDIMKIV EA+
Sbjct: 241 FVTNSAKRITLKEIKNHPWYLKNLPKELLESAQAAYYKRDT-SFSLQSVEDIMKIVGEAR 299
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELE---EDEYEKRVKEAQESGE 347
NP ++ +V G G +E+++E EAE E E EDEYEK VKEAQ E
Sbjct: 300 NPAPSTSAVKSSGSGADEEEEEDVEAEVEEEEDDEDEYEKHVKEAQSCQE 349
>At2g23030 putative protein kinase
Length = 339
Score = 498 bits (1283), Expect = e-141
Identities = 249/339 (73%), Positives = 282/339 (82%), Gaps = 5/339 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
MEKYE+VKD+G GNFG+ARLMR+K T +LVA+K+I+RG+KIDENVAREIINHR+L HPNI
Sbjct: 1 MEKYEMVKDLGFGNFGLARLMRNKQTNELVAVKFIDRGYKIDENVAREIINHRALNHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
+RFKEVVLTPTHLGIVMEYAAGGELF+RI S GRFSE EARYFFQQLI GV Y H++QIC
Sbjct: 61 VRFKEVVLTPTHLGIVMEYAAGGELFERISSVGRFSEAEARYFFQQLICGVHYLHALQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSPAPRLKICDFGYSKSS+LHS PKSTVGTPAYIAPEV R EYDGK
Sbjct: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSVLHSNPKSTVGTPAYIAPEVFCRSEYDGKS 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
DVWSCGV LYVMLVGAYPFED DP+NFRKT+ +IMAV YKIP YVHIS++CR LLSRI
Sbjct: 181 VDVWSCGVALYVMLVGAYPFEDPKDPRNFRKTVQKIMAVNYKIPGYVHISEDCRKLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENP--TYSLQSIEDIMKIVEE 298
FVA+P R T+KEIKSH WFLKNLPREL E AQA+YY++ +S Q +E+IMKIV E
Sbjct: 241 FVANPLHRSTLKEIKSHAWFLKNLPRELKEPAQAIYYQRNVNLINFSPQRVEEIMKIVGE 300
Query: 299 AKNPPQASRSVGGFGWGGEEDDDEINEAEAELEE--DEY 335
A+ P SR V G ++DDDE +A EE D+Y
Sbjct: 301 ARTIPNLSRPVESLG-SDKKDDDEEEYLDANDEEWYDDY 338
>At4g33950 protein kinase like protein
Length = 362
Score = 495 bits (1275), Expect = e-140
Identities = 240/332 (72%), Positives = 287/332 (86%), Gaps = 2/332 (0%)
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNII 61
++YE+VKDIGSGNFGVARLMR K + +LVA+KYIERG KIDENV REIINHRSLRHPNI+
Sbjct: 19 DRYELVKDIGSGNFGVARLMRDKQSNELVAVKYIERGEKIDENVKREIINHRSLRHPNIV 78
Query: 62 RFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICH 121
RFKEV+LTPTHL IVMEYA+GGELF+RIC+AGRFSEDEAR+FFQQLISGVSYCH+MQ+CH
Sbjct: 79 RFKEVILTPTHLAIVMEYASGGELFERICNAGRFSEDEARFFFQQLISGVSYCHAMQVCH 138
Query: 122 RDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKLA 181
RDLKLENTLLDGSPAPRLKICDFGYSKSS+LHS+PKSTVGTPAYIAPEVL ++EYDGK+A
Sbjct: 139 RDLKLENTLLDGSPAPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLLKKEYDGKVA 198
Query: 182 DVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRIF 241
DVWSCGVTLYVMLVGAYPFED ++PKNFRKTI+RI+ VQY IPDYVHIS ECRHL+SRIF
Sbjct: 199 DVWSCGVTLYVMLVGAYPFEDPEEPKNFRKTIHRILNVQYAIPDYVHISPECRHLISRIF 258
Query: 242 VASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAKN 301
VA PA+RI+I EI++H WFLKNLP +L + + + QSIE+IM+I+ EA
Sbjct: 259 VADPAKRISIPEIRNHEWFLKNLPADLMN-DNTMTTQFDESDQPGQSIEEIMQIIAEATV 317
Query: 302 PPQASRSVGGFGWGGEEDDDEINE-AEAELEE 332
PP ++++ + G + DD++ E E++L++
Sbjct: 318 PPAGTQNLNHYLTGSLDIDDDMEEDLESDLDD 349
>At5g66880 protein kinase
Length = 361
Score = 481 bits (1239), Expect = e-136
Identities = 234/332 (70%), Positives = 281/332 (84%), Gaps = 1/332 (0%)
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNII 61
++Y+ VKDIGSGNFGVARLMR K TK+LVA+KYIERG KIDENV REIINHRSLRHPNI+
Sbjct: 20 DRYDFVKDIGSGNFGVARLMRDKLTKELVAVKYIERGDKIDENVQREIINHRSLRHPNIV 79
Query: 62 RFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICH 121
RFKEV+LTPTHL I+MEYA+GGEL++RIC+AGRFSEDEAR+FFQQL+SGVSYCHSMQICH
Sbjct: 80 RFKEVILTPTHLAIIMEYASGGELYERICNAGRFSEDEARFFFQQLLSGVSYCHSMQICH 139
Query: 122 RDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKLA 181
RDLKLENTLLDGSPAPRLKICDFGYSKSS+LHS+PKSTVGTPAYIAPEVL R+EYDGK+A
Sbjct: 140 RDLKLENTLLDGSPAPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLLRQEYDGKIA 199
Query: 182 DVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRIF 241
DVWSCGVTLYVMLVGAYPFED ++P+++RKTI RI++V+Y IPD + IS EC HL+SRIF
Sbjct: 200 DVWSCGVTLYVMLVGAYPFEDPEEPRDYRKTIQRILSVKYSIPDDIRISPECCHLISRIF 259
Query: 242 VASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAKN 301
VA PA RI+I EIK+H WFLKNLP +L + +E P +QS++ IM+I+ EA
Sbjct: 260 VADPATRISIPEIKTHSWFLKNLPADLMNESNTGSQFQE-PEQPMQSLDTIMQIISEATI 318
Query: 302 PPQASRSVGGFGWGGEEDDDEINEAEAELEED 333
P +R + F + DD++++ ++E E D
Sbjct: 319 PAVRNRCLDDFMTDNLDLDDDMDDFDSESEID 350
>At3g50500 protein kinase SPK-2
Length = 362
Score = 476 bits (1226), Expect = e-135
Identities = 232/332 (69%), Positives = 279/332 (83%), Gaps = 1/332 (0%)
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNII 61
++Y+ VKDIGSGNFGVARLM + TK+LVA+KYIERG KIDENV REIINHRSLRHPNI+
Sbjct: 21 DRYDFVKDIGSGNFGVARLMTDRVTKELVAVKYIERGEKIDENVQREIINHRSLRHPNIV 80
Query: 62 RFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICH 121
RFKEV+LTP+HL IVMEYAAGGEL++RIC+AGRFSEDEAR+FFQQLISGVSYCH+MQICH
Sbjct: 81 RFKEVILTPSHLAIVMEYAAGGELYERICNAGRFSEDEARFFFQQLISGVSYCHAMQICH 140
Query: 122 RDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKLA 181
RDLKLENTLLDGSPAPRLKICDFGYSKSS+LHS+PKSTVGTPAYIAPE+L R+EYDGKLA
Sbjct: 141 RDLKLENTLLDGSPAPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEILLRQEYDGKLA 200
Query: 182 DVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRIF 241
DVWSCGVTLYVMLVGAYPFED +P+++RKTI RI++V Y IP+ +H+S ECRHL+SRIF
Sbjct: 201 DVWSCGVTLYVMLVGAYPFEDPQEPRDYRKTIQRILSVTYSIPEDLHLSPECRHLISRIF 260
Query: 242 VASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAKN 301
VA PA RITI EI S WFLKNLP +L + + + + P +QS++ IM+I+ EA
Sbjct: 261 VADPATRITIPEITSDKWFLKNLPGDLMD-ENRMGSQFQEPEQPMQSLDTIMQIISEATI 319
Query: 302 PPQASRSVGGFGWGGEEDDDEINEAEAELEED 333
P +R + F + DD++++ ++E E D
Sbjct: 320 PTVRNRCLDDFMADNLDLDDDMDDFDSESEID 351
>At1g78290 protein kinase 1 like protein
Length = 343
Score = 474 bits (1221), Expect = e-134
Identities = 232/332 (69%), Positives = 276/332 (82%), Gaps = 6/332 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
ME+YE+VKDIGSGNFGVA+L+R K +K+L A+K+IERG KIDE+V REI+NHRSL HPNI
Sbjct: 1 MERYEIVKDIGSGNFGVAKLVRDKFSKELFAVKFIERGQKIDEHVQREIMNHRSLIHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV+LT THL +VMEYAAGGELF RICSAGRFSEDEAR+FFQQLISGV+YCHS+QIC
Sbjct: 61 IRFKEVLLTATHLALVMEYAAGGELFGRICSAGRFSEDEARFFFQQLISGVNYCHSLQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGS APR+KICDFGYSKS +LHS+PK+TVGTPAYIAPEVLS +EYDGK+
Sbjct: 121 HRDLKLENTLLDGSEAPRVKICDFGYSKSGVLHSQPKTTVGTPAYIAPEVLSTKEYDGKI 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED DPK+FRKTI RI+ QY IPDYV +S ECRHLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPSDPKDFRKTIGRILKAQYAIPDYVRVSDECRHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVEEAK 300
FVA+P +RITI+EIK+H WFLKNLP E+ E + + P S Q++E+I+ I+EEA+
Sbjct: 241 FVANPEKRITIEEIKNHSWFLKNLPVEMYEGSLMM----NGP--STQTVEEIVWIIEEAR 294
Query: 301 NPPQASRSVGGFGWGGEEDDDEINEAEAELEE 332
P + + G G G + I + +L++
Sbjct: 295 KPITVATGLAGAGGSGGSSNGAIGSSSMDLDD 326
>At4g40010 putative serine/threonine protein kinase
Length = 350
Score = 466 bits (1199), Expect = e-132
Identities = 227/342 (66%), Positives = 277/342 (80%), Gaps = 4/342 (1%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNI 60
ME+Y++++D+GSGNFGVA+L+R K + A+KYIERG KIDE+V REIINHR L+HPNI
Sbjct: 1 MERYDILRDLGSGNFGVAKLVREKANGEFYAVKYIERGLKIDEHVQREIINHRDLKHPNI 60
Query: 61 IRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQIC 120
IRFKEV +TPTHL IVMEYAAGGELF+RIC+AGRFSEDE RY+F+QLISGVSYCH+MQIC
Sbjct: 61 IRFKEVFVTPTHLAIVMEYAAGGELFERICNAGRFSEDEGRYYFKQLISGVSYCHAMQIC 120
Query: 121 HRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKL 180
HRDLKLENTLLDGSP+ LKICDFGYSKSS+LHS+PKSTVGTPAY+APEVLSR+EY+GK+
Sbjct: 121 HRDLKLENTLLDGSPSSHLKICDFGYSKSSVLHSQPKSTVGTPAYVAPEVLSRKEYNGKI 180
Query: 181 ADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRI 240
ADVWSCGVTLYVMLVGAYPFED +DP+N R TI RI++V Y IPDYV IS EC+HLLSRI
Sbjct: 181 ADVWSCGVTLYVMLVGAYPFEDPEDPRNIRNTIQRILSVHYTIPDYVRISSECKHLLSRI 240
Query: 241 FVASPARRITIKEIKSHPWFLKN---LPRELTEMAQAVYYRKENPTYSLQSIEDIMKIVE 297
FVA P +RIT+ EI+ HPWFLK +P E + V +E QS+E+I+KI+E
Sbjct: 241 FVADPDKRITVPEIEKHPWFLKGPLVVPPEEEKCDNGVEEEEEEEEKCRQSVEEIVKIIE 300
Query: 298 EAKNPPQASRSVGGFGW-GGEEDDDEINEAEAELEEDEYEKR 338
EA+ + + GG G G D D+I++A+ + D+ E+R
Sbjct: 301 EARKGVNGTDNNGGLGLIDGSIDLDDIDDADIYDDVDDDEER 342
>At3g01090 putative SNF1-related protein kinase
Length = 512
Score = 219 bits (557), Expect = 2e-57
Identities = 122/272 (44%), Positives = 160/272 (57%), Gaps = 12/272 (4%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIER----GHKIDENVAREIINHRSLR 56
+ Y++ + +G G+FG ++ H T VA+K + R +++E V REI R
Sbjct: 16 LPNYKLGRTLGIGSFGRVKIAEHALTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLFM 75
Query: 57 HPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHS 116
HP+IIR EV+ TPT + +VMEY GELFD I GR EDEAR FFQQ+ISGV YCH
Sbjct: 76 HPHIIRLYEVIETPTDIYLVMEYVNSGELFDYIVEKGRLQEDEARNFFQQIISGVEYCHR 135
Query: 117 MQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREY 176
+ HRDLK EN LLD +KI DFG S K++ G+P Y APEV+S + Y
Sbjct: 136 NMVVHRDLKPENLLLDSK--CNVKIADFGLSNIMRDGHFLKTSCGSPNYAAPEVISGKLY 193
Query: 177 DGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHL 236
G DVWSCGV LY +L G PF+D++ P F+K I Y +P H+S R L
Sbjct: 194 AGPEVDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGI----YTLPS--HLSPGARDL 247
Query: 237 LSRIFVASPARRITIKEIKSHPWFLKNLPREL 268
+ R+ V P +R+TI EI+ HPWF +LPR L
Sbjct: 248 IPRMLVVDPMKRVTIPEIRQHPWFQAHLPRYL 279
>At3g29160 SNF1-like protein kinase (AKin11)
Length = 512
Score = 215 bits (548), Expect = 3e-56
Identities = 121/272 (44%), Positives = 159/272 (57%), Gaps = 12/272 (4%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIER----GHKIDENVAREIINHRSLR 56
+ Y++ K +G G+FG ++ H T VA+K + R +++E V REI R
Sbjct: 17 LPNYKLGKTLGIGSFGKVKIAEHVVTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLFM 76
Query: 57 HPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHS 116
HP+IIR EV+ T + + +VMEY GELFD I GR EDEAR FFQQ+ISGV YCH
Sbjct: 77 HPHIIRQYEVIETTSDIYVVMEYVKSGELFDYIVEKGRLQEDEARNFFQQIISGVEYCHR 136
Query: 117 MQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREY 176
+ HRDLK EN LLD +KI DFG S K++ G+P Y APEV+S + Y
Sbjct: 137 NMVVHRDLKPENLLLDS--RCNIKIADFGLSNVMRDGHFLKTSCGSPNYAAPEVISGKLY 194
Query: 177 DGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHL 236
G DVWSCGV LY +L G PF+D++ P F+K I Y +P H+S E R L
Sbjct: 195 AGPEVDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGI----YTLPS--HLSSEARDL 248
Query: 237 LSRIFVASPARRITIKEIKSHPWFLKNLPREL 268
+ R+ + P +RITI EI+ H WF +LPR L
Sbjct: 249 IPRMLIVDPVKRITIPEIRQHRWFQTHLPRYL 280
>At4g24400 serine/threonine kinase-like protein
Length = 445
Score = 207 bits (528), Expect = 5e-54
Identities = 116/270 (42%), Positives = 162/270 (59%), Gaps = 21/270 (7%)
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHP 58
KYE+ + IG G F + ++ +T + VAMK ++R K+ + + REI + +RHP
Sbjct: 8 KYELGRTIGEGTFAKVKFAQNTETGESVAMKIVDRSTIIKRKMVDQIKREISIMKLVRHP 67
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
++R EV+ + T + I++EY GGELFD+I GR SE EAR +F QLI GV YCHS
Sbjct: 68 CVVRLYEVLASRTKIYIILEYITGGELFDKIVRNGRLSESEARKYFHQLIDGVDYCHSKG 127
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYSK-----SSLLHSRPKSTVGTPAYIAPEVLSR 173
+ HRDLK EN LLD LKI DFG S ++L K+T GTP Y+APEVLS
Sbjct: 128 VYHRDLKPENLLLDSQ--GNLKISDFGLSALPEQGVTIL----KTTCGTPNYVAPEVLSH 181
Query: 174 REYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQEC 233
+ Y+G +AD+WSCGV LYV++ G PF++ D P + K I ++ P Y +
Sbjct: 182 KGYNGAVADIWSCGVILYVLMAGYLPFDEMDLPTLYSK----IDKAEFSCPSYFALG--A 235
Query: 234 RHLLSRIFVASPARRITIKEIKSHPWFLKN 263
+ L++RI +P RITI EI+ WFLK+
Sbjct: 236 KSLINRILDPNPETRITIAEIRKDEWFLKD 265
>At2g26980 putative protein kinase (At2g26980)
Length = 441
Score = 206 bits (525), Expect = 1e-53
Identities = 115/272 (42%), Positives = 161/272 (58%), Gaps = 23/272 (8%)
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHP 58
KYEV + IG G F + R+ +T + VA+K +++ HK+ E + REI + ++HP
Sbjct: 13 KYEVGRTIGEGTFAKVKFARNSETGEPVALKILDKEKVLKHKMAEQIRREIATMKLIKHP 72
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
N+++ EV+ + T + I++EY GGELFD+I + GR EDEAR +FQQLI V YCHS
Sbjct: 73 NVVQLYEVMASKTKIFIILEYVTGGELFDKIVNDGRMKEDEARRYFQQLIHAVDYCHSRG 132
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYSKSS-------LLHSRPKSTVGTPAYIAPEVL 171
+ HRDLK EN LLD LKI DFG S S LLH ++ GTP Y+APEVL
Sbjct: 133 VYHRDLKPENLLLDS--YGNLKISDFGLSALSQQVRDDGLLH----TSCGTPNYVAPEVL 186
Query: 172 SRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQ 231
+ R YDG AD+WSCGV LYV+L G PF+D N +I + ++ P + +S
Sbjct: 187 NDRGYDGATADMWSCGVVLYVLLAGYLPFDD----SNLMNLYKKISSGEFNCPPW--LSL 240
Query: 232 ECRHLLSRIFVASPARRITIKEIKSHPWFLKN 263
L++RI +P R+T +E+ WF K+
Sbjct: 241 GAMKLITRILDPNPMTRVTPQEVFEDEWFKKD 272
>At5g39440 serine/threonine-specific protein kinase -like protein
Length = 494
Score = 205 bits (522), Expect = 3e-53
Identities = 120/275 (43%), Positives = 159/275 (57%), Gaps = 17/275 (6%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHRSLR 56
+ Y + K +G G+F +L H T VA+K + R I+ V REI R L
Sbjct: 16 LPNYRIGKTLGHGSFAKVKLALHVATGHKVAIKILNRSKIKNMGIEIKVQREIKILRFLM 75
Query: 57 HPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHS 116
HP+IIR EV+ TP + +VMEY GELFD I G+ EDEAR+ FQQ+ISGV YCH
Sbjct: 76 HPHIIRQYEVIETPNDIYVVMEYVKSGELFDYIVEKGKLQEDEARHLFQQIISGVEYCHR 135
Query: 117 MQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRP--KSTVGTPAYIAPEVLSRR 174
I HRDLK EN LLD +KI DFG S+++H K++ G+P Y APEV+S +
Sbjct: 136 NMIVHRDLKPENVLLDSQ--CNIKIVDFGL--SNVMHDGHFLKTSCGSPNYAAPEVISGK 191
Query: 175 EYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECR 234
Y G D+WSCGV LY +L G PF+D++ P F K I Y +P+ H+S R
Sbjct: 192 PY-GPDVDIWSCGVILYALLCGTLPFDDENIPNVFEK----IKRGMYTLPN--HLSHFAR 244
Query: 235 HLLSRIFVASPARRITIKEIKSHPWFLKNLPRELT 269
L+ R+ + P RI+I EI+ HPWF +LP L+
Sbjct: 245 DLIPRMLMVDPTMRISITEIRQHPWFNNHLPLYLS 279
>At1g30270 serine/threonine kinase, putative
Length = 482
Score = 204 bits (519), Expect = 6e-53
Identities = 120/310 (38%), Positives = 170/310 (54%), Gaps = 33/310 (10%)
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHP 58
KYE+ + +G G F + R+ + VA+K I++ +K+ + REI + ++HP
Sbjct: 30 KYELGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNKMIAQIKREISTMKLIKHP 89
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
N+IR EV+ + T + V+E+ GGELFD+I S GR EDEAR +FQQLI+ V YCHS
Sbjct: 90 NVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDEARKYFQQLINAVDYCHSRG 149
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYS-------KSSLLHSRPKSTVGTPAYIAPEVL 171
+ HRDLK EN LLD + A LK+ DFG S + LLH +T GTP Y+APEV+
Sbjct: 150 VYHRDLKPENLLLDANGA--LKVSDFGLSALPQQVREDGLLH----TTCGTPNYVAPEVI 203
Query: 172 SRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQ 231
+ + YDG AD+WSCGV L+V++ G PFED N +I ++ P + S
Sbjct: 204 NNKGYDGAKADLWSCGVILFVLMAGYLPFED----SNLTSLYKKIFKAEFTCPPW--FSA 257
Query: 232 ECRHLLSRIFVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYSLQSIED 291
+ L+ RI +PA RIT E+ + WF K + EN SL ++
Sbjct: 258 SAKKLIKRILDPNPATRITFAEVIENEWFKKGYKAP----------KFENADVSLDDVDA 307
Query: 292 IMKIVEEAKN 301
I E+KN
Sbjct: 308 IFDDSGESKN 317
>At5g35410 serine/threonine protein kinase SOS2 (gb|AAF62923.1)
Length = 446
Score = 202 bits (515), Expect = 2e-52
Identities = 116/270 (42%), Positives = 160/270 (58%), Gaps = 21/270 (7%)
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERG----HKIDENVAREIINHRSLRHP 58
KYEV + IG G F + R+ DT VA+K + + +++ + + REI + +RHP
Sbjct: 10 KYEVGRTIGEGTFAKVKFARNTDTGDNVAIKIMAKSTILKNRMVDQIKREISIMKIVRHP 69
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
NI+R EV+ +P+ + IV+E+ GGELFDRI GR E E+R +FQQL+ V++CH
Sbjct: 70 NIVRLYEVLASPSKIYIVLEFVTGGELFDRIVHKGRLEESESRKYFQQLVDAVAHCHCKG 129
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYSK-----SSLLHSRPKSTVGTPAYIAPEVLSR 173
+ HRDLK EN LLD + LK+ DFG S LL ++T GTP Y+APEVLS
Sbjct: 130 VYHRDLKPENLLLDTN--GNLKVSDFGLSALPQEGVELL----RTTCGTPNYVAPEVLSG 183
Query: 174 REYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQEC 233
+ YDG AD+WSCGV L+V+L G PF + D P +RK I A ++ P + S E
Sbjct: 184 QGYDGSAADIWSCGVILFVILAGYLPFSETDLPGLYRK----INAAEFSCPPW--FSAEV 237
Query: 234 RHLLSRIFVASPARRITIKEIKSHPWFLKN 263
+ L+ RI +P RI I+ IK PWF N
Sbjct: 238 KFLIHRILDPNPKTRIQIQGIKKDPWFRLN 267
>At5g25110 serine/threonine protein kinase-like protein
Length = 488
Score = 201 bits (510), Expect = 6e-52
Identities = 115/273 (42%), Positives = 161/273 (58%), Gaps = 24/273 (8%)
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHK----IDENVAREIINHRSLRHP 58
KYE+ + +G G FG + T + VA+K I + + E + REI R +RHP
Sbjct: 42 KYEMGRLLGKGTFGKVYYGKEITTGESVAIKIINKDQVKREGMMEQIKREISIMRLVRHP 101
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
NI+ KEV+ T T + +MEY GGELF +I G+ ED AR +FQQLIS V +CHS
Sbjct: 102 NIVELKEVMATKTKIFFIMEYVKGGELFSKIVK-GKLKEDSARKYFQQLISAVDFCHSRG 160
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYS-------KSSLLHSRPKSTVGTPAYIAPEVL 171
+ HRDLK EN L+D + LK+ DFG S + LLH++ GTPAY+APEVL
Sbjct: 161 VSHRDLKPENLLVDENGD--LKVSDFGLSALPEQILQDGLLHTQ----CGTPAYVAPEVL 214
Query: 172 SRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQ 231
++ YDG D+WSCG+ LYV+L G PF+D++ K +RK I +++ P + S
Sbjct: 215 RKKGYDGAKGDIWSCGIILYVLLAGFLPFQDENLMKMYRK----IFKSEFEYPPW--FSP 268
Query: 232 ECRHLLSRIFVASPARRITIKEIKSHPWFLKNL 264
E + L+S++ V P +RI+I I PWF KN+
Sbjct: 269 ESKRLISKLLVVDPNKRISIPAIMRTPWFRKNI 301
>At5g01810 serine/threonine protein kinase ATPK10
Length = 421
Score = 200 bits (508), Expect = 1e-51
Identities = 120/284 (42%), Positives = 160/284 (56%), Gaps = 24/284 (8%)
Query: 1 MEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKID----ENVAREIINHRSLR 56
M +YEV K +G G F RH T VA+K I++ + E + REI R LR
Sbjct: 9 MLRYEVGKFLGQGTFAKVYHARHLKTGDSVAIKVIDKERILKVGMTEQIKREISAMRLLR 68
Query: 57 HPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHS 116
HPNI+ EV+ T + + VME+ GGELF+++ S G+ ED AR +FQQL+ V +CHS
Sbjct: 69 HPNIVELHEVMATKSKIYFVMEHVKGGELFNKV-STGKLREDVARKYFQQLVRAVDFCHS 127
Query: 117 MQICHRDLKLENTLLDGSPAPRLKICDFGYSKSS-------LLHSRPKSTVGTPAYIAPE 169
+CHRDLK EN LLD LKI DFG S S LLH+ T GTPAY+APE
Sbjct: 128 RGVCHRDLKPENLLLDEHG--NLKISDFGLSALSDSRRQDGLLHT----TCGTPAYVAPE 181
Query: 170 VLSRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHI 229
V+SR YDG ADVWSCGV L+V+L G PF D N + +I + K P++ +
Sbjct: 182 VISRNGYDGFKADVWSCGVILFVLLAGYLPFRD----SNLMELYKKIGKAEVKFPNW--L 235
Query: 230 SQECRHLLSRIFVASPARRITIKEIKSHPWFLKNLPRELTEMAQ 273
+ + LL RI +P R++ ++I WF K L E+ E +
Sbjct: 236 APGAKRLLKRILDPNPNTRVSTEKIMKSSWFRKGLQEEVKESVE 279
>At2g25090 CBL-interacting protein kinase 16 (CIPK16)
Length = 469
Score = 200 bits (508), Expect = 1e-51
Identities = 117/281 (41%), Positives = 155/281 (54%), Gaps = 31/281 (11%)
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKID-----ENVAREIINHRSLR 56
+KY + + +G+GNF T VA+K I++ H E + REI R LR
Sbjct: 13 DKYNIGRLLGTGNFAKVYHGTEISTGDDVAIKVIKKDHVFKRRGMMEQIEREIAVMRLLR 72
Query: 57 HPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHS 116
HPN++ +EV+ T + VMEY GGELF+ I G+ ED AR +FQQLIS V +CHS
Sbjct: 73 HPNVVELREVMATKKKIFFVMEYVNGGELFEMIDRDGKLPEDLARKYFQQLISAVDFCHS 132
Query: 117 MQICHRDLKLENTLLDGSPAPRLKICDFGY--------------SKSSLLHSRPKSTVGT 162
+ HRD+K EN LLDG LK+ DFG S LLH+R GT
Sbjct: 133 RGVFHRDIKPENLLLDGE--GDLKVTDFGLSALMMPEGLGGRRGSSDDLLHTR----CGT 186
Query: 163 PAYIAPEVLSRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYK 222
PAY+APEVL + YDG +AD+WSCG+ LY +L G PF D+ N +I + +
Sbjct: 187 PAYVAPEVLRNKGYDGAMADIWSCGIVLYALLAGFLPFIDE----NVMTLYTKIFKAECE 242
Query: 223 IPDYVHISQECRHLLSRIFVASPARRITIKEIKSHPWFLKN 263
P + S E + LLSR+ V P +RI++ EIK PWF KN
Sbjct: 243 FPPW--FSLESKELLSRLLVPDPEQRISMSEIKMIPWFRKN 281
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,169,839
Number of Sequences: 26719
Number of extensions: 361909
Number of successful extensions: 4274
Number of sequences better than 10.0: 892
Number of HSP's better than 10.0 without gapping: 385
Number of HSP's successfully gapped in prelim test: 508
Number of HSP's that attempted gapping in prelim test: 2343
Number of HSP's gapped (non-prelim): 1228
length of query: 351
length of database: 11,318,596
effective HSP length: 100
effective length of query: 251
effective length of database: 8,646,696
effective search space: 2170320696
effective search space used: 2170320696
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC134322.21


