

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.19 - phase: 0
(318 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
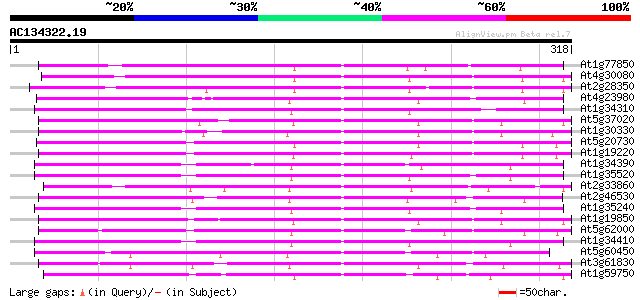
Score E
Sequences producing significant alignments: (bits) Value
At1g77850 putative auxin response factor protein (T32E8.16) 202 2e-52
At4g30080 transcription factor like protein 177 5e-45
At2g28350 unknown protein 173 1e-43
At4g23980 auxin response factor 9 (ARF9) 144 6e-35
At1g34310 hypothetical protein 137 6e-33
At5g37020 auxin response factor 8 (ARF8) 137 1e-32
At1g30330 putative protein 135 2e-32
At5g20730 auxin response factor 7 (ARF7) 135 3e-32
At1g19220 auxin response factor (ARF) like protein 135 4e-32
At1g34390 auxin response factor, putative 134 5e-32
At1g35520 auxin response factor, putative 133 1e-31
At2g33860 auxin response transcription factor 3 (ETTIN/ARF3) 130 9e-31
At2g46530 ARF1 family auxin responsive transcription factor like... 129 2e-30
At1g35240 hypothetical protein 128 4e-30
At1g19850 transcription factor monopteros (MP) 127 6e-30
At5g62000 auxin response factor - like protein 127 1e-29
At1g34410 auxin response factor, putative 125 2e-29
At5g60450 auxin response factor 4 124 5e-29
At3g61830 auxin response factor-like protein 124 9e-29
At1g59750 auxin response factor 1 121 4e-28
>At1g77850 putative auxin response factor protein (T32E8.16)
Length = 585
Score = 202 bits (514), Expect = 2e-52
Identities = 123/342 (35%), Positives = 180/342 (51%), Gaps = 53/342 (15%)
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIIS 76
V P IW+ C GA+VQIP LHSRVYYFPQGH+EH S L CII+
Sbjct: 16 VDPTIWRACAGASVQIPVLHSRVYYFPQGHVEHCCPLLST-------LPSSTSPVPCIIT 68
Query: 77 AVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTNV 136
++ LLADP TDEVFA L+L P+T + + D + + +F +IL ++
Sbjct: 69 SIQLLADPVTDEVFAHLILQPMTQQQFTPTNYSRFGRFDGDVDDNNKVTTFAKILTPSDA 128
Query: 137 SK-HAFYIPRFCAENMFPPLGMEVS---QHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSE 192
+ F +PRFCA+++FP L ++ Q L VTD+HG VW F H+ G +R++ T+
Sbjct: 129 NNGGGFSVPRFCADSVFPLLNFQIDPPVQKLYVTDIHGAVWDFRHIYRGTPRRHLL-TTG 187
Query: 193 WASFVERKKLDVGDAVVFMKNSTGKLFVGIRR----------------------KDAAEQ 230
W+ FV KKL GD+VVFM+ S ++F+G+RR ++
Sbjct: 188 WSKFVNSKKLIAGDSVVFMRKSADEMFIGVRRTPISSSDGGSSYYGGDEYNGYYSQSSVA 247
Query: 231 KKDE---------------LEKAVMEAVKLAEENKPFEIVYYPRGDDWCDFVVDGNIVDE 275
K+D+ +AV +A+ A + PFE+V+YP W +FVV V+
Sbjct: 248 KEDDGSPKKTFRRSGNGKLTAEAVTDAINRASQGLPFEVVFYPAA-GWSEFVVRAEDVES 306
Query: 276 SMKIQWNPRMRVK--MKTDKSSRIP-YQGTITTVSRTSNLWR 314
SM + W P RVK M+T+ SSRI +QG +++ + + WR
Sbjct: 307 SMSMYWTPGTRVKMAMETEDSSRITWFQGIVSSTYQETGPWR 348
>At4g30080 transcription factor like protein
Length = 670
Score = 177 bits (450), Expect = 5e-45
Identities = 116/354 (32%), Positives = 174/354 (48%), Gaps = 62/354 (17%)
Query: 19 PQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIISAV 78
PQ+W C G V++P ++S+V+YFPQGH E+A + P +C + A+
Sbjct: 18 PQLWHACAGGMVRMPPMNSKVFYFPQGHAENAYDCVDFGNL------PIPPMVLCRVLAI 71
Query: 79 DLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTNVSK 138
+AD +DEVFAKL L P+ ++ V + SN + E SF + L ++ +
Sbjct: 72 KYMADAESDEVFAKLRLIPLKDDEYVDHEYGDGEDSNGFESNSEKTPSFAKTLTQSDANN 131
Query: 139 -HAFYIPRFCAENMFPPLGMEVS---QHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSEWA 194
F +PR+CAE +FP L Q +L DVHG+VWKF H+ G +R++ T+ W+
Sbjct: 132 GGGFSVPRYCAETIFPRLDYNAEPPVQTILAKDVHGDVWKFRHIYRGTPRRHLL-TTGWS 190
Query: 195 SFVERKKLDVGDAVVFMKNSTGKLFVGIRRK----------------------------- 225
+FV +KKL GD++VFM+ G L VGIRR
Sbjct: 191 NFVNQKKLVAGDSIVFMRAENGDLCVGIRRAKRGGIGNGPEYSAGWNPIGGSCGYSSLLR 250
Query: 226 ------------DAAEQKKDELEKAVMEAVKLAEENKPFEIVYYPRGDDWCDFVVDGNIV 273
A++K ++V+EA LA +PFE+VYYPR +F V
Sbjct: 251 EDESNSLRRSNCSLADRKGKVTAESVIEAATLAISGRPFEVVYYPRAST-SEFCVKALDA 309
Query: 274 DESMKIQWNPRMRVKM--KTDKSSRIP-YQGTITTVSRTSNL------WRMLQV 318
+M+I W MR KM +T+ SSRI + GT++ V+ + + WR+LQV
Sbjct: 310 RAAMRIPWCSGMRFKMAFETEDSSRISWFMGTVSAVNVSDPIRWPNSPWRLLQV 363
>At2g28350 unknown protein
Length = 693
Score = 173 bits (439), Expect = 1e-43
Identities = 122/369 (33%), Positives = 177/369 (47%), Gaps = 71/369 (19%)
Query: 12 QQPSHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFT 71
+Q + PQ+W C G+ VQIP L+S V+YF QGH EHA + R P
Sbjct: 2 EQEKSLDPQLWHACAGSMVQIPSLNSTVFYFAQGHTEHAHAPP------DFHAPRVPPLI 55
Query: 72 ICIISAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEV----PNCSNDDDVCDEVIDSF 127
+C + +V LAD TDEVFAK+ L P+ N + V P S+ + E SF
Sbjct: 56 LCRVVSVKFLADAETDEVFAKITLLPLPGNDLDLENDAVLGLTPPSSDGNGNGKEKPASF 115
Query: 128 TRILALTNVSK-HAFYIPRFCAENMFPPLGMEVS---QHLLVTDVHGEVWKFHHVCHGFA 183
+ L ++ + F +PR+CAE +FP L Q ++ D+HGE WKF H+ G
Sbjct: 116 AKTLTQSDANNGGGFSVPRYCAETIFPRLDYSAEPPVQTVIAKDIHGETWKFRHIYRGTP 175
Query: 184 KRNVFYTSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRK------------------ 225
+R++ T+ W++FV +KKL GD++VF+++ +G L VGIRR
Sbjct: 176 RRHLL-TTGWSTFVNQKKLIAGDSIVFLRSESGDLCVGIRRAKRGGLGSNAGSDNPYPGF 234
Query: 226 ---------------------------DAAEQKKDELEKAVMEAVKLAEENKPFEIVYYP 258
+AA + +E AV EAV A + FE+VYYP
Sbjct: 235 SGFLRDDESTTTTSKLMMMKRNGNNDGNAAATGRVRVE-AVAEAVARAACGQAFEVVYYP 293
Query: 259 RGDDWCDFVVDGNIVDESMKIQWNPRMRVKM--KTDKSSRIP-YQGTITTVSRTSNL--- 312
R +F V V +M+I+W MR KM +T+ SSRI + GT++ V +
Sbjct: 294 RAST-PEFCVKAADVRSAMRIRWCSGMRFKMAFETEDSSRISWFMGTVSAVQVADPIRWP 352
Query: 313 ---WRMLQV 318
WR+LQV
Sbjct: 353 NSPWRLLQV 361
>At4g23980 auxin response factor 9 (ARF9)
Length = 638
Score = 144 bits (363), Expect = 6e-35
Identities = 98/314 (31%), Positives = 160/314 (50%), Gaps = 23/314 (7%)
Query: 16 HVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSL-DLQRFRPFTICI 74
+++ ++W+ C G V +P+ RVYYFPQGH+E +S+ ++++ L P +C
Sbjct: 8 YLYDELWKLCAGPLVDVPQAQERVYYFPQGHMEQLEASTQQVDLNTMKPLFVLPPKILCN 67
Query: 75 ISAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALT 134
+ V L A+ TDEV+A++ L PV V +P P+ S ++ + SF+++L +
Sbjct: 68 VMNVSLQAEKDTDEVYAQITLIPVGTE--VDEPMS-PDPS-PPELQRPKVHSFSKVLTAS 123
Query: 135 NVSKH-AFYIPRFCAENMFPPLGM---EVSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYT 190
+ S H F + R A PPL M +Q L+ DVHG WKF H+ G +R++ T
Sbjct: 124 DTSTHGGFSVLRKHATECLPPLDMTQQTPTQELVAEDVHGYQWKFKHIFRGQPRRHLL-T 182
Query: 191 SEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQ-------KKDELEKAVMEAV 243
+ W++FV K+L GD VF++ G+L VG+RR + + + V+
Sbjct: 183 TGWSTFVTSKRLVAGDTFVFLRGENGELRVGVRRANLQQSSMPSSVISSHSMHLGVLATA 242
Query: 244 KLAEENKPFEIVYY-PRGDDWCDFVVDGNIVDESMKIQWNPRMRVKMK--TDKSSRIPYQ 300
+ A + K IVYY PR F++ N E+M +++ MR KM+ + S Y
Sbjct: 243 RHATQTKTMFIVYYKPRTS---QFIISLNKYLEAMSNKFSVGMRFKMRFEGEDSPERRYS 299
Query: 301 GTITTVSRTSNLWR 314
GT+ V S W+
Sbjct: 300 GTVIGVKDCSPHWK 313
>At1g34310 hypothetical protein
Length = 591
Score = 137 bits (346), Expect = 6e-33
Identities = 95/312 (30%), Positives = 156/312 (49%), Gaps = 24/312 (7%)
Query: 15 SHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICI 74
S+V+ Q+W+ C G IPKL +VYYFPQGH+E +S+ + C
Sbjct: 22 SYVYEQLWKLCAGPLCDIPKLGEKVYYFPQGHIELVETSTREELNELQPICDLPSKLQCR 81
Query: 75 ISAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALT 134
+ A+ L + ++DE +A++ L P T V + +V + +++ +++SFT++L +
Sbjct: 82 VIAIHLKVENNSDETYAEITLMPDTT---VSENLQVVIPTQNENQFRPLVNSFTKVLTAS 138
Query: 135 NVSKH-AFYIPRFCAENMFPPLGME---VSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYT 190
+ S H F++P+ A P L M +Q LL D+HG W+F+H G +R++ T
Sbjct: 139 DTSAHGGFFVPKKHAIECLPSLDMSQPLPAQELLAIDLHGNQWRFNHNYRGTPQRHLL-T 197
Query: 191 SEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRK-------DAAEQKKDELEKAVMEAV 243
+ W +F KKL GD +VF++ TG+L VGIRR ++ D + V+ +
Sbjct: 198 TGWNAFTTSKKLVAGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVVASA 257
Query: 244 KLAEENK-PFEIVYYPRGDDWCDFVVDGNIVDESMKIQWNPRMRVKMKTDKSSRIPYQGT 302
K A +N+ F +VY P D + D V + K R ++++ D S GT
Sbjct: 258 KHAFDNQCMFTVVYKPSYDKFLDAV--------NNKFNVGSRFTMRLEGDDFSERRCFGT 309
Query: 303 ITTVSRTSNLWR 314
I VS S W+
Sbjct: 310 IIGVSDFSPHWK 321
>At5g37020 auxin response factor 8 (ARF8)
Length = 811
Score = 137 bits (344), Expect = 1e-32
Identities = 98/326 (30%), Positives = 161/326 (49%), Gaps = 32/326 (9%)
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEH-ASSSSSNAYIHSLDLQRFRPFTICII 75
++ ++W C G V +P SRV YFPQGH E A++++ H + P IC +
Sbjct: 19 LNSELWHACAGPLVSLPSSGSRVVYFPQGHSEQVAATTNKEVDGHIPNYPSLPPQLICQL 78
Query: 76 SAVDLLADPHTDEVFAKLLLTPVTNNSCVQD--PHEVPNCSNDDDVCDEVIDSFTRILAL 133
V + AD TDEV+A++ L P+T + P E+ S + F + L
Sbjct: 79 HNVTMHADVETDEVYAQMTLQPLTPEEQKETFVPIELGIPSKQPS------NYFCKTLTA 132
Query: 134 TNVSKH-AFYIPRFCAENMFPPLGMEV---SQHLLVTDVHGEVWKFHHVCHGFAKRNVFY 189
++ S H F +PR AE +FPPL + +Q L+ D+H WKF H+ G KR++
Sbjct: 133 SDTSTHGGFSVPRRAAEKVFPPLDYTLQPPAQELIARDLHDVEWKFRHIFRGQPKRHLL- 191
Query: 190 TSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQ-------KKDELEKAVMEA 242
T+ W+ FV K+L GD+V+F++N +LF+GIR + D + ++ A
Sbjct: 192 TTGWSVFVSAKRLVAGDSVIFIRNEKNQLFLGIRHATRPQTIVPSSVLSSDSMHIGLLAA 251
Query: 243 VKLAE-ENKPFEIVYYPRGDDWCDFVVDGNIVDESM---KIQWNPRMRVKMKTDKSSRIP 298
A N F + ++PR +FV+ + +++ +I R R+ +T++SS
Sbjct: 252 AAHASATNSCFTVFFHPRASQ-SEFVIQLSKYIKAVFHTRISVGMRFRMLFETEESSVRR 310
Query: 299 YQGTITTVSRTSNL------WRMLQV 318
Y GTIT +S ++ WR ++V
Sbjct: 311 YMGTITGISDLDSVRWPNSHWRSVKV 336
>At1g30330 putative protein
Length = 933
Score = 135 bits (341), Expect = 2e-32
Identities = 102/329 (31%), Positives = 163/329 (49%), Gaps = 38/329 (11%)
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEH-ASSSSSNAYIHSLDLQRFRPFTICII 75
++ ++W C G V +P + SRV YFPQGH E A+S++ H + P IC +
Sbjct: 20 LNSELWHACAGPLVSLPPVGSRVVYFPQGHSEQVAASTNKEVDAHIPNYPSLHPQLICQL 79
Query: 76 SAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPH-----EVPNCSNDDDVCDEVIDSFTRI 130
V + AD TDEV+A++ L P+ N +DP+ VP+ + + F +
Sbjct: 80 HNVTMHADVETDEVYAQMTLQPL-NAQEQKDPYLPAELGVPS--------RQPTNYFCKT 130
Query: 131 LALTNVSKH-AFYIPRFCAENMFPPLG---MEVSQHLLVTDVHGEVWKFHHVCHGFAKRN 186
L ++ S H F +PR AE +FPPL +Q L+ D+H WKF H+ G KR+
Sbjct: 131 LTASDTSTHGGFSVPRRAAEKVFPPLDYSQQPPAQELMARDLHDNEWKFRHIFRGQPKRH 190
Query: 187 VFYTSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQ-------KKDELEKAV 239
+ T+ W+ FV K+L GD+V+F+ N +L +GIRR + + D + +
Sbjct: 191 LL-TTGWSVFVSAKRLVAGDSVLFIWNDKNQLLLGIRRANRPQTVMPSSVLSSDSMHLGL 249
Query: 240 M-EAVKLAEENKPFEIVYYPRGDDWCDFVVDGNIVDESM---KIQWNPRMRVKMKTDKSS 295
+ A A N F I Y PR +FV+ +++ ++ R R+ +T++SS
Sbjct: 250 LAAAAHAAATNSRFTIFYNPRASP-SEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESS 308
Query: 296 RIPYQGTITTV-----SRTSNL-WRMLQV 318
Y GTIT + +R +N WR ++V
Sbjct: 309 VRRYMGTITGICDLDPTRWANSHWRSVKV 337
>At5g20730 auxin response factor 7 (ARF7)
Length = 1164
Score = 135 bits (340), Expect = 3e-32
Identities = 96/323 (29%), Positives = 152/323 (46%), Gaps = 26/323 (8%)
Query: 16 HVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICII 75
+++ ++W C G + +P S V YFPQGH E ++S IC++
Sbjct: 20 NINSELWHACAGPLISLPPAGSLVVYFPQGHSEQVAASMQKQTDFIPSYPNLPSKLICML 79
Query: 76 SAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTN 135
V L ADP TDEV+A++ L PV D + + + + F + L ++
Sbjct: 80 HNVTLNADPETDEVYAQMTLQPVNK----YDRDALLASDMGLKLNRQPNEFFCKTLTASD 135
Query: 136 VSKH-AFYIPRFCAENMFPPLGMEVS---QHLLVTDVHGEVWKFHHVCHGFAKRNVFYTS 191
S H F +PR AE +FP L + Q L+ D+H W F H+ G KR++ T+
Sbjct: 136 TSTHGGFSVPRRAAEKIFPALDFSMQPPCQELVAKDIHDNTWTFRHIYRGQPKRHLL-TT 194
Query: 192 EWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQ-------KKDELEKAVMEAVK 244
W+ FV K+L GD+V+F+++ +L +GIRR + + D + V+ A
Sbjct: 195 GWSVFVSTKRLFAGDSVLFIRDGKAQLLLGIRRANRQQPALSSSVISSDSMHIGVLAAAA 254
Query: 245 LAE-ENKPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWNPRMRVKM--KTDKSSRIPYQG 301
A N PF I Y PR +FVV ++M Q + MR +M +T++ Y G
Sbjct: 255 HANANNSPFTIFYNPRAAP-AEFVVPLAKYTKAMYAQVSLGMRFRMIFETEECGVRRYMG 313
Query: 302 TITTVSR------TSNLWRMLQV 318
T+T +S ++ WR LQ+
Sbjct: 314 TVTGISDLDPVRWKNSQWRNLQI 336
>At1g19220 auxin response factor (ARF) like protein
Length = 1086
Score = 135 bits (339), Expect = 4e-32
Identities = 95/322 (29%), Positives = 155/322 (47%), Gaps = 26/322 (8%)
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIIS 76
++ Q+W C G V +P + S V YFPQGH E ++S + IC++
Sbjct: 20 INSQLWHACAGPLVSLPPVGSLVVYFPQGHSEQVAASMQKQTDFIPNYPNLPSKLICLLH 79
Query: 77 AVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTNV 136
+V L AD TDEV+A++ L PV D + + + + F + L ++
Sbjct: 80 SVTLHADTETDEVYAQMTLQPVNK----YDREALLASDMGLKLNRQPTEFFCKTLTASDT 135
Query: 137 SKH-AFYIPRFCAENMFPPLGMEV---SQHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSE 192
S H F +PR AE +FPPL + +Q ++ D+H W F H+ G KR++ T+
Sbjct: 136 STHGGFSVPRRAAEKIFPPLDFSMQPPAQEIVAKDLHDTTWTFRHIYRGQPKRHLL-TTG 194
Query: 193 WASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKD-------AAEQKKDELEKAVMEAVKL 245
W+ FV K+L GD+V+F+++ +L +GIRR + ++ D + ++ A
Sbjct: 195 WSVFVSTKRLFAGDSVLFVRDEKSQLMLGIRRANRQTPTLSSSVISSDSMHIGILAAAAH 254
Query: 246 AEEN-KPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWNPRMRVKM--KTDKSSRIPYQGT 302
A N PF I + PR +FVV ++++ Q + MR +M +T+ Y GT
Sbjct: 255 ANANSSPFTIFFNPRASP-SEFVVPLAKYNKALYAQVSLGMRFRMMFETEDCGVRRYMGT 313
Query: 303 ITTVSR------TSNLWRMLQV 318
+T +S + WR LQV
Sbjct: 314 VTGISDLDPVRWKGSQWRNLQV 335
>At1g34390 auxin response factor, putative
Length = 600
Score = 134 bits (338), Expect = 5e-32
Identities = 92/314 (29%), Positives = 158/314 (50%), Gaps = 26/314 (8%)
Query: 15 SHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICI 74
S+++ Q+W+ C G IPKL ++YYFPQG++E +S+ + C
Sbjct: 22 SYMYEQLWKLCAGPLCDIPKLGEKIYYFPQGNIELVEASTREELNELKPICDLPSKLQCR 81
Query: 75 ISAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALT 134
+ A+ L + ++DE +A++ L P D +V + +++ +++SFT++L +
Sbjct: 82 VIAIQLKVENNSDETYAEITLMP--------DTTQVVIPTQNENQFRPLVNSFTKVLTAS 133
Query: 135 NVSKHAFYIPRFCAENMFPPLGME---VSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYTS 191
+ S F++P+ A PPL M +Q LL TD+HG W+F+H G +R++ T+
Sbjct: 134 DTS-GGFFVPKKHAIECLPPLDMSQPLPTQELLATDLHGNQWRFNHNYRGTPQRHLL-TT 191
Query: 192 EWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQKK---------DELEKAVMEA 242
W +F KKL GD +VF++ TG+L VGIRR A Q+ + + V+ +
Sbjct: 192 GWNAFTTSKKLVAGDVIVFVRGETGELRVGIRR--AGHQQGNIPSSIISIESMRHGVIAS 249
Query: 243 VKLAEENKPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWN--PRMRVKMKTDKSSRIPYQ 300
K A +N+ IV Y F+V + +++ ++N R ++ + D S Y
Sbjct: 250 AKHAFDNQCMFIVVYKPSIRSSQFIVSYDKFLDAVNNKFNVGSRFTMRFEGDDFSERRYF 309
Query: 301 GTITTVSRTSNLWR 314
GTI VS S W+
Sbjct: 310 GTIIGVSDFSPHWK 323
>At1g35520 auxin response factor, putative
Length = 576
Score = 133 bits (335), Expect = 1e-31
Identities = 94/314 (29%), Positives = 157/314 (49%), Gaps = 26/314 (8%)
Query: 15 SHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICI 74
S+++ Q+W+ C G IPKL +VYYFPQG++E +S+ + C
Sbjct: 22 SYMYEQLWKLCAGPLCDIPKLGEKVYYFPQGNIELVEASTREELNELQPICDLPSKLQCR 81
Query: 75 ISAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALT 134
+ A+ L + ++DE +AK+ L P D +V + +++ +++SFT++L +
Sbjct: 82 VIAIHLKVENNSDETYAKITLMP--------DTTQVVIPTQNENQFRPLVNSFTKVLTAS 133
Query: 135 NVSKH-AFYIPRFCAENMFPPLGME---VSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYT 190
++S + F +P+ A PPL M +Q LL D+HG W F H G +R++ T
Sbjct: 134 DISANGVFSVPKKHAIECLPPLDMSQPLPAQELLAIDLHGNQWSFRHSYRGTPQRHLL-T 192
Query: 191 SEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRK-------DAAEQKKDELEKAVMEAV 243
+ W F KKL GD +VF++ TG+L VGIRR ++ D + V+ +
Sbjct: 193 TGWNEFTTSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVIASA 252
Query: 244 KLAEENK-PFEIVYYPRGDDWCDFVVDGNIVDESMKIQWN--PRMRVKMKTDKSSRIPYQ 300
K A +N+ F +VY PR F+V + +++ ++N R ++ + D S Y
Sbjct: 253 KHAFDNQCMFIVVYKPRSS---QFIVSYDKFLDAVNNKFNVGSRFTMRFEGDDLSERRYF 309
Query: 301 GTITTVSRTSNLWR 314
GTI VS S W+
Sbjct: 310 GTIIGVSNFSPHWK 323
>At2g33860 auxin response transcription factor 3 (ETTIN/ARF3)
Length = 608
Score = 130 bits (327), Expect = 9e-31
Identities = 99/326 (30%), Positives = 149/326 (45%), Gaps = 38/326 (11%)
Query: 20 QIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIISAVD 79
++W C G + +PK S V YFPQGHLE A S+ Y P C I V
Sbjct: 54 ELWHACAGPLISLPKRGSLVLYFPQGHLEQAPDFSAAIY-------GLPPHVFCRILDVK 106
Query: 80 LLADPHTDEVFAKLLLTPVTNN---SCVQDPHEVPNCSNDDDVC--DEVIDSFTRILALT 134
L A+ TDEV+A++ L P + + + +V D +V F + L +
Sbjct: 107 LHAETTTDEVYAQVSLLPESEDIERKVREGIIDVDGGEEDYEVLKRSNTPHMFCKTLTAS 166
Query: 135 NVSKH-AFYIPRFCAENMFPPLGM---EVSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYT 190
+ S H F +PR AE+ FPPL SQ LL D+HG W+F H+ G +R++ T
Sbjct: 167 DTSTHGGFSVPRRAAEDCFPPLDYSQPRPSQELLARDLHGLEWRFRHIYRGQPRRHLL-T 225
Query: 191 SEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKD--------AAEQKKDELEKAVMEA 242
+ W++FV +KKL GDAV+F++ GKL +G+RR +A+ ++ E
Sbjct: 226 TGWSAFVNKKKLVSGDAVLFLRGDDGKLRLGVRRASQIEGTAALSAQYNQNMNHNNFSEV 285
Query: 243 VKLAEENKPFEIVYYPRGDDWCDFVVDG----NIVDESMKIQWNPRMRVKMKTDKSSRIP 298
+ F I Y P+ W +F++ +VD I + RV+ + R P
Sbjct: 286 AHAISTHSVFSISYNPKA-SWSNFIIPAPKFLKVVDYPFCIGMRFKARVESEDASERRSP 344
Query: 299 YQGTITTVSRTSNL------WRMLQV 318
G I+ +S + WR L V
Sbjct: 345 --GIISGISDLDPIRWPGSKWRCLLV 368
>At2g46530 ARF1 family auxin responsive transcription factor like
protein
Length = 622
Score = 129 bits (325), Expect = 2e-30
Identities = 93/314 (29%), Positives = 153/314 (48%), Gaps = 28/314 (8%)
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYI-HSLDLQRFRPFTICII 75
++ ++W+ C G V++P+ RV+YFPQGH+E +S++ + + + P +C +
Sbjct: 39 LYTELWKACAGPLVEVPRYGERVFYFPQGHMEQLVASTNQGVVDQEIPVFNLPPKILCRV 98
Query: 76 SAVDLLADPHTDEVFAKLLLTPVTNNS--CVQDPHEVPNCSNDDDVCDEVIDSFTRILAL 133
+V L A+ TDEV+A++ L P + S DP V + +DSF +IL
Sbjct: 99 LSVTLKAEHETDEVYAQITLQPEEDQSEPTSLDPPLV-------EPAKPTVDSFVKILTA 151
Query: 134 TNVSKH-AFYIPRFCAENMFPPLGM---EVSQHLLVTDVHGEVWKFHHVCHGFAKRNVFY 189
++ S H F + R A P L M +Q L+ D+HG W+F H+ G +R++
Sbjct: 152 SDTSTHGGFSVLRKHATECLPSLDMTQPTPTQELVARDLHGYEWRFKHIFRGQPRRHLL- 210
Query: 190 TSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRR--KDAAEQKKDELEKAVMEAVKLAE 247
T+ W++FV K+L GDA VF++ TG L VG+RR K + + M LA
Sbjct: 211 TTGWSTFVTSKRLVAGDAFVFLRGETGDLRVGVRRLAKQQSTMPASVISSQSMRLGVLAT 270
Query: 248 ENKP------FEIVYYPRGDDWCDFVVDGNIVDESMKIQWNPRMRVKMK--TDKSSRIPY 299
+ F + Y PR F++ N +MK ++ MR +M+ ++S +
Sbjct: 271 ASHAVTTTTIFVVFYKPR---ISQFIISVNKYMMAMKNGFSLGMRYRMRFEGEESPERIF 327
Query: 300 QGTITTVSRTSNLW 313
GTI S+ W
Sbjct: 328 TGTIIGSGDLSSQW 341
>At1g35240 hypothetical protein
Length = 615
Score = 128 bits (322), Expect = 4e-30
Identities = 93/314 (29%), Positives = 153/314 (48%), Gaps = 26/314 (8%)
Query: 15 SHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICI 74
S+++ Q+W+ C G IPKL VYYFPQG++E +S+ + C
Sbjct: 22 SYMYEQLWKLCAGPLCDIPKLGENVYYFPQGNIELVDASTREELNELQPICDLPSKLQCR 81
Query: 75 ISAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALT 134
+ A+ L + ++DE +A++ L P D +V + ++ +++SFT++L +
Sbjct: 82 VIAIHLKVENNSDETYAEITLMP--------DTTQVVIPTQSENQFRPLVNSFTKVLTAS 133
Query: 135 NVSKH-AFYIPRFCAENMFPPLGME---VSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYT 190
+ S + F++P+ A PPL M +Q LL D+HG W+F H G +R+ T
Sbjct: 134 DTSAYGGFFVPKKHAIECLPPLDMSQPLPAQELLAKDLHGNQWRFRHSYRGTPQRHSL-T 192
Query: 191 SEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRK-------DAAEQKKDELEKAVMEAV 243
+ W F KKL GD +VF++ TG+L VGIRR ++ D + V+ +
Sbjct: 193 TGWNEFTTSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVIASA 252
Query: 244 KLAEENK-PFEIVYYPRGDDWCDFVVDGNIVDESM--KIQWNPRMRVKMKTDKSSRIPYQ 300
K A +N+ F +VY PR F+V + ++M K R ++ + D S Y
Sbjct: 253 KHALDNQCIFIVVYKPRSS---QFIVSYDKFLDAMNNKFIVGSRFTMRFEGDDFSERRYF 309
Query: 301 GTITTVSRTSNLWR 314
GTI V+ S W+
Sbjct: 310 GTIIGVNDFSPHWK 323
>At1g19850 transcription factor monopteros (MP)
Length = 902
Score = 127 bits (320), Expect = 6e-30
Identities = 97/324 (29%), Positives = 156/324 (47%), Gaps = 28/324 (8%)
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEH-ASSSSSNAYIHSLDLQRFRPFTICII 75
++ ++W C G V +P++ S VYYF QGH E A S+ +A + +C +
Sbjct: 51 INSELWHACAGPLVCLPQVGSLVYYFSQGHSEQVAVSTRRSATTQVPNYPNLPSQLMCQV 110
Query: 76 SAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTN 135
V L AD +DE++A++ L PV + +D VP+ + F + L ++
Sbjct: 111 HNVTLHADKDSDEIYAQMSLQPVHSE---RDVFPVPDFGMLRG-SKHPTEFFCKTLTASD 166
Query: 136 VSKH-AFYIPRFCAENMFPPLGMEV---SQHLLVTDVHGEVWKFHHVCHGFAKRNVFYTS 191
S H F +PR AE +FPPL +Q L+V D+H W F H+ G KR++ T+
Sbjct: 167 TSTHGGFSVPRRAAEKLFPPLDYSAQPPTQELVVRDLHENTWTFRHIYRGQPKRHLL-TT 225
Query: 192 EWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQ-------KKDELEKAVMEAVK 244
W+ FV K+L GD+V+F+++ +L VG+RR + + D + V+ A
Sbjct: 226 GWSLFVGSKRLRAGDSVLFIRDEKSQLMVGVRRANRQQTALPSSVLSADSMHIGVLAAAA 285
Query: 245 LAEENK-PFEIVYYPRGDDWCDFVVDGNIVDESM---KIQWNPRMRVKMKTDKSSRIPYQ 300
A N+ PF I Y PR +FV+ +++ ++ R + +T+ S + Y
Sbjct: 286 HATANRTPFLIFYNPRACP-AEFVIPLAKYRKAICGSQLSVGMRFGMMFETEDSGKRRYM 344
Query: 301 GTITTVSRTSNL------WRMLQV 318
GTI +S L WR LQV
Sbjct: 345 GTIVGISDLDPLRWPGSKWRNLQV 368
>At5g62000 auxin response factor - like protein
Length = 371
Score = 127 bits (318), Expect = 1e-29
Identities = 92/325 (28%), Positives = 157/325 (48%), Gaps = 34/325 (10%)
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIIS 76
++ ++W C G V +P+ RV+YFPQGH+E A S++ A + L +C +
Sbjct: 58 LYRELWHACAGPLVTVPRQDDRVFYFPQGHIEQA--STNQAAEQQMPLYDLPSKLLCRVI 115
Query: 77 AVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTNV 136
VDL A+ TDEV+A++ L P N QD + + + + SF + L ++
Sbjct: 116 NVDLKAEADTDEVYAQITLLPEAN----QDENAIEKEAPLPPPPRFQVHSFCKTLTASDT 171
Query: 137 SKH-AFYIPRFCAENMFPPLGME---VSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSE 192
S H F + R A+ PPL M +Q L+ D+H W+F H+ G +R++ S
Sbjct: 172 STHGGFSVLRRHADECLPPLDMSRQPPTQELVAKDLHANEWRFRHIFRGQPRRHLL-QSG 230
Query: 193 WASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQKKDELEKAVMEAVKL------- 245
W+ FV K+L GDA +F++ G+L VG+RR A +++ + +V+ + +
Sbjct: 231 WSVFVSSKRLVAGDAFIFLRGENGELRVGVRR---AMRQQGNVPSSVISSHSMHLGVLAT 287
Query: 246 ----AEENKPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWNPRMRVKMK--TDKSSRIPY 299
F + Y PR +F+V + ES+K ++ MR KM+ +++ +
Sbjct: 288 AWHAISTGTMFTVYYKPRTSP-SEFIVPFDQYMESVKNNYSIGMRFKMRFEGEEAPEQRF 346
Query: 300 QGTITTVSRT------SNLWRMLQV 318
GTI + + + WR L+V
Sbjct: 347 TGTIVGIEESDPTRWPKSKWRSLKV 371
>At1g34410 auxin response factor, putative
Length = 620
Score = 125 bits (315), Expect = 2e-29
Identities = 89/313 (28%), Positives = 152/313 (48%), Gaps = 22/313 (7%)
Query: 15 SHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICI 74
S+++ Q+W+ C G IPKL VYYFPQG++E +S+ + C
Sbjct: 34 SYMYEQLWKLCAGPLCDIPKLGENVYYFPQGNIELVQASTREELNELQPICDLPSKLQCR 93
Query: 75 ISAVDLLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALT 134
+ A+ L + ++DE++A++ L P D +V + ++ +++SFT++L +
Sbjct: 94 VIAIHLKVENNSDEIYAEITLMP--------DTTQVVIPTQSENRFRPLVNSFTKVLTAS 145
Query: 135 NVSKH-AFYIPRFCAENMFPPLGME---VSQHLLVTDVHGEVWKFHHVCHGFAKRNVFYT 190
+ S + F +P+ A PPL M +Q +L D+H W+F H G +R+ T
Sbjct: 146 DTSAYGGFSVPKKHAIECLPPLDMSQPLPAQEILAIDLHDNQWRFRHNYRGTPQRHSL-T 204
Query: 191 SEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRK-------DAAEQKKDELEKAVMEAV 243
+ W F+ KKL GD +VF++ TG+L VGIRR ++ D + V+ +
Sbjct: 205 TGWNEFITSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVIASA 264
Query: 244 KLAEENKPFEIVYYPRGDDWCDFVVDGNIVDESMKIQWN--PRMRVKMKTDKSSRIPYQG 301
K A +N+ IV Y F+V + +++ ++N R ++ + D S Y G
Sbjct: 265 KHAFDNQCIFIVVYKPSIRSSQFIVSYDKFLDAVNNKFNVGSRFTMRFEGDDFSERRYFG 324
Query: 302 TITTVSRTSNLWR 314
TI VS S W+
Sbjct: 325 TIIGVSDFSPHWK 337
>At5g60450 auxin response factor 4
Length = 788
Score = 124 bits (312), Expect = 5e-29
Identities = 92/316 (29%), Positives = 149/316 (47%), Gaps = 32/316 (10%)
Query: 15 SHVHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRF--RPFTI 72
S ++ ++W C G +PK + V YFPQGHLE + S Y L++ +F P +
Sbjct: 60 SSIYSELWHACAGPLTCLPKKGNVVVYFPQGHLEQDAMVS---YSSPLEIPKFDLNPQIV 116
Query: 73 CIISAVDLLADPHTDEVFAKLLLTPVTNNSCVQ-DPHEVPNCSNDDD-----VCDEVIDS 126
C + V LLA+ TDEV+ ++ L P+ S + + EV +++
Sbjct: 117 CRVVNVQLLANKDTDEVYTQVTLLPLQEFSMLNGEGKEVKELGGEEERNGSSSVKRTPHM 176
Query: 127 FTRILALTNVSKH-AFYIPRFCAENMFPPLG---MEVSQHLLVTDVHGEVWKFHHVCHGF 182
F + L ++ S H F +PR AE+ F PL SQ L+ D+HG WKF H+ G
Sbjct: 177 FCKTLTASDTSTHGGFSVPRRAAEDCFAPLDYKQQRPSQELIAKDLHGVEWKFRHIYRGQ 236
Query: 183 AKRNVFYTSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQKKDELEKAVME- 241
+R++ T+ W+ FV +K L GDAV+F+++ G+L +GIRR A + ++ L +++E
Sbjct: 237 PRRHLL-TTGWSIFVSQKNLVSGDAVLFLRDEGGELRLGIRR---AARPRNGLPDSIIEK 292
Query: 242 ---------AVKLAEENKPFEIVYYPRGDDWCDFVV--DGNIVDESMKIQWNPRMRVKMK 290
F + Y PR +FV+ + I + R R++ +
Sbjct: 293 NSCSNILSLVANAVSTKSMFHVFYSPRATH-AEFVIPYEKYITSIRSPVCIGTRFRMRFE 351
Query: 291 TDKSSRIPYQGTITTV 306
D S G +T V
Sbjct: 352 MDDSPERRCAGVVTGV 367
>At3g61830 auxin response factor-like protein
Length = 602
Score = 124 bits (310), Expect = 9e-29
Identities = 97/325 (29%), Positives = 154/325 (46%), Gaps = 35/325 (10%)
Query: 17 VHPQIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRF--RPFTICI 74
++ ++W+ C G V++P+ RV+YFPQGH+E +S+N I+S ++ F P +C
Sbjct: 22 LYTELWKVCAGPLVEVPRAQERVFYFPQGHMEQL-VASTNQGINSEEIPVFDLPPKILCR 80
Query: 75 ISAVDLLADPHTDEVFAKLLLTPVTNNS--CVQDPHEVPNCSNDDDVCDEVIDSFTRILA 132
+ V L A+ TDEV+A++ L P + S DP V + SF +IL
Sbjct: 81 VLDVTLKAEHETDEVYAQITLQPEEDQSEPTSLDPPIVGPTKQE-------FHSFVKILT 133
Query: 133 LTNVSKH-AFYIPRFCAENMFPPLGM---EVSQHLLVTDVHGEVWKFHHVCHGFAKRNVF 188
++ S H F + R A P L M +Q L+ D+HG W+F H+ G +R++
Sbjct: 134 ASDTSTHGGFSVLRKHATECLPSLDMTQATPTQELVTRDLHGFEWRFKHIFRGQPRRHLL 193
Query: 189 YTSEWASFVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQ-------KKDELEKAVME 241
T+ W++FV K+L GDA VF++ G L VG+RR + + V+
Sbjct: 194 -TTGWSTFVSSKRLVAGDAFVFLRGENGDLRVGVRRLARHQSTMPTSVISSQSMHLGVLA 252
Query: 242 AVKLAEENKPFEIVYY-PRGDDWCDFVVDGNIVDESMK--IQWNPRMRVKMKTDKSSRIP 298
A +V+Y PR F+V N E++K R R++ + ++S
Sbjct: 253 TASHAVRTTTIFVVFYKPR---ISQFIVGVNKYMEAIKHGFSLGTRFRMRFEGEESPERI 309
Query: 299 YQGTITTVSRTS-----NLWRMLQV 318
+ GTI S + WR LQV
Sbjct: 310 FTGTIVGSGDLSSQWPASKWRSLQV 334
>At1g59750 auxin response factor 1
Length = 665
Score = 121 bits (304), Expect = 4e-28
Identities = 85/322 (26%), Positives = 147/322 (45%), Gaps = 33/322 (10%)
Query: 20 QIWQTCTGAAVQIPKLHSRVYYFPQGHLEHASSSSSNAYIHSLDLQRFRPFTICIISAVD 79
++W C G V +P+ RVYYFP+GH+E +S + +C + +
Sbjct: 22 ELWHACAGPLVTLPREGERVYYFPEGHMEQLEASMHQGLEQQMPSFNLPSKILCKVINIQ 81
Query: 80 LLADPHTDEVFAKLLLTPVTNNSCVQDPHEVPNCSNDDDVCDEVIDSFTRILALTNVSKH 139
A+P TDEV+A++ L P + S +P + + C + SF + L ++ S H
Sbjct: 82 RRAEPETDEVYAQITLLPELDQS---EPTSPDAPVQEPEKC--TVHSFCKTLTASDTSTH 136
Query: 140 -AFYIPRFCAENMFPPLGMEVS---QHLLVTDVHGEVWKFHHVCHGFAKRNVFYTSEWAS 195
F + R A++ PPL M Q L+ TD+H W F H+ G +R++ T+ W+
Sbjct: 137 GGFSVLRRHADDCLPPLDMSQQPPWQELVATDLHNSEWHFRHIFRGQPRRHLL-TTGWSV 195
Query: 196 FVERKKLDVGDAVVFMKNSTGKLFVGIRRKDAAEQKKDELEKAVME-----------AVK 244
FV KKL GDA +F++ +L VG+RR +++ + +V+ A
Sbjct: 196 FVSSKKLVAGDAFIFLRGENEELRVGVRRH---MRQQTNIPSSVISSHSMHIGVLATAAH 252
Query: 245 LAEENKPFEIVYYPRGDDWCDFVVDGN--IVDESMKIQWNPRMRVKMKTDKSSRIPYQGT 302
F + Y PR +F+V N + ++ K+ R +++ + +++ + GT
Sbjct: 253 AITTGTIFSVFYKPR-TSRSEFIVSVNRYLEAKTQKLSVGMRFKMRFEGEEAPEKRFSGT 311
Query: 303 ITTVSRTSNL------WRMLQV 318
I V + WR L+V
Sbjct: 312 IVGVQENKSSVWHDSEWRSLKV 333
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,474,683
Number of Sequences: 26719
Number of extensions: 318717
Number of successful extensions: 1160
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 1015
Number of HSP's gapped (non-prelim): 86
length of query: 318
length of database: 11,318,596
effective HSP length: 99
effective length of query: 219
effective length of database: 8,673,415
effective search space: 1899477885
effective search space used: 1899477885
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC134322.19


