

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.18 - phase: 0
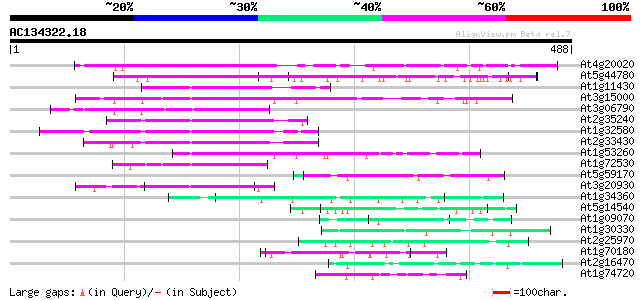
(488 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g20020 DAG-like protein 234 1e-61
At5g44780 unknown protein 172 3e-43
At1g11430 unknown protein 144 8e-35
At3g15000 unknown protein 141 9e-34
At3g06790 DAG protein, putative 134 9e-32
At2g35240 unknown protein 121 8e-28
At1g32580 unknown protein, like plastid protein 120 1e-27
At2g33430 plastid protein 119 4e-27
At1g53260 DAG-like protein emb|CAA16610.1 106 3e-23
At1g72530 DAG-like protein 81 1e-15
At5g59170 cell wall protein 62 7e-10
At3g20930 unknown protein 57 3e-08
At1g34360 transcription factor like protein 55 1e-07
At5g14540 unknown protein 50 2e-06
At1g09070 unknown protein 47 2e-05
At1g30330 putative protein 47 3e-05
At2g25970 pseudogene 46 5e-05
At1g70180 unknown protein 46 5e-05
At2g16470 unknown protein 45 7e-05
At1g74720 hypothetical protein 45 7e-05
>At4g20020 DAG-like protein
Length = 406
Score = 234 bits (596), Expect = 1e-61
Identities = 170/440 (38%), Positives = 219/440 (49%), Gaps = 72/440 (16%)
Query: 57 TATSLSVPITGSISSHFS-LSKSPNVVDGIQSRSF--RSTSIS------LLSSRYGETSE 107
TATS + SISS + S P+V + RS RST ++ L S+R + +
Sbjct: 14 TATSY---VNRSISSSITPASDFPSVSAAVLKRSVIGRSTEVATRAPARLFSTRQYKLYK 70
Query: 108 LSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPP-PEEMIRIYEETCAKGLNISVEEAKKK 166
EI DT+LFEGCDYNHWL DF ++ P PEEM+ YEETCA+GL ISVEEAK++
Sbjct: 71 EGDEITEDTVLFEGCDYNHWLITMDFSKEETPKSPEEMVAAYEETCAQGLGISVEEAKQR 130
Query: 167 IYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRP 226
+YACSTTTY GFQA+MTE+ES+KF+ +PGV+F+LPDSYIDP NK+YGGD+Y G I RP
Sbjct: 131 MYACSTTTYQGFQAIMTEQESEKFKDLPGVVFILPDSYIDPQNKEYGGDKYENGVITHRP 190
Query: 227 PPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASH 286
PP+Q GR R P ++ R G G PQNF +GQ
Sbjct: 191 PPIQSGR----------------ARPRPRFD------RSGGGSGGPQNFQRNTQYGQQ-- 226
Query: 287 IPPQQNIGQQQPNIRIDSTSQRFPPQQSY-----DQASQRYPP-QQSYDQASQGYLPKQN 340
PP Q G + PQQ Y Q +Q PP Q Y+Q + P
Sbjct: 227 -PPMQGGGGS------------YGPQQGYATPGQGQGTQAPPPFQGGYNQGPRSPPPPYQ 273
Query: 341 YGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYA--PQQNFGPPGE 398
G Q +P P Q G P Y QS G+G NY+ PQ + G
Sbjct: 274 AGY-NQGQGSPVPPYQAGYNQVQGSPVPPYQGTQS--SYGQGGSGNYSQGPQGGYNQGGP 330
Query: 399 VDRRNYAPQ--QNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEGGWDFKPSYMEEFE 456
RNY PQ NFGP N P G PG G+ G S P G + ++ + +
Sbjct: 331 ---RNYNPQGAGNFGP--ASGAGNLGPAPGAGNPGYGQ-GYSGP---GQEQNQTFPQADQ 381
Query: 457 QGQKGNNHAEEQKESQQRFP 476
+ + NN+ + +FP
Sbjct: 382 RNRDWNNNNPAGQPGSDQFP 401
>At5g44780 unknown protein
Length = 723
Score = 172 bits (437), Expect = 3e-43
Identities = 150/449 (33%), Positives = 196/449 (43%), Gaps = 91/449 (20%)
Query: 91 RSTSISLLSSRYGETSELS--PEIGPDTIL--FEGCDYNHWLFVCDFPRDNKPPPEEMIR 146
+ST+I+ +R T++ P G D+ + EGCD+NHWL +FP+DN P EEMI
Sbjct: 45 QSTAINRSPARLFSTTQYQYDPYTGEDSFMPDNEGCDFNHWLITMNFPKDNLPSREEMIS 104
Query: 147 IYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYID 206
I+E+TCAKGL IS+EEAKKKIYA TT+Y GFQA MT E +KF +PGV +++PDSYID
Sbjct: 105 IFEQTCAKGLAISLEEAKKKIYAICTTSYQGFQATMTIGEVEKFRDLPGVQYIIPDSYID 164
Query: 207 PVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQ-------------------- 246
NK YGGD+Y G I P P PV + + Q
Sbjct: 165 VENKVYGGDKYENGVITPGPVPVPTKEGFDSLKKESKPEQEEAEIILTPPDEGKTSGQVQ 224
Query: 247 ------LPNNRG------------NPSYNNRDSMPRDGRNYGPPQNFPPQ-QNHGQASHI 287
LP+ R + S+ G+ G + P Q Q+ GQ +
Sbjct: 225 GQGSLTLPDQRSVKERQGTLALVQGQGQRSGMSILGQGQGEGRRMSIPGQWQSRGQGNSF 284
Query: 288 PPQQNIGQQQPNIRIDST--SQRFPPQQSYDQASQRYP------PQQSYDQASQGYLPKQ 339
Q +R T S P Q + Q P Q+S +SQG L +Q
Sbjct: 285 QGSFKQSQGTLPVRKGQTQISDEIPSFQGNVKQRQEMPIHGQGQAQRSQMPSSQGTL-RQ 343
Query: 340 NYGQAPQ------NYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNF 393
Q Q Y+ Q Q Q G +P Y Q+S G+G Y N
Sbjct: 344 GQAQGSQRPSNQVGYNQGQGAQTPPYHQGQGAQTPPY--QESPNNYGQGAFVQY----NQ 397
Query: 394 GPP-GEV-----DRRNYAPQQNFGPPRQGERRNYVPQQNFGPP----GQGE--------R 435
GPP G V ++ N Q N+ P G NY P Q G P GQG+ R
Sbjct: 398 GPPQGNVVQTTQEKYNQMGQGNYAPQSGG---NYSPAQGAGSPRFGYGQGQGGQLLSPYR 454
Query: 436 GNSVPSEG------GWDFKPSYMEEFEQG 458
GN +G G + +PSY F QG
Sbjct: 455 GNYNQGQGTPLPGQGQEGQPSYQMGFSQG 483
Score = 52.4 bits (124), Expect = 6e-07
Identities = 70/261 (26%), Positives = 96/261 (35%), Gaps = 64/261 (24%)
Query: 243 QNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPP-QQNIGQQQPNIR 301
Q +Q+P+++G R G N GQ + PP Q G Q P +
Sbjct: 330 QRSQMPSSQGTLRQGQAQGSQRPSNQVG--------YNQGQGAQTPPYHQGQGAQTPPYQ 381
Query: 302 IDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQ 361
+ Y+Q PPQ + Q +Q + Y Q Q APQ+ NYS Q
Sbjct: 382 ESPNNYGQGAFVQYNQG----PPQGNVVQTTQ-----EKYNQMGQGNYAPQSGGNYSPAQ 432
Query: 362 TYGPASPQYPPQQSFG-----------------PL-GEGERRNYAPQQNFG-------PP 396
G SP++ Q G PL G+G+ + Q F PP
Sbjct: 433 --GAGSPRFGYGQGQGGQLLSPYRGNYNQGQGTPLPGQGQEGQPSYQMGFSQGLGAPVPP 490
Query: 397 GEVDRRNYA-----------PQQNF--GPPR---QGERRNYVPQQ--NFGPPGQGERGNS 438
+V NY PQ NF GP + QG + NY PQ ++GP G+
Sbjct: 491 NQVIPGNYGQWAFVNYNQGPPQGNFLQGPQQNHNQGGQWNYSPQNGGHYGPAQFGQWYPG 550
Query: 439 VPSEGGWDFKPSYMEEFEQGQ 459
P G + P Y + QGQ
Sbjct: 551 PPQGQGIQW-PQYQLSYNQGQ 570
Score = 45.8 bits (107), Expect = 5e-05
Identities = 77/258 (29%), Positives = 97/258 (36%), Gaps = 63/258 (24%)
Query: 217 YIEGQIIPRP-------PPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNY 269
Y +GQ P P P Q G + G N +P N G ++ N N
Sbjct: 457 YNQGQGTPLPGQGQEGQPSYQMGFSQGLGAPVPPNQVIPGNYGQWAFVN--------YNQ 508
Query: 270 GPPQ-NF--PPQQNHGQ----------ASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYD 316
GPPQ NF PQQNH Q H P Q GQ P Q Q SY+
Sbjct: 509 GPPQGNFLQGPQQNHNQGGQWNYSPQNGGHYGPAQ-FGQWYPGPPQGQGIQWPQYQLSYN 567
Query: 317 QASQ-------RYP-----PQQSYDQASQG-YLPKQNYGQ--APQNYSAPQAPQNYSQQQ 361
Q R P Q Y+ QG ++P+Q GQ A +Y A S Q
Sbjct: 568 QGQGTPFSGQCRCPNCGMTSYQGYNNQGQGTHIPEQWEGQDYAVLSYQA-------SYNQ 620
Query: 362 TYGPASPQYP---PQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQ--NFGPPRQG 416
+G +P Y Q + G G+G N+ Q F V+ NY Q N+GPP G
Sbjct: 621 AHGAQAPPYHGNYNQATPGGYGQGTSANF--NQRF----PVNPANYNMQNGGNYGPP-HG 673
Query: 417 ERRNYVPQQNFGPPGQGE 434
N +Q F GQ +
Sbjct: 674 LAGNPGFRQGFSGQGQNQ 691
>At1g11430 unknown protein
Length = 232
Score = 144 bits (364), Expect = 8e-35
Identities = 80/166 (48%), Positives = 99/166 (59%), Gaps = 20/166 (12%)
Query: 115 DTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTT 174
+TI+ GCDYNHWL V +FP+D P ++MI Y T A L S+EEAKK +YA STTT
Sbjct: 78 ETIMLPGCDYNHWLIVMEFPKDPAPSRDQMIDTYLNTLATVLG-SMEEAKKNMYAFSTTT 136
Query: 175 YTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRN 234
YTGFQ + EE S+KF+G+PGV++VLPDSYID NK YGGD+YI G+IIP P
Sbjct: 137 YTGFQCTIDEETSEKFKGLPGVLWVLPDSYIDVKNKDYGGDKYINGEIIPCTYP------ 190
Query: 235 LGGRRDYRQNNQLPNNRGNPSY-NNRDSMPRDGRNYGPPQNFPPQQ 279
P R N Y + R RDG PP+ P+Q
Sbjct: 191 ----------TYQPKQRNNTKYQSKRYERKRDGP--PPPEQRKPRQ 224
>At3g15000 unknown protein
Length = 395
Score = 141 bits (355), Expect = 9e-34
Identities = 128/413 (30%), Positives = 176/413 (41%), Gaps = 69/413 (16%)
Query: 58 ATSLSVPITGSISSHFSLSKSPNVVDGIQSRS----------FRSTSISLLSSRYGETSE 107
A SLS T S +S L+KSP + SRS FR +S+ TS
Sbjct: 15 AKSLSFLFTRSFASSAPLAKSP--ASSLLSRSRPLVAAFSSVFRGGLVSVKGLSTQATSS 72
Query: 108 LSPEIGP--------DTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNIS 159
+ P +TIL +GCD+ HWL V + P +P +E+I Y +T A+ + S
Sbjct: 73 SLNDPNPNWSNRPPKETILLDGCDFEHWLVVVE-PPQGEPTRDEIIDSYIKTLAQIVG-S 130
Query: 160 VEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIE 219
+EA+ KIY+ ST Y F A+++E+ S K + + V +VLPDSY+D NK YGG+ +I+
Sbjct: 131 EDEARMKIYSVSTRCYYAFGALVSEDLSHKLKELSNVRWVLPDSYLDVRNKDYGGEPFID 190
Query: 220 GQIIPRPPPV--QFGRNLGGRRDYRQNNQLP--NNRGNPSYNNRDSMPRDGRNYGPPQNF 275
G+ +P P ++ RN + + N P N+R R++M PP
Sbjct: 191 GKAVPYDPKYHEEWIRNNARANERNRRNDRPRNNDRSRNFERRRENMAGGPPPQRPPMGG 250
Query: 276 PPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGY 335
PP H S PP G P + Q + P + R+PP
Sbjct: 251 PPPPPHIGGSAPPPPHMGGSAPPPPHM---GQNYGPPPPNNMGGPRHPPP---------- 297
Query: 336 LPKQNYGQAPQ-NYSAPQAPQNYSQQQTYGPASPQY---PPQQSFGPLGEGERRNYAPQQ 391
YG PQ N P+ PQNY G P Y PP + G AP
Sbjct: 298 -----YGAPPQNNMGGPRPPQNYG-----GTPPPNYGGAPPANNMGG---------APPP 338
Query: 392 NF--GPP---GEVDRRNY--APQQNFGPPRQGERRNYVPQQNFGPPGQGERGN 437
N+ GPP G V Y AP QN +QG QN PP + GN
Sbjct: 339 NYGGGPPPQYGAVPPPQYGGAPPQNNNYQQQGSGMQQPQYQNNYPPNRDGSGN 391
>At3g06790 DAG protein, putative
Length = 244
Score = 134 bits (338), Expect = 9e-32
Identities = 88/197 (44%), Positives = 115/197 (57%), Gaps = 10/197 (5%)
Query: 36 SLILLRRSLRSLSGIHHSFSATATSLSVPITGSISSHFSLSKSPNVVDGIQSRS--FRST 93
+LI RR+L +L S S+T+ S S P T S S F++ V S + ST
Sbjct: 2 ALISTRRTLSTLLNKTLS-SSTSYSSSFP-TLSSRSRFAMPLIEKVSSSRTSLGPCYIST 59
Query: 94 SISLLSSRYGETSELSPEIG----PDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYE 149
S Y ++ SP +TIL +GCDY HWL V +F D KP EEMI Y
Sbjct: 60 RPKTSGSGYSPLNDPSPNWSNRPPKETILLDGCDYEHWLIVMEFT-DPKPTEEEMINSYV 118
Query: 150 ETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVN 209
+T L EEAKKKIY+ T+TYTGF A+++EE S K + +PGV++VLPDSY+D N
Sbjct: 119 KTLTSVLGWQ-EEAKKKIYSVCTSTYTGFGALISEELSCKVKALPGVLWVLPDSYLDVPN 177
Query: 210 KQYGGDQYIEGQIIPRP 226
K YGGD Y+EG++IPRP
Sbjct: 178 KDYGGDLYVEGKVIPRP 194
>At2g35240 unknown protein
Length = 232
Score = 121 bits (304), Expect = 8e-28
Identities = 74/178 (41%), Positives = 99/178 (55%), Gaps = 18/178 (10%)
Query: 85 IQSRSFRST-SISLLSSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEE 143
I++R RS S S L S + E+ P LF GCDY HWL V + P ++
Sbjct: 59 IRTRMDRSGGSYSPLKSGSNFSDRPPTEMAP---LFPGCDYEHWLIVMEKPGGENAQKQQ 115
Query: 144 MIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDS 203
MI Y +T AK + S EEA+KKIY S Y GF + EE S K EG+PGV+FVLPDS
Sbjct: 116 MIDCYVQTLAKIVG-SEEEARKKIYNVSCERYFGFGCEIDEETSNKLEGLPGVLFVLPDS 174
Query: 204 YIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGN--PSYNNR 259
Y+DP K YG + ++ G+++PRPP Q R+ +L N RG+ P Y++R
Sbjct: 175 YVDPEFKDYGAELFVNGEVVPRPPERQ-----------RRMVELTNQRGSDKPKYHDR 221
>At1g32580 unknown protein, like plastid protein
Length = 229
Score = 120 bits (302), Expect = 1e-27
Identities = 84/243 (34%), Positives = 119/243 (48%), Gaps = 17/243 (6%)
Query: 27 LSTTTAMASSLILLRRSLRSLSGIHHSFSATATSLSVPITGSISSHFSLSKSPNVVDGIQ 86
++ T A +++ + +R + + S S + S P+ S S N I+
Sbjct: 1 MAKTLARSTASRITKRLISTSGATTPSPSYILSRRSTPV---FSHAVGFISSLNRFTTIR 57
Query: 87 SRSFRST-SISLLSSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMI 145
+R RS S S L S + E+ P LF GCDY HWL V D P ++MI
Sbjct: 58 TRMDRSGGSYSPLKSGSNFSDRAPTEMAP---LFPGCDYEHWLIVMDKPGGENATKQQMI 114
Query: 146 RIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYI 205
Y +T AK + S EEAKKKIY S Y GF + EE S K EG+PGV+F+LPDSY+
Sbjct: 115 DCYVQTLAKIIG-SEEEAKKKIYNVSCERYFGFGCEIDEETSNKLEGLPGVLFILPDSYV 173
Query: 206 DPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRD 265
D NK YG + ++ G+I+ RPP Q R+ Q N++ P Y+++ R
Sbjct: 174 DQENKDYGAELFVNGEIVQRPPERQ-------RKIIELTTQRTNDK--PKYHDKTRYVRR 224
Query: 266 GRN 268
N
Sbjct: 225 REN 227
>At2g33430 plastid protein
Length = 219
Score = 119 bits (298), Expect = 4e-27
Identities = 82/221 (37%), Positives = 109/221 (49%), Gaps = 31/221 (14%)
Query: 65 ITGSISSHFSLSKSPNVVDGIQ---SR---SFRSTSISLLSSRYGET----------SEL 108
+T ++ S+ +L P + + SR S R SI ++R G T S+
Sbjct: 11 LTRALLSNVTLMAPPRIPSSVHYGGSRLGCSTRFFSIRCGANRSGSTYSPLNSGSNFSDR 70
Query: 109 SP-EIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKI 167
P E+ P LF GCDY HWL V D P ++MI Y +T AK + S EEAKK+I
Sbjct: 71 PPTEMAP---LFPGCDYEHWLIVMDKPGGEGATKQQMIDCYIQTLAKVVG-SEEEAKKRI 126
Query: 168 YACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPP 227
Y S Y GF + EE S K EG+PGV+FVLPDSY+DP NK YG + ++ G+I+ R P
Sbjct: 127 YNVSCERYLGFGCEIDEETSTKLEGLPGVLFVLPDSYVDPENKDYGAELFVNGEIVQRSP 186
Query: 228 PVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRN 268
R R Q + P YN+R R N
Sbjct: 187 ----------ERQRRVEPQPQRAQDRPRYNDRTRYSRRREN 217
>At1g53260 DAG-like protein emb|CAA16610.1
Length = 358
Score = 106 bits (265), Expect = 3e-23
Identities = 91/303 (30%), Positives = 134/303 (44%), Gaps = 47/303 (15%)
Query: 142 EEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLP 201
+E+I Y +T A+ + S EEA+ KIY+ S Y F A+++E+ S K + +P V +VLP
Sbjct: 66 DEIIDYYIKTLAQVVG-SEEEARMKIYSVSHKCYFAFGALVSEDLSHKIKELPKVKWVLP 124
Query: 202 DSYIDPVNKQYGGDQYIEGQIIPRPPPV--QFGRNLGGRRDYRQNNQLP-------NNRG 252
DSY+D NK YGG+ +I+G+ +P P ++ RN + + + P N+RG
Sbjct: 125 DSYLDGKNKDYGGEPFIDGKAVPYDPKYHEEWIRNNANATNENRRPRRPRNSDGGRNDRG 184
Query: 253 NPSYNNRDSMPRDGRNYGPP---------------------QNF---PPQQNHGQASHIP 288
N R P G PP QN+ PP N Q P
Sbjct: 185 NQDTGYRRPPPNQGMGGAPPPPPHIGNNPNMPPHIQPPNMNQNYRGPPPPPNMNQNYQGP 244
Query: 289 PQQNIGQQQPNIRIDSTSQRF--PPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQ 346
P N+ Q + Q + PP + +Q+ Q PP + +Q+ QG P N G Q
Sbjct: 245 PAPNMNQNYQGPPPSNMGQNYQGPPPPNMNQSYQG-PPPPNMNQSYQG-PPPSNMG---Q 299
Query: 347 NYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAP 406
NY P P Q GP PP + G G ++ QQ G G + ++ Y P
Sbjct: 300 NYRGPSLPPPNMSQNYEGPP----PPNMNGGWSGNYQQNGGYQQQ--GQGGGMQQQPYPP 353
Query: 407 QQN 409
+N
Sbjct: 354 NRN 356
>At1g72530 DAG-like protein
Length = 188
Score = 81.3 bits (199), Expect = 1e-15
Identities = 49/139 (35%), Positives = 77/139 (55%), Gaps = 7/139 (5%)
Query: 90 FRSTSISLLSSRYG----ETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMI 145
FR + +S S G ET+ S I +++ EGCDY HWL + P + P ++
Sbjct: 17 FRLSPLSPFSGNSGSINSETTSWSELIRVPSLV-EGCDYKHWLVLMK-PPNGYPTRNHIV 74
Query: 146 RIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYI 205
+ + ET A L S EEAK+ IY+ ST Y F + E + K +P V +VLPDS+I
Sbjct: 75 QSFVETLAMALG-SEEEAKRSIYSVSTKYYYAFGCRIHEPLTYKIRSLPDVKWVLPDSFI 133
Query: 206 DPVNKQYGGDQYIEGQIIP 224
+ +YGG+ +++G+++P
Sbjct: 134 VDGDNRYGGEPFVDGEVVP 152
>At5g59170 cell wall protein
Length = 288
Score = 62.0 bits (149), Expect = 7e-10
Identities = 45/192 (23%), Positives = 75/192 (38%), Gaps = 20/192 (10%)
Query: 248 PNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQN--IGQQQPNIRIDST 305
P + P + P Y PP +PP PP ++ + P
Sbjct: 41 PPFKWGPKFPYSPPKPPPIEKYPPPVQYPPPIKKYPP---PPYEHPPVKYPPPIKTYPHP 97
Query: 306 SQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAP-QAP---QNYSQQQ 361
++PP + Y ++YPP + Y + Y P + Y + Y P Q P + Y +
Sbjct: 98 PVKYPPPEQYPPPIKKYPPPEQYPPPIKKYPPPEQYSPPFKKYPPPEQYPPPIKKYPPPE 157
Query: 362 TYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQ---GER 418
Y P +YPPQ+ + P + Y P + + PP + Y P + + PP +
Sbjct: 158 HYPPPIKKYPPQEQYPP----PIKKYPPPEKYPPP----IKKYPPPEQYPPPIKKYPPPI 209
Query: 419 RNYVPQQNFGPP 430
+ Y P + + PP
Sbjct: 210 KKYPPPEEYPPP 221
Score = 57.0 bits (136), Expect = 2e-08
Identities = 41/182 (22%), Positives = 74/182 (40%), Gaps = 17/182 (9%)
Query: 256 YNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQN---IGQQQPNIRIDSTSQRFPPQ 312
Y + P + Y PP+ +PP + PP+Q + P + +++PP
Sbjct: 101 YPPPEQYPPPIKKYPPPEQYPPPIK----KYPPPEQYSPPFKKYPPPEQYPPPIKKYPPP 156
Query: 313 QSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPP 372
+ Y ++YPPQ+ Y + Y P + Y + Y P+ Q + Y P +YPP
Sbjct: 157 EHYPPPIKKYPPQEQYPPPIKKYPPPEKYPPPIKKYPPPE--QYPPPIKKYPPPIKKYPP 214
Query: 373 QQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQ----GERRNYVPQQNFG 428
+ + P + Y PP + P + + PP++ E + P++ +
Sbjct: 215 PEEYPP----PIKTYPHPPVKYPPPPYKTYPHPPIKTYPPPKECPPPPEHYPWPPKKKYP 270
Query: 429 PP 430
PP
Sbjct: 271 PP 272
Score = 38.5 bits (88), Expect = 0.008
Identities = 45/200 (22%), Positives = 73/200 (36%), Gaps = 33/200 (16%)
Query: 194 PGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPN--NR 251
P V + P+ Y P+ K +QY I PPP Q+ + Y Q P +
Sbjct: 97 PPVKYPPPEQYPPPIKKYPPPEQY-PPPIKKYPPPEQYSPPF---KKYPPPEQYPPPIKK 152
Query: 252 GNP---------SYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQN---IGQQQPN 299
P Y ++ P + Y PP+ +PP + PP+Q I + P
Sbjct: 153 YPPPEHYPPPIKKYPPQEQYPPPIKKYPPPEKYPPPIK----KYPPPEQYPPPIKKYPPP 208
Query: 300 IRIDSTSQRFPPQ-QSYDQASQRYPPQ----------QSYDQASQGYLPKQNYGQAPQNY 348
I+ + +PP ++Y +YPP ++Y + P ++Y P+
Sbjct: 209 IKKYPPPEEYPPPIKTYPHPPVKYPPPPYKTYPHPPIKTYPPPKECPPPPEHYPWPPKKK 268
Query: 349 SAPQAPQNYSQQQTYGPASP 368
P + Y PA P
Sbjct: 269 YPPPVEYPSPPYKKYPPADP 288
>At3g20930 unknown protein
Length = 374
Score = 56.6 bits (135), Expect = 3e-08
Identities = 47/162 (29%), Positives = 75/162 (46%), Gaps = 16/162 (9%)
Query: 58 ATSLSVPITGSISSH------FSLSKSPNVVDGIQSRSFRSTSISLLSSRYGETSELSPE 111
+TS VPI+ S SSH F SPN+ G +FR TS+S + ++ +P
Sbjct: 7 STSFFVPISNSSSSHIINNRFFPSFYSPNLNFG----TFRKTSLSSSHLVFSSSAISAPP 62
Query: 112 IGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACS 171
T+L ++W+ + D P M+ Y E AK L + ++A+ IY S
Sbjct: 63 --SSTVLTRN---SYWMVLLDKPPHWVSSKSAMVDYYVEILAKVLG-NEKDAQVSIYDAS 116
Query: 172 TTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYG 213
T+ GF + E+ S++ +PGV+ + P+ K YG
Sbjct: 117 FDTHFGFCCHIDEDASRQLASLPGVVSIRPEQDYSSEKKNYG 158
Score = 44.3 bits (103), Expect = 2e-04
Identities = 32/115 (27%), Positives = 54/115 (46%), Gaps = 3/115 (2%)
Query: 118 LFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTG 177
LF+ HW+ D P +M+ + +K L + ++A+ +Y S + G
Sbjct: 167 LFDHGTVKHWMVRIDKPGVGIVTKAQMVDHCVQLLSKVL-WNEKDAQMCLYHVSWQSDFG 225
Query: 178 FQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGD--QYIEGQIIPRPPPVQ 230
F + E + + G+PGV+ V+PD+ + +NK Y GD Q Q PPV+
Sbjct: 226 FCCDLDERSAVELAGVPGVLAVVPDNSFESLNKDYEGDSTQDSRDQDDSESPPVK 280
>At1g34360 transcription factor like protein
Length = 520
Score = 54.7 bits (130), Expect = 1e-07
Identities = 67/257 (26%), Positives = 90/257 (34%), Gaps = 27/257 (10%)
Query: 180 AVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQY----IEGQIIPRPPPVQFGRNL 235
A + EE K GV V I+ ++ +Q + R P +F
Sbjct: 271 ARVKEESPKPDSSKAGVATVDDQEDIEKSEPRFSVEQAQPVKFQNAYAKREPSSEFS--- 327
Query: 236 GGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQ 295
GGR R Q PN NP R N P F PQ + S P+
Sbjct: 328 GGRDASRFEPQSPNQHVNPQ---RPRFSNQAPNQQPTGRFDPQSPNQPPS--APRPQFPN 382
Query: 296 QQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQA-- 353
QQP R D PP+ + P Q+ +Q S G Q Q P S PQ+
Sbjct: 383 QQPTGRFDPQFPSQPPRPQF--------PNQAPNQQSTGRFNPQFPNQRP---SPPQSRF 431
Query: 354 PQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGP-PGEVDRRNYAPQQNFGP 412
P QQ GP+ ++P +Q P + + N P F P P R PQ P
Sbjct: 432 PDQAPNQQPSGPSPNRHPDRQGPPPRFQNQAPNQQPTGRFEPQPPNPPRAPPRPQTRL-P 490
Query: 413 PRQGERRNYVPQQNFGP 429
+ P ++ GP
Sbjct: 491 NETSNEQPTAPGRSSGP 507
Score = 42.7 bits (99), Expect = 4e-04
Identities = 69/267 (25%), Positives = 99/267 (36%), Gaps = 40/267 (14%)
Query: 139 PPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIF 198
PP + +++ ++ K + E K ++ G V +E+ +K E V
Sbjct: 254 PPKKGGVKLMKDIDIKSARVKEESPKPD------SSKAGVATVDDQEDIEKSEPRFSVEQ 307
Query: 199 VLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQN---NQLPNNRGNP- 254
P + + K+ ++ G+ R P +++ +R N NQ P R +P
Sbjct: 308 AQPVKFQNAYAKREPSSEFSGGRDASRFEPQSPNQHVNPQRPRFSNQAPNQQPTGRFDPQ 367
Query: 255 SYNNRDSMPRDG-RNYGPPQNFPPQ--------QNHGQASHIPPQQNIGQ---QQPNIRI 302
S N S PR N P F PQ Q QA P QQ+ G+ Q PN R
Sbjct: 368 SPNQPPSAPRPQFPNQQPTGRFDPQFPSQPPRPQFPNQA---PNQQSTGRFNPQFPNQRP 424
Query: 303 DSTSQRFPPQQSYDQAS----QRYPPQQSYDQASQGYLPKQN-----YGQAPQNYSAPQA 353
RFP Q Q S R+P +Q Q P Q Q P AP
Sbjct: 425 SPPQSRFPDQAPNQQPSGPSPNRHPDRQGPPPRFQNQAPNQQPTGRFEPQPPNPPRAPPR 484
Query: 354 PQNYSQQQTYGPASPQYP--PQQSFGP 378
PQ +T S + P P +S GP
Sbjct: 485 PQTRLPNET----SNEQPTAPGRSSGP 507
>At5g14540 unknown protein
Length = 530
Score = 50.4 bits (119), Expect = 2e-06
Identities = 56/174 (32%), Positives = 66/174 (37%), Gaps = 40/174 (22%)
Query: 271 PPQNF--PPQQNHG------QASHIP----PQQN-----IGQQQPNIRIDSTSQRFPPQQ 313
PPQ F PP HG Q +P PQQ GQ QP I Q PP Q
Sbjct: 245 PPQQFIQPPASQHGLSPPSLQLPQLPNQFSPQQEPYFPPSGQSQPPPTIQPPYQPPPPTQ 304
Query: 314 SYDQASQRYPPQQ-SYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPP 372
S Q + PPQQ Y Q L Q P Y+ + P QQ+Y P P+ PP
Sbjct: 305 SLHQPPYQPPPQQPQYPQQPPPQL------QHPSGYNPEEPPY---PQQSYPPNPPRQPP 355
Query: 373 QQSFGPLGEGERRNY------APQQNFGPPGEVDR---RNYAPQQ--NFGPPRQ 415
S P G + Y P GP G + Y+P+ GPP Q
Sbjct: 356 --SHPPPGSAPSQQYYNAPPTPPSMYDGPGGRSNSGFPSGYSPESYPYTGPPSQ 407
Score = 45.8 bits (107), Expect = 5e-05
Identities = 50/200 (25%), Positives = 67/200 (33%), Gaps = 16/200 (8%)
Query: 245 NQLPNNRGN---PSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIR 301
NQ P P+ S+P+ + P + Q Q PP G P+++
Sbjct: 206 NQQPETHSTHVEPTAQPPASLPQPPASAAAPPSLTQQGLPPQQFIQPPASQHGLSPPSLQ 265
Query: 302 IDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQ 361
+ +F PQQ + Y P Q P Y P S Q P QQ
Sbjct: 266 LPQLPNQFSPQQ------EPYFPPSGQSQPPPTIQPP--YQPPPPTQSLHQPPYQPPPQQ 317
Query: 362 TYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNY 421
P PQ PP Q P G PQQ++ PP + P P +Q
Sbjct: 318 ---PQYPQQPPPQLQHPSGYNPEEPPYPQQSY-PPNPPRQPPSHPPPGSAPSQQYYNAPP 373
Query: 422 VPQQNFGPPGQGERGNSVPS 441
P + PG G + PS
Sbjct: 374 TPPSMYDGPG-GRSNSGFPS 392
>At1g09070 unknown protein
Length = 324
Score = 47.0 bits (110), Expect = 2e-05
Identities = 38/125 (30%), Positives = 49/125 (38%), Gaps = 13/125 (10%)
Query: 313 QSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQ-APQNYSQQQTYGPASPQYP 371
++ DQ YPP A + G PQ + Y QQ Y + YP
Sbjct: 157 KTMDQPVTAYPPGHGAPSAYPAPPAGPSSGYPPQGHDDKHDGVYGYPQQAGYPAGTGGYP 216
Query: 372 PQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPG 431
P ++ +G Y PQQ G PG Y PQ +G P+QG Y PQ +G P
Sbjct: 217 PPGAYPQ--QGGYPGYPPQQQGGYPG------YPPQGPYGYPQQG----YPPQGPYGYPQ 264
Query: 432 QGERG 436
Q G
Sbjct: 265 QQAHG 269
Score = 45.4 bits (106), Expect = 7e-05
Identities = 35/115 (30%), Positives = 44/115 (37%), Gaps = 13/115 (11%)
Query: 270 GPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQAS--QRYPPQQS 327
GP +PPQ H + + + +PP +Y Q YPPQQ
Sbjct: 182 GPSSGYPPQ------GHDDKHDGVYGYPQQAGYPAGTGGYPPPGAYPQQGGYPGYPPQQQ 235
Query: 328 YDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEG 382
GY P+ YG Q Y PQ P Y QQQ +G PQ P + G G
Sbjct: 236 --GGYPGYPPQGPYGYPQQGYP-PQGPYGYPQQQAHG--KPQKPKKHGKAGAGMG 285
Score = 33.5 bits (75), Expect = 0.27
Identities = 41/140 (29%), Positives = 55/140 (39%), Gaps = 34/140 (24%)
Query: 306 SQRFPPQQSYDQASQRYPP------QQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQ 359
S +F + +Y +S + P ++ DQ Y P AP Y AP A
Sbjct: 131 SFKFGEKYTYGSSSGPHAPVPSAMDHKTMDQPVTAYPPGHG---APSAYPAPPA------ 181
Query: 360 QQTYGPASPQYPPQQSFGPLGEGERRNYA---PQQNFGPPGEVDRRNYAPQQNFGPPRQG 416
GP+S YPPQ G ++ + PQQ P G Y P + P+QG
Sbjct: 182 ----GPSSG-YPPQ------GHDDKHDGVYGYPQQAGYPAGT---GGYPPPGAY--PQQG 225
Query: 417 ERRNYVPQQNFGPPGQGERG 436
Y PQQ G PG +G
Sbjct: 226 GYPGYPPQQQGGYPGYPPQG 245
>At1g30330 putative protein
Length = 933
Score = 46.6 bits (109), Expect = 3e-05
Identities = 53/213 (24%), Positives = 77/213 (35%), Gaps = 24/213 (11%)
Query: 272 PQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQA 331
P PQ Q S QQ QQQ ++ Q+ QQ Q SQ+ Q S Q
Sbjct: 474 PSLVQPQMLQQQLSQ-QQQQLSQQQQQQQQLSQQQQQQLSQQQQQQLSQQQQQQLSQQQQ 532
Query: 332 SQGYLPKQNYGQAPQNYSAPQAPQNYSQQQ-----TYGPASPQYPPQQSFGPLGEGERRN 386
Q YL Q PQ+ + Q+ + SQQQ + P++ + G + N
Sbjct: 533 QQAYLGVPETHQ-PQSQAQSQSNNHLSQQQQQVVDNHNPSASSAAVVSAMSQFGSASQPN 591
Query: 387 YAPQQNFGPPGEVDRRNYAPQQNFGPPRQGER---RNYVPQQNFGPPGQGE-----RGNS 438
+P Q+ + QQ+F G + NF + R NS
Sbjct: 592 TSPLQSM--------TSLCHQQSFSDTNGGNNPISPLHTLLSNFSQDESSQLLHLTRTNS 643
Query: 439 VPSEGGWDFK-PSYMEEFEQGQKGNNHAEEQKE 470
+ GW K P+ F+ GNN+ + E
Sbjct: 644 AMTSSGWPSKRPAVDSSFQHSGAGNNNTQSVLE 676
>At2g25970 pseudogene
Length = 632
Score = 45.8 bits (107), Expect = 5e-05
Identities = 65/245 (26%), Positives = 87/245 (34%), Gaps = 51/245 (20%)
Query: 252 GNPSYNNRDSMPRDGRNYGPPQ-------NFPPQQNHG--QASHIPPQQNIGQ------- 295
G P+ + G GPPQ ++P Q + G S +PP Q Q
Sbjct: 335 GPPAQPGYGGYMQPGAYPGPPQYGQSPYGSYPQQTSAGYYDQSSVPPSQQSAQGEYDYYG 394
Query: 296 QQPNIRIDSTSQRFPPQQS-----------YDQASQRYPP----------QQSYDQASQG 334
QQ + + S PP + Y QA Q Y Q Y QA+ G
Sbjct: 395 QQQSQQPSSGGSSAPPTDTTGYNYYQHASGYGQAGQGYQQDGYGAYNASQQSGYGQAA-G 453
Query: 335 YLPKQNYGQAP---QNYSAPQA-PQNYSQ--QQTYGPASPQYPPQQSFGPLGEGERRNYA 388
Y + YG Q A QA P + +Q Q YG Q P Q S G G G
Sbjct: 454 YDQQGGYGSTTNPSQEEDASQAAPPSSAQSGQAGYGTTGQQPPAQGSTGQAGYGAPPTSQ 513
Query: 389 PQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQ--GERGNSVPSEGGWD 446
+ P + AP PP G+ QQ+ G PG + G + P+ G+
Sbjct: 514 AGYSSQPAAAYNSGYGAPPPASKPPTYGQ-----SQQSPGAPGSYGSQSGYAQPAASGYG 568
Query: 447 FKPSY 451
P+Y
Sbjct: 569 QPPAY 573
Score = 35.4 bits (80), Expect = 0.072
Identities = 39/117 (33%), Positives = 46/117 (38%), Gaps = 21/117 (17%)
Query: 269 YGPPQNFPPQQNH-GQASH-IPPQQNIG-QQQPNIRIDSTSQRFPPQQ---SYDQASQR- 321
YG PP Q GQA + PP G QP +S PP +Y Q+ Q
Sbjct: 488 YGTTGQQPPAQGSTGQAGYGAPPTSQAGYSSQPAAAYNSGYGAPPPASKPPTYGQSQQSP 547
Query: 322 -----YPPQQSYDQ-ASQGY--LPKQNYGQAPQNYSA------PQAPQNYSQQQTYG 364
Y Q Y Q A+ GY P YGQAPQ Y + P A YS + G
Sbjct: 548 GAPGSYGSQSGYAQPAASGYGQPPAYGYGQAPQGYGSYGGYTQPAAGGGYSSDGSAG 604
>At1g70180 unknown protein
Length = 460
Score = 45.8 bits (107), Expect = 5e-05
Identities = 45/139 (32%), Positives = 69/139 (49%), Gaps = 26/139 (18%)
Query: 223 IPRPP-PVQFGRNL-GGRRDYRQNNQLPNNRGNPSYNNRD-SMPRD-GRNYGPPQNFPPQ 278
IPR P P + GR + G RD P + G + RD S PRD GR +GPP++ P
Sbjct: 189 IPREPSPPRNGRRMIGSARD----RSPPRSDGRIIGSPRDISPPRDAGRRFGPPRDQSPP 244
Query: 279 QNHGQASHIP----PQQNIGQQQPNIRIDSTSQRFPPQ--QSYDQASQRYPPQQ---SYD 329
+N G+ + P P +N G+ R+ + PP+ +S+ S+ P + SY
Sbjct: 245 RNAGRVTGSPRDRSPPRNAGR-----RMGPPRDQSPPRSTRSFSSNSRALSPARNMGSYM 299
Query: 330 QASQGYLPKQNYGQAPQNY 348
+SQG+ P +N P++Y
Sbjct: 300 SSSQGFSPPRN----PRSY 314
Score = 43.1 bits (100), Expect = 3e-04
Identities = 51/183 (27%), Positives = 77/183 (41%), Gaps = 31/183 (16%)
Query: 219 EGQIIPRP----PPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRD-SMPRD-GRNYGPP 272
+G+II P PP GR G RD P N G + + RD S PR+ GR GPP
Sbjct: 215 DGRIIGSPRDISPPRDAGRRFGPPRD----QSPPRNAGRVTGSPRDRSPPRNAGRRMGPP 270
Query: 273 QN-FPPQQNHGQASH---IPPQQNIGQQQPNIRIDSTSQRFPP---QQSYDQASQRYPPQ 325
++ PP+ +S+ + P +N+G S+SQ F P +SY +S+ PP+
Sbjct: 271 RDQSPPRSTRSFSSNSRALSPARNMGSYM------SSSQGFSPPRNPRSYMGSSRGSPPR 324
Query: 326 QSYDQASQGYLPKQNYGQAPQ------NYSAPQAPQNYSQQQTYGPASPQY--PPQQSFG 377
+ + N +P N AP + +++ P + PP
Sbjct: 325 SNIEDFHGRSRMLDNMRASPYPVRGVLNGQAPSSGAPFARPMLPPPVPNPHPLPPLSQLP 384
Query: 378 PLG 380
PLG
Sbjct: 385 PLG 387
Score = 37.4 bits (85), Expect = 0.019
Identities = 44/219 (20%), Positives = 77/219 (35%), Gaps = 15/219 (6%)
Query: 267 RNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQ 326
R+ PP++ + + P + + + I S+S P + + +R P +
Sbjct: 113 RSEQPPRSLDTRPRMAEPRDRPLSSSRTARGSSQMISSSSSH--PAWALEDLRRRSP--E 168
Query: 327 SYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRN 386
+ S+G P +N G P P+N ++ G A + PP+ +G
Sbjct: 169 RFVDTSRGRSPPRNAGSFIGIPREPSPPRN--GRRMIGSARDRSPPRSDGRIIGS----- 221
Query: 387 YAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEGGWD 446
++ PP + RR + P ++ PPR R P+ P G R +
Sbjct: 222 ---PRDISPPRDAGRR-FGPPRDQSPPRNAGRVTGSPRDRSPPRNAGRRMGPPRDQSPPR 277
Query: 447 FKPSYMEEFEQGQKGNNHAEEQKESQQRFPPPGPGNFTG 485
S+ N SQ PP P ++ G
Sbjct: 278 STRSFSSNSRALSPARNMGSYMSSSQGFSPPRNPRSYMG 316
>At2g16470 unknown protein
Length = 670
Score = 45.4 bits (106), Expect = 7e-05
Identities = 54/210 (25%), Positives = 82/210 (38%), Gaps = 24/210 (11%)
Query: 278 QQNHGQASHIPPQQN----IGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQ 333
Q +HG S+ P QN G P++ + FPP S+ A P Q+ QA
Sbjct: 409 QTDHG-GSNTPSSQNNSTSYGTPSPSVLPSQSQPGFPPSDSWKVAVPSQPNAQA--QAQW 465
Query: 334 GYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNF 393
G N QN + PQAP N + G +P GP G N+
Sbjct: 466 G----MNMVNNNQNSAQPQAPANQNSSWGQGTVNPN---MGWVGPAQTGVNVNWG---GS 515
Query: 394 GPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPG--QGERGNSVPSEGGWDFKPSY 451
P V ++ + P QG+ + Y P +GP G Q + + V ++ G +
Sbjct: 516 SVPSTVQGITHS---GWVAPVQGQTQAY-PNPGWGPTGHPQSQSQSQVQAQAG-TTGSGW 570
Query: 452 MEEFEQGQKGNNHAEEQKESQQRFPPPGPG 481
M+ + Q GN++ ++Q P G G
Sbjct: 571 MQPGQGIQSGNSNQNWGTQNQTAIPSGGSG 600
>At1g74720 hypothetical protein
Length = 1081
Score = 45.4 bits (106), Expect = 7e-05
Identities = 43/139 (30%), Positives = 61/139 (42%), Gaps = 26/139 (18%)
Query: 267 RNYGPPQNFPPQQNHGQASHIPPQQ--NIGQQQPNIRIDSTSQRFPPQQS--YDQASQRY 322
+ + PPQ +Q H Q H PPQQ NI ++PN+ + + F QS Y + Q+
Sbjct: 169 QQFHPPQQEADEQQHQQQFHPPPQQMMNIPPEKPNVVVVEEGRVFESAQSQRYTETHQQ- 227
Query: 323 PPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEG 382
PP +++ P Q+ Q P N + P N+ Q+ P SP PP G
Sbjct: 228 PPVVIVEES-----PPQHVMQGP-NDNHPHRNDNHPQR----PPSPPPPP-------SAG 270
Query: 383 ERRNYAPQ----QNFGPPG 397
E Y P+ Q PPG
Sbjct: 271 EVHYYPPEVRKMQVGRPPG 289
Score = 31.6 bits (70), Expect = 1.0
Identities = 34/128 (26%), Positives = 46/128 (35%), Gaps = 33/128 (25%)
Query: 380 GEGERRNYAPQQNFGPP-GEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQGERGNS 438
G G ++ QQ F PP E D + + QQ F PP PQQ P E+ N
Sbjct: 158 GGGGQQQQQQQQQFHPPQQEADEQQH--QQQFHPP---------PQQMMNIP--PEKPNV 204
Query: 439 VPSEGGWDFKPSYMEEFEQGQK-------------------GNNHAEEQKESQQRFPPPG 479
V E G F+ + + + + + +NH QR P P
Sbjct: 205 VVVEEGRVFESAQSQRYTETHQQPPVVIVEESPPQHVMQGPNDNHPHRNDNHPQRPPSPP 264
Query: 480 PGNFTGEV 487
P GEV
Sbjct: 265 PPPSAGEV 272
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,333,573
Number of Sequences: 26719
Number of extensions: 702247
Number of successful extensions: 2175
Number of sequences better than 10.0: 221
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 171
Number of HSP's that attempted gapping in prelim test: 1576
Number of HSP's gapped (non-prelim): 451
length of query: 488
length of database: 11,318,596
effective HSP length: 103
effective length of query: 385
effective length of database: 8,566,539
effective search space: 3298117515
effective search space used: 3298117515
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC134322.18


