

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.16 - phase: 0
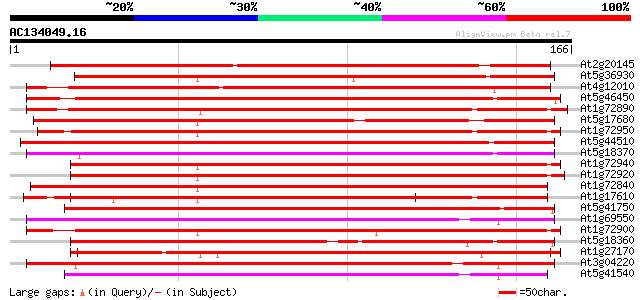
(166 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g20145 unknown protein 148 1e-36
At5g36930 disease resistance like protein 132 1e-31
At4g12010 like disease resistance protein (TMV N-like) 131 2e-31
At5g46450 disease resistance protein-like 129 9e-31
At1g72890 hypothetical protein 129 9e-31
At5g17680 disease resistance protein RPP1-WsB - like protein 128 2e-30
At1g72950 hypothetical protein 128 2e-30
At5g44510 disease resistance protein-like 127 2e-30
At5g18370 disease resistance protein -like 127 2e-30
At1g72940 unknown protein 127 3e-30
At1g72920 hypothetical protein 127 3e-30
At1g72840 hypothetical protein 127 3e-30
At1g17610 disease resistance protein, putative 127 3e-30
At5g41750 disease resistance protein-like 127 4e-30
At1g69550 putative disease resistance protein 126 5e-30
At1g72900 unknown protein 126 6e-30
At5g18360 disease resistance protein -like 125 1e-29
At1g27170 disease resistance protein, putative 125 1e-29
At3g04220 putative disease resistance protein 123 4e-29
At5g41540 disease resistance protein-like 123 5e-29
>At2g20145 unknown protein
Length = 187
Score = 148 bits (374), Expect = 1e-36
Identities = 72/148 (48%), Positives = 106/148 (70%), Gaps = 4/148 (2%)
Query: 13 NNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEE 72
N++P KH+VFISFR +DTR+NF SHL G L+R I+T++ ++L + + +L +AIE
Sbjct: 2 NSSPKMKHDVFISFRSKDTRDNFVSHLCGCLRRKRIKTFLYDELPADERYEESL-KAIEV 60
Query: 73 AKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFA 132
+K+SVIVFS+N+ S+WCL+E++ IL+ K+ GQIV+PV Y VDP D+ NQ GS+ +AFA
Sbjct: 61 SKISVIVFSENFGDSRWCLDEVVAILKCKEKFGQIVIPVLYHVDPLDIENQTGSFGDAFA 120
Query: 133 KHENNFEGKIKVQEWRNGLLEAANYAGW 160
K + E ++QEW++ EA N GW
Sbjct: 121 KRRDKAE---QLQEWKDSFTEAINLPGW 145
>At5g36930 disease resistance like protein
Length = 1164
Score = 132 bits (331), Expect = 1e-31
Identities = 65/144 (45%), Positives = 100/144 (69%), Gaps = 3/144 (2%)
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTLVRAIEEAKLSVI 78
++VF+SFRG D R NF SHL +L+R I T++D+ +L G+ I L+ AIE +K+ ++
Sbjct: 14 YDVFVSFRGADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPELLNAIETSKILIV 73
Query: 79 VFSKNYAVSKWCLEELMKILEI-KKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENN 137
V +K+YA S WCL+EL+ I++ K +V P+F VDPSD+R Q+GSYA++F+KH+N+
Sbjct: 74 VLTKDYASSAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQGSYAKSFSKHKNS 133
Query: 138 FEGKIKVQEWRNGLLEAANYAGWD 161
K+++WR L + AN +GWD
Sbjct: 134 HPLN-KLKDWREALTKVANISGWD 156
>At4g12010 like disease resistance protein (TMV N-like)
Length = 1219
Score = 131 bits (329), Expect = 2e-31
Identities = 74/157 (47%), Positives = 97/157 (61%), Gaps = 9/157 (5%)
Query: 6 SSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTT 65
SSS SS + +VF+SFRG DTRNNFT HL AL+ I ++ID+ L GD + T
Sbjct: 3 SSSPSS------AEFDVFLSFRGFDTRNNFTGHLQKALRLRGIDSFIDDRLRRGDNL-TA 55
Query: 66 LVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRG 125
L IE++K+++IVFS NYA S WCL EL+KILE + Q+VVP+FY VD SDV QR
Sbjct: 56 LFDRIEKSKIAIIVFSTNYANSAWCLRELVKILECRNSNQQLVVPIFYKVDKSDVEKQRN 115
Query: 126 SYAEAFAKHENNFEGKI--KVQEWRNGLLEAANYAGW 160
S+A F E F G ++ W+ L A+N G+
Sbjct: 116 SFAVPFKLPELTFPGVTPEEISSWKAALASASNILGY 152
>At5g46450 disease resistance protein-like
Length = 1123
Score = 129 bits (323), Expect = 9e-31
Identities = 71/161 (44%), Positives = 99/161 (61%), Gaps = 8/161 (4%)
Query: 6 SSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTT 65
SSSSSS N + ++VF SF GED R F SH L+R I T+ DN++ I
Sbjct: 3 SSSSSSRNWS----YDVFPSFSGEDVRKTFLSHFLRELERKSIITFKDNEMERSQSIAPE 58
Query: 66 LVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRG 125
LV AI++++++VIVFSKNYA S WCL EL++I+ K GQ V+PVFY +DPS +R Q G
Sbjct: 59 LVEAIKDSRIAVIVFSKNYASSSWCLNELLEIMRCNKYLGQQVIPVFYYLDPSHLRKQSG 118
Query: 126 SYAEAFAKHENNFEGKIKVQEWRNGLLEAANYAGW---DCN 163
+ EAF K N ++K +W+ L + +N G+ +CN
Sbjct: 119 EFGEAFKKTCQNQTEEVK-NQWKQALTDVSNILGYHSKNCN 158
>At1g72890 hypothetical protein
Length = 438
Score = 129 bits (323), Expect = 9e-31
Identities = 71/161 (44%), Positives = 106/161 (65%), Gaps = 6/161 (3%)
Query: 6 SSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPT 64
SSSSSSF +K +VF+SFRG+DTR NF S L AL IRT+ D++ L G IP
Sbjct: 28 SSSSSSFTP---QKFDVFLSFRGKDTRKNFVSFLYKALVSKGIRTFKDDEELERGRPIPP 84
Query: 65 TLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQR 124
L +AI+ ++++V+V S Y S WCLEEL +IL+++K+ V+P+FY+++PSDVR Q
Sbjct: 85 ELRQAIKGSRIAVVVVSVTYPASSWCLEELREILKLEKLGLLTVIPIFYEINPSDVRRQS 144
Query: 125 GSYAEAFAKHENNFEGKIKVQEWRNGLLEAANYAGWDCNVN 165
G ++ F KHE + + +V+ WR L + A+ +G +C+ N
Sbjct: 145 GVVSKQFKKHEKR-QSRERVKSWREALTKLASLSG-ECSKN 183
>At5g17680 disease resistance protein RPP1-WsB - like protein
Length = 1294
Score = 128 bits (321), Expect = 2e-30
Identities = 68/155 (43%), Positives = 98/155 (62%), Gaps = 8/155 (5%)
Query: 8 SSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTL 66
SSSS +++ + K +VF+SFRGED R F SHL R+ I+ + D+ DL G I L
Sbjct: 6 SSSSSSSSTVWKTDVFVSFRGEDVRKTFVSHLFCEFDRMGIKAFRDDLDLQRGKSISPEL 65
Query: 67 VRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGS 126
+ AI+ ++ +++V S+NYA S WCL+EL+KI+E K +VP+FY+VDPSDVR QRGS
Sbjct: 66 IDAIKGSRFAIVVVSRNYAASSWCLDELLKIMECNK---DTIVPIFYEVDPSDVRRQRGS 122
Query: 127 YAEAFAKHENNFEGKIKVQEWRNGLLEAANYAGWD 161
+ E H + K KV +W+ L + A +G D
Sbjct: 123 FGEDVESHSD----KEKVGKWKEALKKLAAISGED 153
>At1g72950 hypothetical protein
Length = 379
Score = 128 bits (321), Expect = 2e-30
Identities = 74/156 (47%), Positives = 97/156 (61%), Gaps = 5/156 (3%)
Query: 9 SSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTLV 67
S S N P K +VF+SFRG DTR +F S L L R +IRT+ D+ +L +G I L+
Sbjct: 2 SDSSNTLP--KFDVFLSFRGLDTRRSFISFLYKELIRRNIRTFKDDKELKNGRRITPELI 59
Query: 68 RAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSY 127
RAIE ++ +V+V S NYA S+WCLEEL+KI++ + M V+P+FY VDP VR Q G
Sbjct: 60 RAIEGSRFAVVVVSVNYAASRWCLEELVKIMDFENMGSLKVMPIFYGVDPCHVRRQIGEV 119
Query: 128 AEAFAKHENNFEGKIKVQEWRNGLLEAANYAGWDCN 163
AE F KHE E KV WR L A+ +G DC+
Sbjct: 120 AEQFKKHEGR-EDHEKVLSWRQALTNLASISG-DCS 153
>At5g44510 disease resistance protein-like
Length = 1187
Score = 127 bits (320), Expect = 2e-30
Identities = 67/158 (42%), Positives = 97/158 (60%), Gaps = 1/158 (0%)
Query: 4 SYSSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIP 63
S SSSS + + H VF+SFRGED R SH+ +R I +IDN++ G I
Sbjct: 24 SLSSSSPPSSLSQNWLHPVFLSFRGEDVRKGLLSHIQKEFQRNGITPFIDNEMKRGGSIG 83
Query: 64 TTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQ 123
L++AI +K+++I+ S+NY SKWCL+EL++I++ ++ GQ V+ VFYDVDPSDVR Q
Sbjct: 84 PELLQAIRGSKIAIILLSRNYGSSKWCLDELVEIMKCREELGQTVMTVFYDVDPSDVRKQ 143
Query: 124 RGSYAEAFAKHENNFEGKIKVQEWRNGLLEAANYAGWD 161
+G + + F K ++ VQ W+ L AAN G D
Sbjct: 144 KGDFGKVFKKTCVGRPEEM-VQRWKQALTSAANILGED 180
>At5g18370 disease resistance protein -like
Length = 1210
Score = 127 bits (320), Expect = 2e-30
Identities = 69/164 (42%), Positives = 97/164 (59%), Gaps = 9/164 (5%)
Query: 6 SSSSSSFNNAPLEK--------HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLN 57
SSSSSS ++ P H+VF SFRGED R F SH+ K I +ID+++
Sbjct: 39 SSSSSSLSSPPSLSSPISRTWTHQVFPSFRGEDVRKGFLSHIQKEFKSKGIVPFIDDEMK 98
Query: 58 SGDEIPTTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDP 117
G+ I L +AI E+K+++++ SKNYA S WCL EL++I+ ++ GQ V+ VFY VDP
Sbjct: 99 RGESIGPGLFQAIRESKIAIVLLSKNYASSSWCLNELVEIMNCREEIGQTVMTVFYQVDP 158
Query: 118 SDVRNQRGSYAEAFAKHENNFEGKIKVQEWRNGLLEAANYAGWD 161
SDVR Q G + +AF K ++K Q W L++ AN G D
Sbjct: 159 SDVRKQTGDFGKAFKKTCVGKTQEVK-QRWSRALMDVANILGQD 201
>At1g72940 unknown protein
Length = 371
Score = 127 bits (319), Expect = 3e-30
Identities = 69/146 (47%), Positives = 90/146 (61%), Gaps = 2/146 (1%)
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTLVRAIEEAKLSV 77
K++VF+SFRG DTR NF S L R IRT+ D+ +L +G I L RAIEE+K +V
Sbjct: 8 KYDVFLSFRGVDTRRNFISFLYKEFVRRKIRTFKDDKELENGRRISPELKRAIEESKFAV 67
Query: 78 IVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENN 137
+V S NYA S WCL+EL+KI++ + V+P+FY VDP +R Q G AE F KHE
Sbjct: 68 VVVSVNYAASPWCLDELVKIMDFENKGSITVMPIFYGVDPCHLRRQIGDVAEQFKKHEAR 127
Query: 138 FEGKIKVQEWRNGLLEAANYAGWDCN 163
E KV WR L A+ +G DC+
Sbjct: 128 EEDHEKVASWRRALTSLASISG-DCS 152
>At1g72920 hypothetical protein
Length = 275
Score = 127 bits (319), Expect = 3e-30
Identities = 69/147 (46%), Positives = 92/147 (61%), Gaps = 2/147 (1%)
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTLVRAIEEAKLSV 77
K++VF+SFRG DTR NF S L L R IRT+ D+ +L +G I L RAIEE+K +V
Sbjct: 8 KYDVFLSFRGLDTRRNFISFLYKELVRRKIRTFKDDKELENGRRISPELKRAIEESKFAV 67
Query: 78 IVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENN 137
+V S NYA S WCL+EL+KI++ + V+P+FY VDP +R Q G AE F KHE
Sbjct: 68 VVVSVNYAASPWCLDELVKIMDFENKGSITVMPIFYGVDPCHLRRQIGDVAEQFKKHEAR 127
Query: 138 FEGKIKVQEWRNGLLEAANYAGWDCNV 164
E KV WR L A+ +G +C++
Sbjct: 128 EEDHEKVASWRRALTSLASISG-ECSL 153
>At1g72840 hypothetical protein
Length = 1183
Score = 127 bits (319), Expect = 3e-30
Identities = 68/154 (44%), Positives = 98/154 (63%), Gaps = 1/154 (0%)
Query: 7 SSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTT 65
+SSSS + L ++VF+SFRG DTR SHL AL+ + T+ D+ L GD I
Sbjct: 2 ASSSSSSATRLRHYDVFLSFRGVDTRQTIVSHLYVALRNNGVLTFKDDRKLEIGDTIADG 61
Query: 66 LVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRG 125
LV+AI+ + +V++ S+NYA S WCLEEL I+++ + V+P+FY V PSDVR Q G
Sbjct: 62 LVKAIQTSWFAVVILSENYATSTWCLEELRLIMQLHSEEQIKVLPIFYGVKPSDVRYQEG 121
Query: 126 SYAEAFAKHENNFEGKIKVQEWRNGLLEAANYAG 159
S+A AF ++E + E + KV +WR L + AN +G
Sbjct: 122 SFATAFQRYEADPEMEEKVSKWRRALTQVANLSG 155
>At1g17610 disease resistance protein, putative
Length = 836
Score = 127 bits (319), Expect = 3e-30
Identities = 70/156 (44%), Positives = 101/156 (63%), Gaps = 4/156 (2%)
Query: 5 YSSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIP 63
YSSSSSS +A +K +VF+SFRG+DTR F S L L + IRT+ D+ +L SG I
Sbjct: 458 YSSSSSS--SATPQKFDVFLSFRGKDTRRTFISFLYKELIEMGIRTFKDDVELQSGRRIA 515
Query: 64 TTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQ 123
+ L+ AIE +K++V++ SKNY+ S WCL+EL+ I++++K IV+P+FY+V+P+ VR Q
Sbjct: 516 SDLLAAIENSKIAVVIISKNYSASPWCLQELVMIMDVEKKGSIIVMPIFYNVEPAHVRRQ 575
Query: 124 RGSYAEAFAKHENNFEGKIKVQEWRNGLLEAANYAG 159
A+ F KHE E V WR L A+ +G
Sbjct: 576 IEQVAKQFRKHEGR-ENYETVVSWRQALTNLASISG 610
Score = 39.7 bits (91), Expect = 8e-04
Identities = 30/105 (28%), Positives = 52/105 (48%), Gaps = 7/105 (6%)
Query: 19 KHEVFISFRGE---DTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEEAKL 75
K +VF+SF G+ D + NG +K ++ ++ I + A+ E+K+
Sbjct: 22 KLDVFLSFSGKIALDVDFGYDLSRNG-IKAFKSESWKESSFKP---IDLRTLEALTESKV 77
Query: 76 SVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDV 120
+V++ S S LEEL+ I+E ++ + V+PVF P DV
Sbjct: 78 AVVMTSDEEVSSVGFLEELIVIIEFQEKRSLTVIPVFLTKHPLDV 122
>At5g41750 disease resistance protein-like
Length = 1068
Score = 127 bits (318), Expect = 4e-30
Identities = 64/150 (42%), Positives = 92/150 (60%), Gaps = 6/150 (4%)
Query: 17 LEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEEAKLS 76
+++++VF SF G D R F SHL+ I T+ D ++ G I L++ I EA++S
Sbjct: 10 IKRYQVFSSFHGPDVRKGFLSHLHSVFASKGITTFNDQKIDRGQTIGPELIQGIREARVS 69
Query: 77 VIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHEN 136
++V SK YA S WCL+EL++IL+ K+ GQIV+ VFY+VDPSDV+ Q G + EAF K
Sbjct: 70 IVVLSKKYASSSWCLDELVEILKCKEALGQIVMTVFYEVDPSDVKKQSGVFGEAFEKTCQ 129
Query: 137 NFEGKIKVQEWRNGLLEAANYAG-----WD 161
++K++ WRN L A AG WD
Sbjct: 130 GKNEEVKIR-WRNALAHVATIAGEHSLNWD 158
>At1g69550 putative disease resistance protein
Length = 1398
Score = 126 bits (317), Expect = 5e-30
Identities = 64/158 (40%), Positives = 95/158 (59%), Gaps = 5/158 (3%)
Query: 6 SSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTT 65
SS+S + + H VF SFRG+D R NF SH+ +R I +IDN++ G+ I
Sbjct: 64 SSTSPPSSLSCTGTHHVFPSFRGDDVRRNFLSHIQKEFRRKGITPFIDNEIRRGESIGPE 123
Query: 66 LVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRG 125
L++AI E+K+++++ S+NYA SKWCLEEL++I++ KK G V +FY+VDPS V+ G
Sbjct: 124 LIKAIRESKIAIVLLSRNYASSKWCLEELVEIMKCKKEFGLTVFAIFYEVDPSHVKKLTG 183
Query: 126 SYAEAFAKHENNFEGKIK--VQEWRNGLLEAANYAGWD 161
+ F K +G+ K + WR E A AG+D
Sbjct: 184 EFGAVFQK---TCKGRTKENIMRWRQAFEEVATIAGYD 218
>At1g72900 unknown protein
Length = 363
Score = 126 bits (316), Expect = 6e-30
Identities = 71/160 (44%), Positives = 103/160 (64%), Gaps = 9/160 (5%)
Query: 6 SSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPT 64
SS+++++N ++VF+SFRG DTR F S L L DIRT+ D+ +L +G I
Sbjct: 2 SSATATYN------YDVFLSFRGPDTRRKFISFLYKELVGRDIRTFKDDKELENGQMISP 55
Query: 65 TLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQI-VVPVFYDVDPSDVRNQ 123
L+ AIE+++ +V+V S NYA S WCL+EL+KI++I+K KG I V+P+FY V+P +R Q
Sbjct: 56 ELILAIEDSRFAVVVVSVNYAASSWCLDELVKIMDIQKNKGSITVMPIFYGVNPCHLRRQ 115
Query: 124 RGSYAEAFAKHENNFEGKIKVQEWRNGLLEAANYAGWDCN 163
G AE F KHE + KV +WR L A+ +G DC+
Sbjct: 116 IGDVAEQFKKHEAREKDLEKVLKWRQALAALADISG-DCS 154
>At5g18360 disease resistance protein -like
Length = 900
Score = 125 bits (314), Expect = 1e-29
Identities = 69/149 (46%), Positives = 96/149 (64%), Gaps = 11/149 (7%)
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEEAKLSVI 78
+H VF SF G+D R F SHL +R IRT+IDND+ I + LVRAI E++++V+
Sbjct: 15 RHHVFPSFSGKDVRRTFLSHLLKEFRRKGIRTFIDNDIKRSQMISSELVRAIRESRIAVV 74
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKH-ENN 137
V S+ YA S WCL EL +EIKK+ Q+++PVFY+VDPSDVR + G + +AF + E
Sbjct: 75 VLSRTYASSSWCLNEL---VEIKKV-SQMIMPVFYEVDPSDVRKRTGEFGKAFEEACERQ 130
Query: 138 FEGKIKVQEWRNGLLEAANYAG-----WD 161
+ ++K Q+WR L+ AN AG WD
Sbjct: 131 PDEEVK-QKWREALVYIANIAGESSQNWD 158
>At1g27170 disease resistance protein, putative
Length = 1560
Score = 125 bits (314), Expect = 1e-29
Identities = 66/147 (44%), Positives = 95/147 (63%), Gaps = 3/147 (2%)
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSV 77
K++VF+SFRG DTR+NF HL ALK +R + DN+ + GDEI ++L +E++ SV
Sbjct: 160 KYDVFLSFRGADTRDNFGDHLYKALKD-KVRVFRDNEGMERGDEISSSLKAGMEDSAASV 218
Query: 78 IVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENN 137
IV S+NY+ S+WCL+EL + ++K + ++P+FY VDPS VR Q + F +H+
Sbjct: 219 IVISRNYSGSRWCLDELAMLCKMKSSLDRRILPIFYHVDPSHVRKQSDHIKKDFEEHQVR 278
Query: 138 F-EGKIKVQEWRNGLLEAANYAGWDCN 163
F E K KVQEWR L N AG+ C+
Sbjct: 279 FSEEKEKVQEWREALTLVGNLAGYVCD 305
Score = 102 bits (254), Expect = 9e-23
Identities = 53/141 (37%), Positives = 84/141 (58%), Gaps = 3/141 (2%)
Query: 21 EVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGD-EIPTTLVRAIEEAKLSVIV 79
+VF+SF+ D R+ FT L L + +R + ++D+ G+ E+ +LV A+E++ V+V
Sbjct: 17 DVFLSFQ-RDARHKFTERLYEVLVKEQVRVWNNDDVERGNHELGASLVEAMEDSVALVVV 75
Query: 80 FSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNFE 139
S NYA S WCLEEL + ++K G++V+P+FY+V+P +R Q G Y F +H F
Sbjct: 76 LSPNYAKSHWCLEELAMLCDLKSSLGRLVLPIFYEVEPCMLRKQNGPYEMDFEEHSKRFS 135
Query: 140 GKIKVQEWRNGLLEAANYAGW 160
+ K+Q WR L N G+
Sbjct: 136 EE-KIQRWRRALNIIGNIPGF 155
>At3g04220 putative disease resistance protein
Length = 896
Score = 123 bits (309), Expect = 4e-29
Identities = 64/164 (39%), Positives = 99/164 (60%), Gaps = 11/164 (6%)
Query: 6 SSSSSSFNNAPLE------KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSG 59
SSS+ + PL K++VF SFRGED R +F SH+ +R I ++DN++ G
Sbjct: 43 SSSTQPSPSPPLSSLSLTRKYDVFPSFRGEDVRKDFLSHIQKEFQRQGITPFVDNNIKRG 102
Query: 60 DEIPTTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSD 119
+ I L+RAI +K+++I+ SKNYA S WCL+EL++I++ K+ GQ V+ +FY VDPS
Sbjct: 103 ESIGPELIRAIRGSKIAIILLSKNYASSSWCLDELVEIIKCKEEMGQTVIVIFYKVDPSL 162
Query: 120 VRNQRGSYAEAFAKHENNFEGKIK--VQEWRNGLLEAANYAGWD 161
V+ G + + F N +GK + ++ WR + A AG+D
Sbjct: 163 VKKLTGDFGKVF---RNTCKGKERENIERWREAFKKVATIAGYD 203
>At5g41540 disease resistance protein-like
Length = 1038
Score = 123 bits (308), Expect = 5e-29
Identities = 65/145 (44%), Positives = 88/145 (59%), Gaps = 5/145 (3%)
Query: 17 LEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEEAKLS 76
+ K+ VF SF G D R F SHL I + D ++ G I LV+AI E+++S
Sbjct: 8 VRKYHVFPSFHGSDVRRKFLSHLRFHFAIKGIVAFKDQEIERGQRIGPELVQAIRESRVS 67
Query: 77 VIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHEN 136
++V SKNY S WCL+EL++IL+ K+ + QIV+P+FY++DPSDVR Q G + +AF K
Sbjct: 68 LVVLSKNYPSSSWCLDELVEILKCKEDQEQIVMPIFYEIDPSDVRKQSGDFGKAFGK--- 124
Query: 137 NFEGKIK--VQEWRNGLLEAANYAG 159
GK K Q W N L EAAN G
Sbjct: 125 TCVGKTKEVKQRWTNALTEAANIGG 149
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,000,210
Number of Sequences: 26719
Number of extensions: 166973
Number of successful extensions: 727
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 138
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 458
Number of HSP's gapped (non-prelim): 168
length of query: 166
length of database: 11,318,596
effective HSP length: 92
effective length of query: 74
effective length of database: 8,860,448
effective search space: 655673152
effective search space used: 655673152
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC134049.16


