

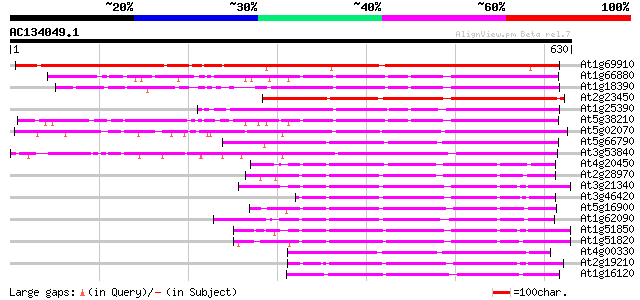
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.1 - phase: 0
(630 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g69910 protein kinase like protein 715 0.0
At1g66880 wall-associated kinase like protein 299 4e-81
At1g18390 wall-associated kinase, putative 292 4e-79
At2g23450 protein kinase like protein 275 7e-74
At1g25390 putative protein 268 9e-72
At5g38210 protein kinase - like protein 265 8e-71
At5g02070 putative protein kinase 259 2e-69
At5g66790 unknown protein 256 3e-68
At3g53840 protein kinase-like protein 247 2e-65
At4g20450 receptor protein kinase - like protein 224 9e-59
At2g28970 putative receptor-like protein kinase 218 8e-57
At3g21340 serine/threonine-specific protein kinase, putative 218 1e-56
At3g46420 hypothetical protein 215 7e-56
At5g16900 receptor protein kinase -like protein 214 9e-56
At1g62090 hypothetical protein 214 9e-56
At1g51850 Putative protein kinase 214 9e-56
At1g51820 Putative protein kinase 214 9e-56
At4g00330 unknown protein 214 1e-55
At2g19210 putative receptor-like protein kinase 214 1e-55
At1g16120 214 1e-55
>At1g69910 protein kinase like protein
Length = 636
Score = 715 bits (1845), Expect = 0.0
Identities = 366/628 (58%), Positives = 459/628 (72%), Gaps = 31/628 (4%)
Query: 7 LPFLVLLLFPTSTHTCPTNNTTKTHPSPCPPFQSTPPFPFSTTPGCGHPSFQLTCSTPHS 66
L VL +F T + T+ ++ H CPPF S+PP+PFS +PGCGHP+FQ+ CS+ +
Sbjct: 11 LLIFVLTIFSTKPSSASTSCSSSFH---CPPFNSSPPYPFSASPGCGHPNFQIQCSSSRA 67
Query: 67 FITINNLTFSILSYKPNSTSIILSPHNPISQQNNNTKCPTTSSIPNKPINLSNTPFTLSD 126
ITI NLTFSIL Y S+S+ LSP ++ NN SS PN+ I+L+ +PF +SD
Sbjct: 68 TITIKNLTFSILHYSSISSSLTLSPITNTNRNTNNCSSLRFSSSPNRFIDLTGSPFRVSD 127
Query: 127 ETCSRLSFLQPCSPPNLPNCSHCPWQCKLIKNPSEIFKSCRSMHHSVSDNEPSCQSDVLV 186
+CSRLS L+PCSP LPNCS CPW CKL+KNP I C S H S+S E CQ D+L
Sbjct: 128 SSCSRLSLLRPCSPFTLPNCSRCPWDCKLLKNPGRILHGCESTHGSLS--EQGCQGDLLG 185
Query: 187 YLNEILIQ-GIELEWDEALTQDTYFTNCKECINNNNGFCGFNSSDTKKQFVCYHFHSKST 245
+L + + G E+EWDE +QD YF C++C NG CGFNS+ + F+C+H S+S
Sbjct: 186 FLQDFFTRFGFEVEWDE--SQDPYFIKCRDC-QIKNGVCGFNSTHPNQDFICFH-KSRSE 241
Query: 246 LSPPWIHKMKPSKIAVFAIVIAFTSLILFLSVVISILRSRK---VNTTVEEDPTAVFLHN 302
L + + + IAV +++ A T L+L SV ++I RSR+ +++ EEDP A+FL +
Sbjct: 242 LVTQRDNNKRVNHIAVLSLIFALTCLLLVFSVAVAIFRSRRASFLSSINEEDPAALFLRH 301
Query: 303 HRNANLLPPVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHN--- 359
HR+A LLPPVFT++EL +TN FDPKRKIGDGGFGSVYLG L DG+L AVK LH H+
Sbjct: 302 HRSAALLPPVFTFEELESATNKFDPKRKIGDGGFGSVYLGQLSDGQLLAVKFLHHHHGAT 361
Query: 360 -----HTAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLH 414
H AFS KSFCNEILILSSI+HPNLVKLHGYCSDPRGL+LV+DYV NGTLA+HLH
Sbjct: 362 AAATEHCKAFSMKSFCNEILILSSINHPNLVKLHGYCSDPRGLLLVHDYVTNGTLADHLH 421
Query: 415 GSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGD 474
G +G MTW+ RL+IA+QTALAMEYLHF + PP+VHRDITSSNIF+EKDM+IKVGD
Sbjct: 422 G-----RGPKMTWRVRLDIALQTALAMEYLHFDIVPPVVHRDITSSNIFVEKDMKIKVGD 476
Query: 475 FGLSRLLVLQESN-QTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLEL 533
FGLSRLLV E+ + +S +V TGPQGTPGYLDPDYHRSFRLTEKSDVYS+GVVL+EL
Sbjct: 477 FGLSRLLVFSETTVNSATSSDYVCTGPQGTPGYLDPDYHRSFRLTEKSDVYSYGVVLMEL 536
Query: 534 ISGLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDA----VGAVAE 589
I+G+KAVD R+KR+MALAD+VVS+I G L +V+DP+L L D+ A + V AVAE
Sbjct: 537 ITGMKAVDQRREKRDMALADLVVSKIQMGLLDQVIDPLLALDGDDVAAVSDGFGVAAVAE 596
Query: 590 LAFRCVASDKDDRPDSKEVVGELKRVRS 617
LAFRCVA+DKDDRPD+KE+V EL+R+RS
Sbjct: 597 LAFRCVATDKDDRPDAKEIVQELRRIRS 624
>At1g66880 wall-associated kinase like protein
Length = 848
Score = 299 bits (765), Expect = 4e-81
Identities = 220/636 (34%), Positives = 326/636 (50%), Gaps = 100/636 (15%)
Query: 43 PFPFSTTPGCGHPSFQLT-CSTPHSFITINNLTFSILSYKPNSTSIILSPHNPISQQNNN 101
PF S CGHP F+L CS + ++I+++ F IL+ S I L I
Sbjct: 198 PFWTSGREDCGHPDFKLDDCSGRFAELSISSVKFRILASVYGSNIIRLGRSEYIGDL--- 254
Query: 102 TKCPTTSSIPNKPINLSNTPFTLSDETCSRLSFLQPCS---PPNLPNC--------SHCP 150
CP N P + S PF + E L+ CS P + N S
Sbjct: 255 --CPQDPI--NAPFSESVLPFAPNTEL---LTIFYNCSRDFPQQVTNFGDFACGDDSDDD 307
Query: 151 WQCKLIKNPSEIFKSCRSMHHSVSDNEPSCQSDVLVYL-------------NEILIQGIE 197
+ +N S F ++ + D SC +V + N+ L + +E
Sbjct: 308 RSYYVTRNLS--FPPLSEINDLLYDFSQSCDRNVSIPASGSTLNILQSTPSNDNLKKALE 365
Query: 198 LEWDEALTQDTYFTNCKECINNNNGFCGFNSSDTKKQFVCYHFHSKSTLSPPWIHKMKPS 257
++ L QD C+ CI++ G CG+ S T +FVCY T +PP + K +
Sbjct: 366 YGFELELNQD-----CRTCIDSK-GACGY--SQTSSRFVCYSIEEPQTPTPPNPTRNKDT 417
Query: 258 KIAVFA---IVIAFTS---LILFLSVVISILRSRKVNTT----------------VEEDP 295
+++ A I +A S ++L + + I R RK +P
Sbjct: 418 SLSIGAKAGIAVASVSGLAILLLAGLFLCIRRRRKTQDAQYTSKSLPITSYSSRDTSRNP 477
Query: 296 TAVFLHNHRNANLLPP--------------VFTYDELNISTNNFDPKRKIGDGGFGSVYL 341
T+ + + N +LLP VF+Y+EL +T NF R++GDGGFG+VY
Sbjct: 478 TSTTISSSSNHSLLPSISNLANRSDYCGVQVFSYEELEEATENFS--RELGDGGFGTVYY 535
Query: 342 GNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDHPNLVKLHGYCS-DPRGLILV 400
G L+DG+ AVK L+ + + F NEI IL S+ HPNLV L+G S R L+LV
Sbjct: 536 GVLKDGRAVAVKRLYERSLKRV---EQFKNEIEILKSLKHPNLVILYGCTSRHSRELLLV 592
Query: 401 YDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSS 460
Y+Y+ NGTLAEHLHG++++ + + W TRL IAI+TA A+ +LH I+HRDI ++
Sbjct: 593 YEYISNGTLAEHLHGNRAEARP--LCWSTRLNIAIETASALSFLHIK---GIIHRDIKTT 647
Query: 461 NIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEK 520
NI ++ + ++KV DFGLSRL + +++ + T PQGTPGY+DP+Y++ ++L EK
Sbjct: 648 NILLDDNYQVKVADFGLSRLFPMDQTH--------ISTAPQGTPGYVDPEYYQCYQLNEK 699
Query: 521 SDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDA 580
SDVYSFGVVL ELIS +AVD R + ++ LA+M VS+I L E++D L ND +
Sbjct: 700 SDVYSFGVVLTELISSKEAVDITRHRHDINLANMAVSKIQNNALHELVDSSLGYDNDPEV 759
Query: 581 LDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVR 616
+ AVAELAFRC+ ++D RP E+V L+ ++
Sbjct: 760 RRKMMAVAELAFRCLQQERDVRPAMDEIVEILRGIK 795
>At1g18390 wall-associated kinase, putative
Length = 605
Score = 292 bits (747), Expect = 4e-79
Identities = 206/589 (34%), Positives = 299/589 (49%), Gaps = 98/589 (16%)
Query: 52 CGHPSFQLTCSTPHSFITINNLTFSILSYKPNSTSIILSPHNPISQQNNNTKCPTTSSIP 111
CG+PSF+LTC + S Y + S + ++ + ++ CP P
Sbjct: 59 CGYPSFELTCDDEEKLPVLG---ISGEEYVIKNISYLTQSFQVVNSKASHDPCPR----P 111
Query: 112 NKPINLSNTPFTLSDETCSRLSFLQPCSPPNLPNCSHCPWQC----------------KL 155
+ L TPF ++ + + L CS L + P C KL
Sbjct: 112 LNNLTLHRTPFFVNPSHIN-FTILYNCSDHLLEDFRTYPLTCARNTSLLRSFGVFDRKKL 170
Query: 156 IKNPSEIFKSCRSMHHS--VSDNEPSCQSDVLVYLNEILIQGIELEWDEALTQDTYFTNC 213
K SC+ + ++ NE V EIL +G L W +C
Sbjct: 171 GKEKQIASMSCQKLVDVPVLASNESDVMGMTYV---EILKRGFVLNWTA--------NSC 219
Query: 214 KECINNNNGFCGFNSSDTKKQFVCYHFHSKSTLSP--PWIHKMKPSKIAVFAIVIAFTSL 271
CI + G CG + +++FVC L P P +H
Sbjct: 220 FRCITSG-GRCGTD----QQEFVC--------LCPDGPKLH------------------- 247
Query: 272 ILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLLPPV--FTYDELNISTNNFDPKR 329
+ L R +++ DP+A + LL V F+Y+EL +TNNFDP +
Sbjct: 248 ----DTCTNALLPRNISS----DPSAKSFDIEKAEELLVGVHIFSYEELEEATNNFDPSK 299
Query: 330 KIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDHPNLVKLHG 389
++GDGGFG+VY G L+DG+ AVK L+ +N A + F NE+ IL+ + HPNLV L G
Sbjct: 300 ELGDGGFGTVYYGKLKDGRSVAVKRLYDNNFKRA---EQFRNEVEILTGLRHPNLVALFG 356
Query: 390 YCS-DPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSV 448
S R L+LVY+YV NGTLA+HLHG ++ + W RL+IA++TA A++YLH S
Sbjct: 357 CSSKQSRDLLLVYEYVANGTLADHLHGPQANPSS--LPWSIRLKIAVETASALKYLHAS- 413
Query: 449 KPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLD 508
I+HRD+ S+NI ++++ +KV DFGLSRL + +++ V T PQGTPGY+D
Sbjct: 414 --KIIHRDVKSNNILLDQNFNVKVADFGLSRLFPMDKTH--------VSTAPQGTPGYVD 463
Query: 509 PDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLKEVL 568
PDYH ++L+ KSDVYSF VVL+ELIS L AVD R ++E+ L++M V +I +L++++
Sbjct: 464 PDYHLCYQLSNKSDVYSFAVVLMELISSLPAVDITRPRQEINLSNMAVVKIQNHELRDMV 523
Query: 569 DPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVRS 617
DP L D V AVAELAF+C+ SDKD RP V L R+++
Sbjct: 524 DPSLGFDTDTRVRQTVIAVAELAFQCLQSDKDLRPCMSHVQDTLTRIQN 572
>At2g23450 protein kinase like protein
Length = 708
Score = 275 bits (702), Expect = 7e-74
Identities = 152/339 (44%), Positives = 211/339 (61%), Gaps = 17/339 (5%)
Query: 285 RKVNTTVEEDPTAVFLHNHRNANLLPPVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNL 344
R+ +T + +A L + N F Y E+ +T+ F K+K+G G +G+VY G L
Sbjct: 308 RRRSTPLRSHLSAKRLLSEAAGNSSVAFFPYKEIEKATDGFSEKQKLGIGAYGTVYRGKL 367
Query: 345 RDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYV 404
++ + A+K L H + S NEI +LSS+ HPNLV+L G C + +LVY+Y+
Sbjct: 368 QNDEWVAIKRLR---HRDSESLDQVMNEIKLLSSVSHPNLVRLLGCCIEQGDPVLVYEYM 424
Query: 405 PNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFI 464
PNGTL+EHL + +G + W RL +A QTA A+ YLH S+ PPI HRDI S+NI +
Sbjct: 425 PNGTLSEHLQ----RDRGSGLPWTLRLTVATQTAKAIAYLHSSMNPPIYHRDIKSTNILL 480
Query: 465 EKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVY 524
+ D KV DFGLSRL + + S+ + T PQGTPGYLDP YH+ F L++KSDVY
Sbjct: 481 DYDFNSKVADFGLSRLGMTESSH--------ISTAPQGTPGYLDPQYHQCFHLSDKSDVY 532
Query: 525 SFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAV 584
SFGVVL E+I+GLK VD+ R E+ LA + V +I +G + E++DP+LDL D L ++
Sbjct: 533 SFGVVLAEIITGLKVVDFTRPHTEINLAALAVDKIGSGCIDEIIDPILDLDLDAWTLSSI 592
Query: 585 GAVAELAFRCVASDKDDRPDSKEVVGELKRVRSRISGGI 623
VAELAFRC+A D RP EV EL+++ R+SG I
Sbjct: 593 HTVAELAFRCLAFHSDMRPTMTEVADELEQI--RLSGWI 629
>At1g25390 putative protein
Length = 629
Score = 268 bits (684), Expect = 9e-72
Identities = 161/407 (39%), Positives = 233/407 (56%), Gaps = 27/407 (6%)
Query: 212 NCKECINNNNGFCGFNSSDTKKQFVCYHFHSKSTLSPPWIHKMKPSKIAVFAIVIAFTSL 271
NC C +NN G C + K + C +++ P H + + VI L
Sbjct: 189 NCFRC-HNNGGEC----TKVKNNYRCVGANTE----PNNYHAEMRLGLGIGGSVILIIIL 239
Query: 272 ILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLLPPVFTYDELNISTNNFDPKRKI 331
+ +V+ R RK + + D + + P+F+Y EL +T+NF R +
Sbjct: 240 VALFAVIHRNYR-RKDGSELSRDNSKSDVE-FSQVFFKIPIFSYKELQAATDNFSKDRLL 297
Query: 332 GDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDHPNLVKLHGYC 391
GDGGFG+VY G +RDG+ AVK L+ HN+ + F NEI IL+ + H NLV L+G
Sbjct: 298 GDGGFGTVYYGKVRDGREVAVKRLYEHNYRRL---EQFMNEIEILTRLHHKNLVSLYGCT 354
Query: 392 SD-PRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKP 450
S R L+LVY+++PNGT+A+HL+G + +G++ TW RL IAI+TA A+ YLH S
Sbjct: 355 SRRSRELLLVYEFIPNGTVADHLYGENTPHQGFL-TWSMRLSIAIETASALAYLHAS--- 410
Query: 451 PIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLDPD 510
I+HRD+ ++NI ++++ +KV DFGLSRLL S V T PQGTPGY+DP+
Sbjct: 411 DIIHRDVKTTNILLDRNFGVKVADFGLSRLL--------PSDVTHVSTAPQGTPGYVDPE 462
Query: 511 YHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDP 570
YHR + LT+KSDVYSFGVVL+ELIS AVD R K E+ L+ + +++I E++D
Sbjct: 463 YHRCYHLTDKSDVYSFGVVLVELISSKPAVDISRCKSEINLSSLAINKIQNHATHELIDQ 522
Query: 571 VLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVRS 617
L + VAELAF+C+ D RP ++VV ELK +++
Sbjct: 523 NLGYATNEGVRKMTTMVAELAFQCLQQDNTMRPTMEQVVHELKGIQN 569
>At5g38210 protein kinase - like protein
Length = 686
Score = 265 bits (676), Expect = 8e-71
Identities = 215/666 (32%), Positives = 332/666 (49%), Gaps = 104/666 (15%)
Query: 9 FLVLLLFPTSTHTCPTNNTTKTHPSPCPPF----QSTPPFPF---STTPGCGHPSFQLTC 61
FLVLL F + H P + + PC T FPF + CGHPS L C
Sbjct: 15 FLVLLFFLSYIHFLPCAQSQR---EPCDTLFRCGDLTAGFPFWGVARPQPCGHPSLGLHC 71
Query: 62 S--TPHSFITINNLTFSILSYKPNSTSIILSPHNPISQQNNNTKCPTTSSIPNKPINLSN 119
T + + I++L + +L ++++ L + Q + C + S L+
Sbjct: 72 QKQTNSTSLIISSLMYRVLEVNTTTSTLKL-----VRQDFSGPFCSASFS----GATLTP 122
Query: 120 TPFTLSDETCSRLSFLQPCSPP-NLPNCSHCPWQCKLIKNPSEIFKS-CRSMHHSVSDNE 177
F L + LS C+P + P CP + + +++ + C + +
Sbjct: 123 ELFELLPDY-KTLSAYYLCNPSLHYPAKFICPNKGVGSIHQDDLYHNHCGGIFNITVPIG 181
Query: 178 PSCQSDVL--VYLNEILIQGIELEWDEALTQDTYFTNCKECINNNNGFCGFNSSDTKKQF 235
+ + L L +L +G E++ L+ D C+EC N G C ++ +
Sbjct: 182 YAPEEGALNVTNLESVLKKGFEVK----LSIDE--RPCQEC-KTNGGICAYHVATP---- 230
Query: 236 VCYHFHSKSTLSPPWIHKMKPSKIAVFA-------IVIAFTSLILFLSVV------ISIL 282
VC +S S ++ M PS + A I I F S L +++ I I
Sbjct: 231 VCCKTNSSSEVN---CTPMMPSGSSAHAGLSKKGKIGIGFASGFLGATLIGGCLLCIFIR 287
Query: 283 RSRKV-----------------NTTVEEDPTAVFLHNHRNANLLPP-------------- 311
R +K+ N T+ PT+ + N +L+P
Sbjct: 288 RRKKLATQYTNKGLSTTTPYSSNYTMSNTPTSTTISGS-NHSLVPSISNLGNGSVYSGIQ 346
Query: 312 VFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCN 371
VF+Y+EL +T NF +++GDGGFG+VY G L+DG+ AVK L + + F N
Sbjct: 347 VFSYEELEEATENFS--KELGDGGFGTVYYGTLKDGRAVAVKRLFERSLKRV---EQFKN 401
Query: 372 EILILSSIDHPNLVKLHGYCS-DPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTR 430
EI IL S+ HPNLV L+G + R L+LVY+Y+ NGTLAEHLHG++++ + + W R
Sbjct: 402 EIDILKSLKHPNLVILYGCTTRHSRELLLVYEYISNGTLAEHLHGNQAQSRP--ICWPAR 459
Query: 431 LEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTT 490
L+IAI+TA A+ YLH S I+HRD+ ++NI ++ + ++KV DFGLSRL + +++
Sbjct: 460 LQIAIETASALSYLHAS---GIIHRDVKTTNILLDSNYQVKVADFGLSRLFPMDQTH--- 513
Query: 491 SSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMA 550
+ T PQGTPGY+DP+Y++ +RL EKSDVYSFGVVL ELIS +AVD R + ++
Sbjct: 514 -----ISTAPQGTPGYVDPEYYQCYRLNEKSDVYSFGVVLSELISSKEAVDITRHRHDIN 568
Query: 551 LADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVG 610
LA+M +S+I + E+ D L D + +VAELAFRC+ ++D RP E+V
Sbjct: 569 LANMAISKIQNDAVHELADLSLGFARDPSVKKMMSSVAELAFRCLQQERDVRPSMDEIVE 628
Query: 611 ELKRVR 616
L+ ++
Sbjct: 629 VLRVIQ 634
>At5g02070 putative protein kinase
Length = 657
Score = 259 bits (663), Expect = 2e-69
Identities = 201/677 (29%), Positives = 325/677 (47%), Gaps = 86/677 (12%)
Query: 6 LLPFLVLLLFPTSTHTCPTNNTTK---THPSPCPPF-QSTPPFPFSTTPGCGHPSFQLTC 61
L+P L+ + + H+C + K P CP P+P ST P CG ++++ C
Sbjct: 11 LIPTLLTVWLACAGHSCARHAKAKPPMAGPPRCPNCGPMVVPYPLSTGPTCGDQAYRINC 70
Query: 62 -----------STPHSFITINNLTFSILSYKPNSTSIILSPHNPISQQNNNTKCPTTSSI 110
+ + +IN++T I+ P S + +S+Q
Sbjct: 71 VGGKLYFGALHGSSYVITSINSVTQRIVLRPPGLASSVSCISADVSKQGLELD------- 123
Query: 111 PNKPINL--SNTPFTLSDETCSRLSFLQP--CSPPNL---------PNCSHCPWQCKLIK 157
P+ P ++ SNT L+ CS+ P CSP +L CS P C
Sbjct: 124 PHLPFSITSSNTILLLN---CSQAMLQAPIDCSPTSLCYSYIKNNASPCSKAPLCCTFRT 180
Query: 158 NPSEIFKSCRSMHHSVSDNEPSC---QSDVLVYLNEILIQ--------GIELEWDEALTQ 206
+ S+ +++ N C QS V + N+ + G+EL+W AL +
Sbjct: 181 DGSQTA-------YTIRINGGGCLAYQSFVGLNPNKEVPPPGKKWPDTGLELQW--ALPK 231
Query: 207 DTYFTNCKECINNN----NGFC-------GFNSSDTKKQFVCYHFHSKSTLSPPWIHKMK 255
+ CK ++ N C G KK ++ H K
Sbjct: 232 EPV---CKTDVDCNLLLGKSKCLPDPTSLGLKRCSCKKGLEWDPVNAICGKCRHGKHCKK 288
Query: 256 PSKIAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHR------NANLL 309
K VFA A + + L++ ++++ ++ + V++D + +
Sbjct: 289 KKKTVVFAGA-AVAVVGVTLAIAVAVIGTKHSHQKVKKDIHKNIVKEREEMLSANSTGKS 347
Query: 310 PPVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSF 369
+FT E+ +TNNF IG GGFG V+ L DG + A+K +N +
Sbjct: 348 SRIFTGREITKATNNFSKDNLIGTGGFGEVFKAVLEDGTITAIKRAKLNNTKG---TDQI 404
Query: 370 CNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQT 429
NE+ IL ++H +LV+L G C D +L+Y+++PNGTL EHLHGS S R +TW+
Sbjct: 405 LNEVRILCQVNHRSLVRLLGCCVDLELPLLIYEFIPNGTLFEHLHGS-SDRTWKPLTWRR 463
Query: 430 RLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQT 489
RL+IA QTA + YLH + +PPI HRD+ SSNI +++ + KV DFGLSRL+ L E T
Sbjct: 464 RLQIAYQTAEGLAYLHSAAQPPIYHRDVKSSNILLDEKLNAKVSDFGLSRLVDLTE---T 520
Query: 490 TSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREM 549
++ ++TG QGT GYLDP+Y+R+F+LT+KSDVYSFGVVLLE+++ KA+D+ R++ ++
Sbjct: 521 ANNESHIFTGAQGTLGYLDPEYYRNFQLTDKSDVYSFGVVLLEMVTSKKAIDFTREEEDV 580
Query: 550 ALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVV 609
L + + +L E +DP+L + + + + LA C+ + +RP KEV
Sbjct: 581 NLVMYINKMMDQERLTECIDPLLKKTANKIDMQTIQQLGNLASACLNERRQNRPSMKEVA 640
Query: 610 GELKRVRSRISGGITRS 626
E++ + + +S +T +
Sbjct: 641 DEIEYIINILSQEVTET 657
>At5g66790 unknown protein
Length = 622
Score = 256 bits (653), Expect = 3e-68
Identities = 147/382 (38%), Positives = 216/382 (56%), Gaps = 16/382 (4%)
Query: 240 FHSKSTLSPPWIHKMKPSKIAVFAIVIAFTSLILFLSVVISILRS-----RKVNTTVEED 294
F K+ P H++ + + +V+ T+ IL +VI +L + K + + E
Sbjct: 224 FSGKAFTVPGGCHRLVYKRKGLHKLVVLGTAGILVGVLVIVVLIATYFFRNKQSASSERA 283
Query: 295 PTAVFLHNHRNANLLPPVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKH 354
A L N P +TY E+ +T++F K +G G +G+VY G + A+K
Sbjct: 284 SIANRLLCELAGNSSVPFYTYKEIEKATDSFSDKNMLGTGAYGTVYAGEFPNSSCVAIKR 343
Query: 355 LHRHNHTAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLH 414
L H S NEI +LSS+ HPNLV+L G C LVY+++PNGTL +HL
Sbjct: 344 L---KHKDTTSIDQVVNEIKLLSSVSHPNLVRLLGCCFADGEPFLVYEFMPNGTLYQHLQ 400
Query: 415 GSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGD 474
+ + ++WQ RL IA QTA A+ +LH SV PPI HRDI SSNI ++ + K+ D
Sbjct: 401 HERGQPP---LSWQLRLAIACQTANAIAHLHSSVNPPIYHRDIKSSNILLDHEFNSKISD 457
Query: 475 FGLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELI 534
FGLSRL T + T PQGTPGYLDP YH+ F+L++KSDVYSFGVVL+E+I
Sbjct: 458 FGLSRL-----GMSTDFEASHISTAPQGTPGYLDPQYHQDFQLSDKSDVYSFGVVLVEII 512
Query: 535 SGLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRC 594
SG K +D+ R E+ LA + V RI G++ +++DP L+ + ++ +AELAFRC
Sbjct: 513 SGFKVIDFTRPYSEVNLASLAVDRIGRGRVVDIIDPCLNKEINPKMFASIHNLAELAFRC 572
Query: 595 VASDKDDRPDSKEVVGELKRVR 616
++ ++ RP E+ +L R++
Sbjct: 573 LSFHRNMRPTMVEITEDLHRIK 594
>At3g53840 protein kinase-like protein
Length = 640
Score = 247 bits (630), Expect = 2e-65
Identities = 206/666 (30%), Positives = 312/666 (45%), Gaps = 86/666 (12%)
Query: 1 MSLTTLLPFLVLLLFPTST----HTCPTNNTTKTHPSPCPPFQSTPPFPFSTTPGCGHPS 56
+SLTT L LL++ +ST CP +T+ P+P ST CG P
Sbjct: 6 LSLTTFT--LSLLIYFSSTTQAFKRCPNCGSTRV------------PYPLSTGLDCGDPG 51
Query: 57 FQLTCSTPHS--FITINNLTFSILSYKPNSTSIILSPHNPISQQNNNTKCPTTSSIPNKP 114
+++ C S F T+N T I + P+ +L P P +QN KC + I
Sbjct: 52 YRIRCDNYGSLWFDTLNGSTNPIKTIDPSGQRFVLRP--PGFEQN---KCVSVD-IKYHG 105
Query: 115 INLS-NTPFTLSDETCSRLSFLQPCSPPNLP-------NCSHCPWQCKLIKNPSEIFKSC 166
I L N PF +S CS + C+ L NCS K + E +C
Sbjct: 106 IQLDLNLPFNVS---CSNTVIIMNCTKDGLDAYSSQGFNCSDNSLCHKFLNANLEARGNC 162
Query: 167 RSMH-------------HSVSDNEPSCQSDVLVYLNEILIQGIELEWDEALTQDTYFTN- 212
R + + V P S ++N L + +W E + +
Sbjct: 163 RGVTSCCWYKTGASVNTYKVYRARPDMCSAYQSFMNLDLTIPVS-KWGEPAVEILWEAPR 221
Query: 213 ---CK---ECINNNNGFCGFNSSDT-KKQFVC---YHFHSKSTL--SPPWIHKMKPSK-- 258
CK +C + N C +S++ +K+ C + + S + + P K K K
Sbjct: 222 EPVCKSQGDCRDLLNSVCSNDSTNLGQKRCFCKKGFQWDSVNAVCEGPVRCSKRKSCKRW 281
Query: 259 --IAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRN-----ANLLPP 311
+ + + IL + + S++ A HRN + L
Sbjct: 282 SNLPLLGGLAGGVGAILIAGFITKTIVSKQNRRIAGNQSWASVRKLHRNLLSINSTGLDR 341
Query: 312 VFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCN 371
+FT E+ +T+NF +G GGFG V+ GNL DG AVK N + + N
Sbjct: 342 IFTGKEIVKATDNFAKSNLLGFGGFGEVFKGNLDDGTTVAVKRAKLGNEKSIYQ---IVN 398
Query: 372 EILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYM--MTWQT 429
E+ IL + H NLVKL G C + +LVY++VPNGTL EH++G G + +
Sbjct: 399 EVQILCQVSHKNLVKLLGCCIELEMPVLVYEFVPNGTLFEHIYGGGGGGGGLYDHLPLRR 458
Query: 430 RLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQT 489
RL IA QTA ++YLH S PPI HRD+ SSNI +++++ +KV DFGLSRL V S+ T
Sbjct: 459 RLMIAHQTAQGLDYLHSSSSPPIYHRDVKSSNILLDENLDVKVADFGLSRLGVSDVSHVT 518
Query: 490 TSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREM 549
T + QGT GYLDP+Y+ +F+LT+KSDVYSFGVVL EL++ KA+D+ R++ ++
Sbjct: 519 TCA--------QGTLGYLDPEYYLNFQLTDKSDVYSFGVVLFELLTCKKAIDFNREEEDV 570
Query: 550 ALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVV 609
L V + G+L +V+DPV+ +G ++++ A+ LA CV + RP +
Sbjct: 571 NLVVFVRKALKEGRLMDVIDPVIGIGATEKEIESMKALGVLAELCVKETRQCRPTMQVAA 630
Query: 610 GELKRV 615
E++ +
Sbjct: 631 KEIENI 636
>At4g20450 receptor protein kinase - like protein
Length = 866
Score = 224 bits (572), Expect = 9e-59
Identities = 132/343 (38%), Positives = 198/343 (57%), Gaps = 31/343 (9%)
Query: 271 LILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLLPPVFTYDELNISTNNFDPKRK 330
+ + L V I +LR +K + A N R+ +TY+E+ + TNNF+ R
Sbjct: 518 IAVLLLVNILLLRKKKPSKASRSSMVA----NKRS-------YTYEEVAVITNNFE--RP 564
Query: 331 IGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDHPNLVKLHGY 390
+G+GGFG VY GN+ D + AVK L + ++A K F E+ +L + H NLV L GY
Sbjct: 565 LGEGGFGVVYHGNVNDNEQVAVKVL---SESSAQGYKQFKAEVDLLLRVHHINLVTLVGY 621
Query: 391 CSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKP 450
C + + L+L+Y+Y+ NG L +HL G S+ ++W+ RL IA +TA +EYLH KP
Sbjct: 622 CDEGQHLVLIYEYMSNGNLKQHLSGENSRSP---LSWENRLRIAAETAQGLEYLHIGCKP 678
Query: 451 PIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQGTPGYLDPD 510
P++HRDI S NI ++ + + K+GDFGLSR + S V T G+PGYLDP+
Sbjct: 679 PMIHRDIKSMNILLDNNFQAKLGDFGLSR-------SFPVGSETHVSTNVAGSPGYLDPE 731
Query: 511 YHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDP 570
Y+R+ LTEKSDV+SFGVVLLE+I+ +D R+K + + V ++ G +K ++DP
Sbjct: 732 YYRTNWLTEKSDVFSFGVVLLEIITSQPVIDQTREKSH--IGEWVGFKLTNGDIKNIVDP 789
Query: 571 VLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELK 613
++ D+ +L ELA CV+ RP+ +V EL+
Sbjct: 790 SMNGDYDSSSL---WKALELAMSCVSPSSSGRPNMSQVANELQ 829
>At2g28970 putative receptor-like protein kinase
Length = 786
Score = 218 bits (555), Expect = 8e-57
Identities = 137/358 (38%), Positives = 204/358 (56%), Gaps = 29/358 (8%)
Query: 265 VIAFTSLILFLSVVIS---ILRSRKVNTTVEEDPTA----VFLHNHRNANLLPPV--FTY 315
VIA S +L + VV++ +LR +K+ + + P+ V H ++ + F Y
Sbjct: 412 VIASISSVLIVIVVVALFFVLRKKKMPSDAQAPPSLPVEDVGQAKHSESSFVSKKIRFAY 471
Query: 316 DELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILI 375
E+ TNNF +R +G+GGFG VY G + + AVK L + +++ K F E+ +
Sbjct: 472 FEVQEMTNNF--QRVLGEGGFGVVYHGCVNGTQQVAVKLL---SQSSSQGYKHFKAEVEL 526
Query: 376 LSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAI 435
L + H NLV L GYC + L L+Y+Y+PNG L +HL G KR G++++W++RL +A+
Sbjct: 527 LMRVHHKNLVSLVGYCDEGDHLALIYEYMPNGDLKQHLSG---KRGGFVLSWESRLRVAV 583
Query: 436 QTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGF 495
AL +EYLH KPP+VHRDI S+NI +++ + K+ DFGLSR + T +
Sbjct: 584 DAALGLEYLHTGCKPPMVHRDIKSTNILLDERFQAKLADFGLSR-------SFPTENETH 636
Query: 496 VWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMV 555
V T GTPGYLDP+Y+++ LTEKSDVYSFG+VLLE+I+ + R+K L + V
Sbjct: 637 VSTVVAGTPGYLDPEYYQTNWLTEKSDVYSFGIVLLEIITNRPIIQQSREKPH--LVEWV 694
Query: 556 VSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELK 613
+ TG + ++DP L D + +V ELA CV RP +VV +LK
Sbjct: 695 GFIVRTGDIGNIVDPNLHGAYD---VGSVWKAIELAMSCVNISSARRPSMSQVVSDLK 749
>At3g21340 serine/threonine-specific protein kinase, putative
Length = 880
Score = 218 bits (554), Expect = 1e-56
Identities = 134/374 (35%), Positives = 205/374 (53%), Gaps = 30/374 (8%)
Query: 258 KIAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLLPPVFTYDE 317
K+ V ++A + ++ L ++ K T +A+ N R FTY E
Sbjct: 516 KMNVVIPIVASVAFVVVLGSALAFFFIFKKKKTSNSQESAIMTKNRR--------FTYSE 567
Query: 318 LNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILS 377
+ TNNF+ R +G GGFG VY G + + + AVK L +H+++ K F E+ +L
Sbjct: 568 VVTMTNNFE--RVLGKGGFGMVYHGTVNNTEQVAVKML---SHSSSQGYKEFKAEVELLL 622
Query: 378 SIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQT 437
+ H NLV L GYC + L L+Y+Y+ NG L EH+ G KR G ++ W+TRL+I +++
Sbjct: 623 RVHHKNLVGLVGYCDEGENLALIYEYMANGDLREHMSG---KRGGSILNWETRLKIVVES 679
Query: 438 ALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVW 497
A +EYLH KPP+VHRD+ ++NI + + + K+ DFGLSR ++ V
Sbjct: 680 AQGLEYLHNGCKPPMVHRDVKTTNILLNEHLHAKLADFGLSRSFPIEGETH-------VS 732
Query: 498 TGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVS 557
T GTPGYLDP+Y+R+ L EKSDVYSFG+VLLE+I+ ++ R+K +A+ V
Sbjct: 733 TVVAGTPGYLDPEYYRTNWLNEKSDVYSFGIVLLEIITNQLVINQSREKPH--IAEWVGL 790
Query: 558 RIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKRVRS 617
+ G ++ ++DP L D D+ +V ELA C+ RP +VV EL S
Sbjct: 791 MLTKGDIQNIMDP--KLYGDYDS-GSVWRAVELAMSCLNPSSARRPTMSQVVIELNECLS 847
Query: 618 --RISGGITRSMST 629
GG +++M++
Sbjct: 848 YENARGGTSQNMNS 861
>At3g46420 hypothetical protein
Length = 838
Score = 215 bits (547), Expect = 7e-56
Identities = 124/292 (42%), Positives = 171/292 (58%), Gaps = 20/292 (6%)
Query: 322 TNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDH 381
TNNF +R +G+GGFG VY G L + AVK L + ++ K F E+ +L + H
Sbjct: 530 TNNF--QRALGEGGFGVVYHGYLNGSEQVAVKLL---SQSSVQGYKEFKAEVELLLRVHH 584
Query: 382 PNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAM 441
NLV L GYC D L LVY+Y+ NG L HL G + G++++W TRL+IA+ AL +
Sbjct: 585 INLVSLVGYCDDRNHLALVYEYMSNGDLKHHLSG---RNNGFVLSWSTRLQIAVDAALGL 641
Query: 442 EYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQ 501
EYLH +P +VHRD+ S+NI + + K+ DFGLSR + + N + T
Sbjct: 642 EYLHIGCRPSMVHRDVKSTNILLGEQFTAKMADFGLSRSFQIGDENH-------ISTVVA 694
Query: 502 GTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHT 561
GTPGYLDP+Y+R+ RL EKSD+YSFG+VLLE+I+ A+D R K + D VVS I
Sbjct: 695 GTPGYLDPEYYRTSRLAEKSDIYSFGIVLLEMITSQHAIDRTRVKHH--ITDWVVSLISR 752
Query: 562 GQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELK 613
G + ++DP L GN N +V ELA C + RP+ +VV +LK
Sbjct: 753 GDITRIIDPNLQ-GNYNSR--SVWRALELAMSCANPTSEKRPNMSQVVIDLK 801
>At5g16900 receptor protein kinase -like protein
Length = 851
Score = 214 bits (546), Expect = 9e-56
Identities = 131/349 (37%), Positives = 199/349 (56%), Gaps = 30/349 (8%)
Query: 270 SLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNANLL----PPVFTYDELNISTNNF 325
++ +F++V++ I+ +++ P+++ + ANL TY E+ + TNNF
Sbjct: 507 AIAIFIAVLVLII------VFIKKRPSSIRALHPSRANLSLENKKRRITYSEILLMTNNF 560
Query: 326 DPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNEILILSSIDHPNLV 385
+ R IG+GGFG VY G L D + AVK L + +++ K F E+ +L + H NLV
Sbjct: 561 E--RVIGEGGFGVVYHGYLNDSEQVAVKVL---SPSSSQGYKEFKAEVELLLRVHHINLV 615
Query: 386 KLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLH 445
L GYC + L L+Y+Y+ NG L HL G K ++ W+ RL IA++TAL +EYLH
Sbjct: 616 SLVGYCDEQAHLALIYEYMANGDLKSHLSG---KHGDCVLKWENRLSIAVETALGLEYLH 672
Query: 446 FSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSSGGFVWTGPQGTPG 505
KP +VHRD+ S NI +++ + K+ DFGLSR + E + + TG GTPG
Sbjct: 673 SGCKPLMVHRDVKSMNILLDEHFQAKLADFGLSRSFSVGEESHVS-------TGVVGTPG 725
Query: 506 YLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLK 565
YLDP+Y+R++RLTEKSDVYSFG+VLLE+I+ ++ + R +A+ V + + +
Sbjct: 726 YLDPEYYRTYRLTEKSDVYSFGIVLLEIITNQPVLEQANENRH--IAERVRTMLTRSDIS 783
Query: 566 EVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGELKR 614
++DP L D+ +V +LA CV RPD VV ELK+
Sbjct: 784 TIVDPNLIGEYDS---GSVRKALKLAMSCVDPSPVARPDMSHVVQELKQ 829
>At1g62090 hypothetical protein
Length = 1123
Score = 214 bits (546), Expect = 9e-56
Identities = 141/386 (36%), Positives = 209/386 (53%), Gaps = 35/386 (9%)
Query: 230 DTKKQFVCYHFHSKSTLSPPWIHKMKPSK-IAVFAIVIAFTSLILFLSVVISILRSRKVN 288
D +K+ + FH + L H+ K +AV A A +++ L V+ LR + +
Sbjct: 297 DRRKEGLDLKFHGNTKLYEFISHRKKNFPLVAVVASASAIAIVLVVLGVLFICLRRKPPS 356
Query: 289 TTVEEDPTAVFLHNHRNANLLPPVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGK 348
E P H + + FTY E+ TNNF K+ IG+GGFG +Y G+L DG+
Sbjct: 357 ---REAP-------HSSIETVKRRFTYSEVMAMTNNF--KKIIGEGGFGIIYHGHLNDGQ 404
Query: 349 LAAVKHLHRHNHTAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGT 408
AVK L + +++ K F E+ +L + H NLV L GYC D L L+Y+++ N
Sbjct: 405 QVAVKVL---SESSSQGYKQFKAEVELLMRVHHVNLVNLAGYCDDSNHLALIYEFMENRD 461
Query: 409 LAEHLHGSKSKRKGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDM 468
L EHL G K + W RL+IA + AL +EYLH KPP++HRD+ S+NI + +D
Sbjct: 462 LKEHLSG---KEGSSFLDWPCRLKIAAEAALGLEYLHTGCKPPMIHRDVKSTNILLNEDF 518
Query: 469 RIKVGDFGLSRLLVLQESNQTTSSGG--FVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSF 526
+ K+GDFGLSR + GG V T GT G+LDP+Y+++ RL+EKSDVYSF
Sbjct: 519 QAKLGDFGLSRSFPI---------GGETHVSTVVVGTHGFLDPEYYQTQRLSEKSDVYSF 569
Query: 527 GVVLLELISGLKAVDYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGA 586
G+VLLE+I+ +D R++ +A+ V + G ++ V+DP L D+ +
Sbjct: 570 GIVLLEIITNKLVIDQTRERPH--IAEWVRYMLSIGDIESVMDPNLKGKYDS---SSAWK 624
Query: 587 VAELAFRCVASDKDDRPDSKEVVGEL 612
V ELA C +RP+ ++V EL
Sbjct: 625 VLELAMLCSKLSLAERPNMAQIVHEL 650
>At1g51850 Putative protein kinase
Length = 875
Score = 214 bits (546), Expect = 9e-56
Identities = 141/386 (36%), Positives = 211/386 (54%), Gaps = 38/386 (9%)
Query: 252 HKMKPSKIAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPT------AVFLHNHRN 305
HK K + V A + + LI L V+ ILR +K + VE+ + A+ N R
Sbjct: 501 HKKKSVIVPVVASIASIAVLIGAL-VLFFILRKKK-SPKVEDGRSPRSSEPAIVTKNRR- 557
Query: 306 ANLLPPVFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFS 365
FTY ++ I TNNF +R +G GGFG VY G + + AVK L +H+++
Sbjct: 558 -------FTYSQVAIMTNNF--QRILGKGGFGMVYHGFVNGTEQVAVKIL---SHSSSQG 605
Query: 366 SKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMM 425
K F E+ +L + H NLV L GYC + + L+Y+Y+ NG L EH+ G++++ + +
Sbjct: 606 YKEFKAEVELLLRVHHKNLVGLVGYCDEGENMALIYEYMANGDLKEHMSGTRNR---FTL 662
Query: 426 TWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQE 485
W TRL+I +++A +EYLH KPP+VHRD+ ++NI + + + K+ DFGLSR ++
Sbjct: 663 NWGTRLKIVVESAQGLEYLHNGCKPPMVHRDVKTTNILLNEHFQAKLADFGLSRSFPIEG 722
Query: 486 SNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRD 545
V T GTPGYLDP+Y+++ LTEKSDVYSFG+VLLELI+ +D R+
Sbjct: 723 ETH-------VSTVVAGTPGYLDPEYYKTNWLTEKSDVYSFGIVLLELITNRPVIDKSRE 775
Query: 546 KREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDS 605
K +A+ V + G + ++DP +L D D+ +V ELA C+ RP
Sbjct: 776 KPH--IAEWVGVMLTKGDINSIMDP--NLNEDYDS-GSVWKAVELAMSCLNPSSARRPTM 830
Query: 606 KEVVGELKR--VRSRISGGITRSMST 629
+VV EL GG +R M +
Sbjct: 831 SQVVIELNECIASENSRGGASRDMDS 856
>At1g51820 Putative protein kinase
Length = 883
Score = 214 bits (546), Expect = 9e-56
Identities = 137/391 (35%), Positives = 210/391 (53%), Gaps = 38/391 (9%)
Query: 252 HKMK----PSKIAVFAIVIAFTSLILFLSVVISILRSRKVNTTVEEDPTAVFLHNHRNAN 307
HK K P ++ +I + +L+LFL ILR ++ P+ + + R
Sbjct: 499 HKKKSVIVPVVASIASIAVLIGALVLFL-----ILRKKRSPKVEGPPPSYMQASDGRLPR 553
Query: 308 LLPPV-------FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNH 360
P F+Y ++ I TNNF +R +G GGFG VY G + + AVK L +H
Sbjct: 554 SSEPAIVTKNRRFSYSQVVIMTNNF--QRILGKGGFGMVYHGFVNGTEQVAVKIL---SH 608
Query: 361 TAAFSSKSFCNEILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKR 420
+++ K F E+ +L + H NLV L GYC + L L+Y+Y+ NG L EH+ G++++
Sbjct: 609 SSSQGYKQFKAEVELLLRVHHKNLVGLVGYCDEGDNLALIYEYMANGDLKEHMSGTRNR- 667
Query: 421 KGYMMTWQTRLEIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRL 480
+++ W TRL+I I++A +EYLH KPP+VHRD+ ++NI + + K+ DFGLSR
Sbjct: 668 --FILNWGTRLKIVIESAQGLEYLHNGCKPPMVHRDVKTTNILLNEHFEAKLADFGLSRS 725
Query: 481 LVLQESNQTTSSGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAV 540
+++ V T GTPGYLDP+YHR+ LTEKSDVYSFG++LLE+I+ +
Sbjct: 726 FLIEGETH-------VSTVVAGTPGYLDPEYHRTNWLTEKSDVYSFGILLLEIITNRHVI 778
Query: 541 DYCRDKREMALADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKD 600
D R+K + + V + G ++ ++DP L N++ +V ELA C+
Sbjct: 779 DQSREKPH--IGEWVGVMLTKGDIQSIMDPSL---NEDYDSGSVWKAVELAMSCLNHSSA 833
Query: 601 DRPDSKEVVGELKR--VRSRISGGITRSMST 629
RP +VV EL GG +R M +
Sbjct: 834 RRPTMSQVVIELNECLASENARGGASRDMES 864
>At4g00330 unknown protein
Length = 411
Score = 214 bits (545), Expect = 1e-55
Identities = 122/296 (41%), Positives = 176/296 (59%), Gaps = 12/296 (4%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKS-FCN 371
FT+DE+ +T NF P +IG GGFG+VY LRDGK AVK + H + + F +
Sbjct: 107 FTFDEIYDATKNFSPSFRIGQGGFGTVYKVKLRDGKTFAVKRAKKSMHDDRQGADAEFMS 166
Query: 372 EILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRL 431
EI L+ + H +LVK +G+ ILV +YV NGTL +HL ++G + TRL
Sbjct: 167 EIQTLAQVTHLSLVKYYGFVVHNDEKILVVEYVANGTLRDHL----DCKEGKTLDMATRL 222
Query: 432 EIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTS 491
+IA A A+ YLH +PPI+HRDI SSNI + ++ R KV DFG +RL + T S
Sbjct: 223 DIATDVAHAITYLHMYTQPPIIHRDIKSSNILLTENYRAKVADFGFARL-----APDTDS 277
Query: 492 SGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMAL 551
V T +GT GYLDP+Y +++LTEKSDVYSFGV+L+EL++G + ++ R ++E
Sbjct: 278 GATHVSTQVKGTAGYLDPEYLTTYQLTEKSDVYSFGVLLVELLTGRRPIELSRGQKERIT 337
Query: 552 ADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKE 607
+ + +G VLDP L+ + N+ A+ V E+AF+C+A + RP K+
Sbjct: 338 IRWAIKKFTSGDTISVLDPKLEQNSANNL--ALEKVLEMAFQCLAPHRRSRPSMKK 391
>At2g19210 putative receptor-like protein kinase
Length = 881
Score = 214 bits (545), Expect = 1e-55
Identities = 126/311 (40%), Positives = 183/311 (58%), Gaps = 21/311 (6%)
Query: 313 FTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCNE 372
+ Y E+ TNNF+ R +G GGFG VY G L D ++A VK L + ++A K F E
Sbjct: 566 YKYSEVVKVTNNFE--RVLGQGGFGKVYHGVLNDDQVA-VKIL---SESSAQGYKEFRAE 619
Query: 373 ILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRLE 432
+ +L + H NL L GYC + + + L+Y+++ NGTL ++L G KS Y+++W+ RL+
Sbjct: 620 VELLLRVHHKNLTALIGYCHEGKKMALIYEFMANGTLGDYLSGEKS----YVLSWEERLQ 675
Query: 433 IAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTSS 492
I++ A +EYLH KPPIV RD+ +NI I + ++ K+ DFGLSR + L +NQ T
Sbjct: 676 ISLDAAQGLEYLHNGCKPPIVQRDVKPANILINEKLQAKIADFGLSRSVALDGNNQDT-- 733
Query: 493 GGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKRE-MAL 551
T GT GYLDP+YH + +L+EKSD+YSFGVVLLE++SG + R E + +
Sbjct: 734 -----TAVAGTIGYLDPEYHLTQKLSEKSDIYSFGVVLLEVVSGQPVIARSRTTAENIHI 788
Query: 552 ADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGE 611
D V + TG ++ ++DP LG DA + + E+A C +S +RP VV E
Sbjct: 789 TDRVDLMLSTGDIRGIVDP--KLGERFDA-GSAWKITEVAMACASSSSKNRPTMSHVVAE 845
Query: 612 LKRVRSRISGG 622
LK SR G
Sbjct: 846 LKESVSRARAG 856
>At1g16120
Length = 730
Score = 214 bits (545), Expect = 1e-55
Identities = 126/306 (41%), Positives = 179/306 (58%), Gaps = 17/306 (5%)
Query: 312 VFTYDELNISTNNFDPKRKIGDGGFGSVYLGNLRDGKLAAVKHLHRHNHTAAFSSKSFCN 371
+F+ EL +T+NF R +G GG G+VY G L DG + AVK R + F N
Sbjct: 416 IFSSKELRKATDNFSIDRVLGQGGQGTVYKGMLVDGSIVAVK---RSKVVDEDKMEEFIN 472
Query: 372 EILILSSIDHPNLVKLHGYCSDPRGLILVYDYVPNGTLAEHLHGSKSKRKGYMMTWQTRL 431
EI++LS I+H N+VKL G C + ILVY+Y+PNG L + LH Y MTW+ RL
Sbjct: 473 EIVLLSQINHRNIVKLLGCCLETEVPILVYEYIPNGDLFKRLHDESDD---YTMTWEVRL 529
Query: 432 EIAIQTALAMEYLHFSVKPPIVHRDITSSNIFIEKDMRIKVGDFGLSRLLVLQESNQTTS 491
IAI+ A A+ Y+H + PI HRDI ++NI +++ R KV DFG SR + L +++ TT
Sbjct: 530 RIAIEIAGALTYMHSAASFPIFHRDIKTTNILLDEKYRAKVSDFGTSRSVTLDQTHLTTL 589
Query: 492 SGGFVWTGPQGTPGYLDPDYHRSFRLTEKSDVYSFGVVLLELISGLKAVDYCRDKREMAL 551
GT GY+DP+Y S + T KSDVYSFGVVL+ELI+G K + R + L
Sbjct: 590 VA--------GTFGYMDPEYFLSSQYTHKSDVYSFGVVLVELITGEKPLSRVRSEEGRGL 641
Query: 552 ADMVVSRIHTGQLKEVLDPVLDLGNDNDALDAVGAVAELAFRCVASDKDDRPDSKEVVGE 611
A + + ++ +++D + D L+ V AVA+LA +C+ +RP+ KEV E
Sbjct: 642 ATHFLEAMKENRVIDIIDIRI---KDESKLEQVMAVAKLARKCLNRKGKNRPNMKEVSNE 698
Query: 612 LKRVRS 617
L+R+RS
Sbjct: 699 LERIRS 704
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,336,660
Number of Sequences: 26719
Number of extensions: 701124
Number of successful extensions: 6106
Number of sequences better than 10.0: 989
Number of HSP's better than 10.0 without gapping: 849
Number of HSP's successfully gapped in prelim test: 140
Number of HSP's that attempted gapping in prelim test: 2300
Number of HSP's gapped (non-prelim): 1207
length of query: 630
length of database: 11,318,596
effective HSP length: 105
effective length of query: 525
effective length of database: 8,513,101
effective search space: 4469378025
effective search space used: 4469378025
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC134049.1


