

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133864.7 - phase: 0
(563 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
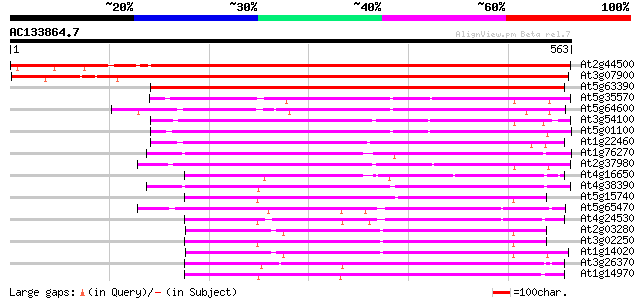
Score E
Sequences producing significant alignments: (bits) Value
At2g44500 axi 1 protein-like protein 769 0.0
At3g07900 putative auxin-independent growth promoter 696 0.0
At5g63390 auxin-independent growth promoter-like protein 541 e-154
At5g35570 unknown protein 259 2e-69
At5g64600 auxin-independent growth promoter-like protein 255 5e-68
At3g54100 unknown protein 254 1e-67
At5g01100 unknown protein 253 2e-67
At1g22460 Similar to auxin-independent growth promoter 253 3e-67
At1g76270 putative auxin-independent growth promoter (At1g76270) 247 1e-65
At2g37980 similar to axi 1 protein from Nicotiana tabacum 245 5e-65
At4g16650 growth regulator like protein 240 2e-63
At4g38390 putative growth regulator protein 237 1e-62
At5g15740 unknown protein 235 6e-62
At5g65470 unknown protein 231 8e-61
At4g24530 PsRT17-1 like protein 230 2e-60
At2g03280 axi 1 -like protein 229 3e-60
At3g02250 putative auxin-independent growth promoter 228 7e-60
At1g14020 growth regulator like protein 223 3e-58
At3g26370 unknown protein 215 6e-56
At1g14970 auxin-independent growth promoter, putative 212 5e-55
>At2g44500 axi 1 protein-like protein
Length = 573
Score = 770 bits (1987), Expect = 0.0
Identities = 395/577 (68%), Positives = 460/577 (79%), Gaps = 25/577 (4%)
Query: 1 MAKSKN------NAKKLSYISVPSQIINSISSSSLQSLIDSPKKSSKTT----IFTRKST 50
MA SKN N KK+SYISVPSQIINS+SSSSLQSL+ SPKKSS++T R
Sbjct: 1 MALSKNSNSNSFNKKKVSYISVPSQIINSLSSSSLQSLLVSPKKSSRSTNRFSFSYRNPR 60
Query: 51 LWFLALFLVGFFGMFKFGFNPDT---PFPQKPCSTTLQKFSISNH-HSFSKLGFGSVLLQ 106
+WF LFLV FGM K GFN D PF + PCSTT Q S ++ S LG L
Sbjct: 61 IWFFTLFLVSLFGMLKLGFNVDPISLPFSRYPCSTTQQPLSFDGEQNAASHLG-----LA 115
Query: 107 KERKTSLKNDHNDEFLQHGSEFLVSHVQLQAKGSSEFWKKPNGLGYKPCLSFSNDYRRQS 166
+E S + +++ +Q + L +G +FWK+P+GLG+KPCL F++ YR+ S
Sbjct: 116 QEPILSTGSSNSNAIIQLNGG---KNETLLTEG--DFWKQPDGLGFKPCLGFTSQYRKDS 170
Query: 167 ERVLKDRRKYLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDI 226
+LK+R KYL+VVVSGGMNQQRNQIVDAVVIARILGA+LVVPVLQVNVIWGDESEF DI
Sbjct: 171 NSILKNRWKYLLVVVSGGMNQQRNQIVDAVVIARILGASLVVPVLQVNVIWGDESEFADI 230
Query: 227 FDLEHFKRVLANDVRVVSALPSTHIMTRPVEG-RPPLHATPSWIRARYLRRLNREGVLLL 285
FDLEHFK VLA+DV +VS+LPSTH+MTRPVE R PLHA+P WIRA YL+R+NRE VLLL
Sbjct: 231 FDLEHFKDVLADDVHIVSSLPSTHVMTRPVEEKRTPLHASPQWIRAHYLKRINRERVLLL 290
Query: 286 RGLDSRLSKDLPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSKGPYLALHLRMEKD 345
RGLDSRLSKDLPSDLQKLRCKVAF ALRF+ I ELGNK+A RM+++G YL+LHLRMEKD
Sbjct: 291 RGLDSRLSKDLPSDLQKLRCKVAFQALRFSPRILELGNKLASRMRNQGQYLSLHLRMEKD 350
Query: 346 VWVRTGCLPGLTPEYDEVINNERIQRPELLTARSNMTYHQRKMAGLCPLNVMEVMRLLKA 405
VWVRTGCLPGLTPEYDE++N+ER + PELLT RSNMTYH+RK+AGLCPL +EV RLLKA
Sbjct: 351 VWVRTGCLPGLTPEYDEIVNSERERHPELLTGRSNMTYHERKLAGLCPLTALEVTRLLKA 410
Query: 406 LGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDY 465
L APKDARIYWAGG+PLGGKE L PL +EFP FYNK DLALPGEL+PF KAS+MAAIDY
Sbjct: 411 LEAPKDARIYWAGGEPLGGKEVLEPLTKEFPQFYNKHDLALPGELEPFANKASVMAAIDY 470
Query: 466 IVSEKSDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNKRHMLPYFLNSSLSEEEFNRIIK 525
IV EKSDVF+PSHGGNMG A+QG RA+ GHKKYITPNKR MLPYF+NSSL E +FNRI+K
Sbjct: 471 IVCEKSDVFIPSHGGNMGHALQGQRAYAGHKKYITPNKRQMLPYFMNSSLPESDFNRIVK 530
Query: 526 ELHQDSLGQPELRTNKNGRDVTKYPVPECMCNDSHAQ 562
+LH++SLGQPELR +K G+DVTK+PVPECMC+D Q
Sbjct: 531 DLHRESLGQPELRMSKAGKDVTKHPVPECMCSDRQQQ 567
>At3g07900 putative auxin-independent growth promoter
Length = 579
Score = 696 bits (1796), Expect = 0.0
Identities = 355/572 (62%), Positives = 439/572 (76%), Gaps = 16/572 (2%)
Query: 3 KSKNNAKKLSYISVPSQIINSISSSSLQSLID---SPKKSSKTTIFTRKSTLWFLALFLV 59
K+ N ++SYISVP+QIINS+SSSSL +D S K +SK R LW +LFL+
Sbjct: 7 KNSNKKNRVSYISVPAQIINSVSSSSLHKFLDNKSSKKNTSKFFFNLRNPKLWAFSLFLL 66
Query: 60 GFFGM-FKFGFNPDTPFPQKPCSTTLQKFSISNHHSFSKLGFGSVLLQ---------KER 109
G+ + G + F + LQ S SN S +GF + K+R
Sbjct: 67 SILGISLRLGLCL-SHFGSGDHESQLQS-SDSNGSPKSHMGFAYIRSSTTQVEISNAKDR 124
Query: 110 KTSLKNDHNDEFLQHGSEFLVSHVQLQAKGSSEFWKKPNGLGYKPCLSFSNDYRRQSERV 169
+ + N+ F H S ++ + + +FWK+P+GLGYKPCL FS +YRR+S+++
Sbjct: 125 SLDMGVEKNETFGGHQSHLIIPNGNGHDDKNYDFWKQPDGLGYKPCLDFSIEYRRESKKI 184
Query: 170 LKDRRKYLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDL 229
L +RRKYLMVVVSGG+NQQ+ QIVDAVVIARILGA LVVP+LQ+N+IWGDESEF DIFDL
Sbjct: 185 LVERRKYLMVVVSGGLNQQKIQIVDAVVIARILGAVLVVPILQINLIWGDESEFSDIFDL 244
Query: 230 EHFKRVLANDVRVVSALPSTHIMTRPVE-GRPPLHATPSWIRARYLRRLNREGVLLLRGL 288
E FK VLANDV++VS LP++ +MTRP E G P +A+P WIR+ Y +R NREGVLLLR L
Sbjct: 245 EQFKSVLANDVKIVSLLPASKVMTRPSEDGSMPFNASPQWIRSHYPKRFNREGVLLLRRL 304
Query: 289 DSRLSKDLPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSKGPYLALHLRMEKDVWV 348
DSRLSKDLPSDLQKLRCKVAF AL+F+ + E+G K+A+RM+SKGPY+ALHLRMEKDVWV
Sbjct: 305 DSRLSKDLPSDLQKLRCKVAFEALKFSPRVMEMGTKLAERMRSKGPYIALHLRMEKDVWV 364
Query: 349 RTGCLPGLTPEYDEVINNERIQRPELLTARSNMTYHQRKMAGLCPLNVMEVMRLLKALGA 408
RTGCL GL+ +YDE++N ERI+RPELLTA+S+MT ++RK+AGLCPLN EV RLL+ALGA
Sbjct: 365 RTGCLSGLSSKYDEIVNIERIKRPELLTAKSSMTSNERKLAGLCPLNAKEVTRLLRALGA 424
Query: 409 PKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVS 468
P+DARIYWAGG+PLGGKEAL PL EFP+ YNK D+ALP EL+PF ++AS+MAAIDYIV
Sbjct: 425 PRDARIYWAGGEPLGGKEALKPLTSEFPHLYNKYDIALPLELKPFAKRASIMAAIDYIVC 484
Query: 469 EKSDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNKRHMLPYFLNSSLSEEEFNRIIKELH 528
++SDVFM SHGGNMG AIQGHRA+ GHKK ITPNKRHMLPYF+NSSL+E EF ++IK+LH
Sbjct: 485 KESDVFMASHGGNMGHAIQGHRAYEGHKKIITPNKRHMLPYFVNSSLTETEFEKMIKKLH 544
Query: 529 QDSLGQPELRTNKNGRDVTKYPVPECMCNDSH 560
+ SLGQPELR +K GRDVTKYPVPECMCN S+
Sbjct: 545 RQSLGQPELRISKAGRDVTKYPVPECMCNQSN 576
>At5g63390 auxin-independent growth promoter-like protein
Length = 559
Score = 541 bits (1395), Expect = e-154
Identities = 260/416 (62%), Positives = 328/416 (78%), Gaps = 1/416 (0%)
Query: 142 EFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQRNQIVDAVVIARI 201
EFWK+PNG GYKPCL FS +Y+++S V K+++++L+VVVSGG+NQQRNQIVDAVVIA I
Sbjct: 133 EFWKQPNGEGYKPCLDFSLEYKKKSASVSKEKKRFLVVVVSGGLNQQRNQIVDAVVIAMI 192
Query: 202 LGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPSTHIMTRP-VEGRP 260
L AALVVPVLQVN +WGDESEF D+FD+EHFK+ L +DVR+VS+LPSTH+M+R +E +
Sbjct: 193 LEAALVVPVLQVNRVWGDESEFSDLFDVEHFKKTLRSDVRIVSSLPSTHLMSRQTIENQI 252
Query: 261 PLHATPSWIRARYLRRLNREGVLLLRGLDSRLSKDLPSDLQKLRCKVAFNALRFAEPIEE 320
P +P WIRA+Y ++LN EG+L+L+GLDS+L+K+LP DLQKLRCKVAF+ALRFA PIE
Sbjct: 253 PWDVSPVWIRAKYFKQLNEEGLLVLKGLDSKLAKNLPPDLQKLRCKVAFHALRFAAPIEN 312
Query: 321 LGNKIADRMKSKGPYLALHLRMEKDVWVRTGCLPGLTPEYDEVINNERIQRPELLTARSN 380
LGNK+ RM +GPY+ALHLR+EKDVWVRTGCL GL E+D +I R +P LT R N
Sbjct: 313 LGNKLTRRMWIEGPYIALHLRLEKDVWVRTGCLTGLGSEFDRIIAETRTSQPRYLTGRLN 372
Query: 381 MTYHQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFYN 440
+TY +R++AG CPLNV E+ RLLKALGAP +A IY AGG+P GG AL PL +EF N
Sbjct: 373 LTYTERRLAGFCPLNVYEIARLLKALGAPSNASIYIAGGEPFGGSRALEPLAKEFSNLVT 432
Query: 441 KEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAFTGHKKYIT 500
KE LA GEL P+ ++S +AAIDYIVS SDVF+PSHGGNM +A+QG+RA+ GH+K+I
Sbjct: 433 KETLAHKGELLPYTNRSSALAAIDYIVSLSSDVFIPSHGGNMAKAMQGNRAYVGHRKFIM 492
Query: 501 PNKRHMLPYFLNSSLSEEEFNRIIKELHQDSLGQPELRTNKNGRDVTKYPVPECMC 556
PNKR MLP NSS+S+ E + + ++LH+ S G PE R + RDV YPVPECMC
Sbjct: 493 PNKRAMLPLMENSSVSDAELSFLTRKLHRKSQGYPESRRGRRDRDVIAYPVPECMC 548
>At5g35570 unknown protein
Length = 652
Score = 259 bits (663), Expect = 2e-69
Identities = 159/437 (36%), Positives = 239/437 (54%), Gaps = 28/437 (6%)
Query: 141 SEFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQRNQIVDAVVIAR 200
S W KP + C+ D R +++ + YL++ +GG+NQ R I D V +A+
Sbjct: 223 SGVWAKPESGNFSRCI----DSSRSRKKLGANTNGYLLINANGGLNQMRFGICDMVAVAK 278
Query: 201 ILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPSTHIMTRPVEGRP 260
I+ A LV+P L + W D+S F D+FD +HF L +D+ +V LPS + G
Sbjct: 279 IMKATLVLPSLDHSSYWADDSGFKDLFDWQHFIEELKDDIHIVEMLPS------ELAGIE 332
Query: 261 PLHATP-SWIRARYLRR-----LNREGVLLLRGLDSRL-SKDLPSDLQKLRCKVAFNALR 313
P TP SW + Y ++ L + V+ L DSRL + DLP +QKLRC+V + AL+
Sbjct: 333 PFVKTPISWSKVGYYKKEVLPLLKQHIVMYLTHTDSRLANNDLPDSVQKLRCRVNYRALK 392
Query: 314 FAEPIEELGNKIADRMK-SKGPYLALHLRMEKDVWVRTGCLPGLTPEYDEVINNERIQRP 372
++ PIEELGN + RM+ +GPYLALHLR EKD+ TGC LT E DE + R +
Sbjct: 393 YSAPIEELGNVLVSRMRQDRGPYLALHLRYEKDMLAFTGCSHSLTAEEDEELRQMRYEVS 452
Query: 373 ELLTARSNMTYHQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLPLI 432
N T +R++ G CPL E LL+AL P +RIY G+ G ++ PL
Sbjct: 453 HWKEKEINGT--ERRLQGGCPLTPRETSLLLRALEFPSSSRIYLVAGEAY-GNGSMDPLN 509
Query: 433 QEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAF 492
+FPN ++ LA EL PF +++A +DYIV+ +S+VF+ ++ GNM +A+QGHR F
Sbjct: 510 TDFPNIFSHSILATKEELSPFNNHQNMLAGLDYIVALQSEVFLYTYDGNMAKAVQGHRRF 569
Query: 493 TGHKKYITPNKRH---MLPYFLNSSLSEEEFNRIIKELHQDSLGQPELRTN----KNGRD 545
KK I P+K + ++ +S ++F+ +K+LH+D G P R + K
Sbjct: 570 EDFKKTINPDKMNFVKLVDALDEGRISWKKFSSKVKKLHKDRNGAPYNRESGEFPKLEES 629
Query: 546 VTKYPVPECMCNDSHAQ 562
P+P C+C ++ +
Sbjct: 630 FYANPLPGCICENTEEE 646
>At5g64600 auxin-independent growth promoter-like protein
Length = 539
Score = 255 bits (651), Expect = 5e-68
Identities = 171/489 (34%), Positives = 240/489 (48%), Gaps = 48/489 (9%)
Query: 103 VLLQKERKTSLKNDHNDEFLQHGSE----------FLVSHVQLQAK-GSSEFWKKPNGLG 151
VL+ R +DH QH + +L LQ +++ W P G
Sbjct: 58 VLVFSSRNIQTDDDHTKHQQQHHRDLIDSESFPPPYLPPRKNLQKPYENTQLWTPPFSFG 117
Query: 152 YKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVL 211
PC+ + Y+ SE Y+ V +GG+NQ R I D V +A I+ A LV+P L
Sbjct: 118 LHPCVKPTPKYKEFSES-----DHYITVKSNGGLNQMRTGIADIVAVAHIMNATLVIPEL 172
Query: 212 QVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPSTHIMTRPVEGRPPL--HATPSWI 269
W D S F DIFD E F + L DV+V+ LP + VE P H T SW
Sbjct: 173 DKRSFWQDSSVFSDIFDEEQFIKSLRRDVKVIKKLP------KEVESLPRARKHFT-SWS 225
Query: 270 RARYLRRLNR----EGVLLLRGLDSRLSK-DLPSDLQKLRCKVAFNALRFAEPIEELGNK 324
Y + V+ + DSRL+ DLP D+Q+LRC+V + L F+ IE LG K
Sbjct: 226 SVGYYEEMTHLWKEYKVIHVAKSDSRLANNDLPIDVQRLRCRVLYRGLCFSPAIESLGQK 285
Query: 325 IADRMKSK-GPYLALHLRMEKDVWVRTGCLPGLTPEYDEVINNERIQRPELLTARSNMTY 383
+ +R+KS+ G Y+ALHLR EKD+ TGC GLT E + R N T
Sbjct: 286 LVERLKSRAGRYIALHLRYEKDMLAFTGCTYGLTDAESEELRVMRESTSHWKIKSINST- 344
Query: 384 HQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKED 443
+++ GLCPL EV LK LG + IY A G+ GG + L L FPN KE
Sbjct: 345 -EQREEGLCPLTPKEVGIFLKGLGYSQSTVIYIAAGEIYGGDDRLSELKSRFPNLVFKET 403
Query: 444 LALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNK 503
LA EL+ F A+ AA+DYI+S +SDVF+PSH GNM RA++GHR F GH++ ITP++
Sbjct: 404 LAGNEELKGFTGHATKTAALDYIISVESDVFVPSHSGNMARAVEGHRRFLGHRRTITPDR 463
Query: 504 RHMLPYFLNSSLSE----EEFNRIIKELHQDSLGQPELRTN-----------KNGRDVTK 548
+ ++ F+ + + + + ++H+D G P R + +
Sbjct: 464 KGLVKLFVKMERGQLKEGPKLSNFVNQMHKDRQGAPRRRKGPTQGIKGRARFRTEEAFYE 523
Query: 549 YPVPECMCN 557
P PEC+C+
Sbjct: 524 NPYPECICS 532
>At3g54100 unknown protein
Length = 638
Score = 254 bits (648), Expect = 1e-67
Identities = 153/436 (35%), Positives = 238/436 (54%), Gaps = 28/436 (6%)
Query: 142 EFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQRNQIVDAVVIARI 201
E W+KP Y+ C++ +Y R + + YL+V +GG+NQ R I D V +A+I
Sbjct: 195 EIWQKPESGNYRQCVTRPKNYTR----LQRQTNGYLVVHANGGLNQMRTGICDMVAVAKI 250
Query: 202 LGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPSTHIMTRPVEGRPP 261
+ A LV+P+L W D S F DIFD +F VL +DV +V LP + +P+ P
Sbjct: 251 MNATLVLPLLDHESFWTDPSTFKDIFDWRNFMNVLKHDVDIVEYLPPQYAAMKPLLKAPV 310
Query: 262 LHATPSWIRARYLRRLNREGVLLLRGLDSRLSKD-LPSDLQKLRCKVAFNALRFAEPIEE 320
+ S+ R+ L L R VL DSRL+ + LP +Q+LRC+ + AL + + IE+
Sbjct: 311 SWSKASYYRSEMLPLLKRHKVLKFTLTDSRLANNGLPPSIQRLRCRANYQALLYTKEIED 370
Query: 321 LGNKIADRMKSK-GPYLALHLRMEKDVWVRTGCLPGLTPEYDEVINNERIQRPELL-TAR 378
LG + +R+++ PY+ALHLR EKD+ TGC LT E E + RI R +
Sbjct: 371 LGKILVNRLRNNTEPYIALHLRYEKDMLAFTGCNHNLTTEEAEEL---RIMRYSVKHWKE 427
Query: 379 SNMTYHQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLPLIQEFPNF 438
+ +R++ G CP++ E LKA+G P +Y G+ + G E++ E+PN
Sbjct: 428 KEIDSRERRIQGGCPMSPREAAIFLKAMGYPSSTTVYIVAGE-IYGSESMDAFRAEYPNV 486
Query: 439 YNKEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAFTGHKKY 498
++ LA EL+PF + + +AA+DYIV+ +SDVF+ ++ GNM +A+QGHR F G +K
Sbjct: 487 FSHSTLATEEELEPFSQYQNRLAALDYIVALESDVFVYTYDGNMAKAVQGHRKFEGFRKS 546
Query: 499 ITPNKRH---MLPYFLNSSLSEEEFNRIIKELHQDSLGQ---------PELRTNKNGRDV 546
I P++ + ++ +F +S EEF+ +K L++D +G P L N
Sbjct: 547 INPDRLNFVRLIDHFDEGIISWEEFSSEVKRLNRDRIGAAYGRLPAALPRLEENFYAN-- 604
Query: 547 TKYPVPECMCNDSHAQ 562
P P+C+CN SH +
Sbjct: 605 ---PQPDCICNKSHPE 617
>At5g01100 unknown protein
Length = 631
Score = 253 bits (646), Expect = 2e-67
Identities = 152/434 (35%), Positives = 234/434 (53%), Gaps = 20/434 (4%)
Query: 142 EFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQRNQIVDAVVIARI 201
E W +P Y+ C++ R +++R +K YL+V +GG+NQ R I D V IA+I
Sbjct: 190 EIWNQPEVGNYQKCVA-----RPKNQRPIKQTNGYLLVHANGGLNQMRTGICDMVAIAKI 244
Query: 202 LGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPSTHIMTRPVEGRPP 261
+ A LV+P L + W D S F DIFD +HF +VLA DV +V LP +P+E P
Sbjct: 245 MNATLVLPFLDHSSFWSDPSSFKDIFDWKHFIKVLAEDVNIVEYLPQEFASIKPLEKNPV 304
Query: 262 LHATPSWIRARYLRRLNREGVLLLRGLDSRLSKDL-PSDLQKLRCKVAFNALRFAEPIEE 320
+ S+ R + L + V++ DSRL+ + P +Q+LRC+ + ALR++E IE
Sbjct: 305 SWSKSSYYRNSISKLLKKHKVIVFNHTDSRLANNSPPPSIQRLRCRANYEALRYSEDIEN 364
Query: 321 LGNKIADRMKSKG-PYLALHLRMEKDVWVRTGCLPGLTPEYDEVINNERIQRPELLTARS 379
L N ++ R++ PYLALHLR EKD+ TGC L+ E + R P
Sbjct: 365 LSNVLSSRLRENNEPYLALHLRYEKDMLAFTGCNHSLSNEESIDLEKMRFSIPHWKEKVI 424
Query: 380 NMTYHQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFY 439
N T +R++ G CP+ E LKA+G P IY GK + G+ ++ +EFPN +
Sbjct: 425 NGT--ERRLEGNCPMTPREAAVFLKAMGFPSTTNIYIVAGK-IYGQNSMTAFHEEFPNVF 481
Query: 440 NKEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAFTGHKKYI 499
LA EL + +AA+DY ++ +SD+F ++ GNM +A+QGHR F G +K I
Sbjct: 482 FHNTLATEEELSTIKPYQNRLAALDYNLALESDIFAYTYDGNMAKAVQGHRRFEGFRKTI 541
Query: 500 TPNKRHMLPYF--LNSSL-SEEEFNRIIKELHQDSLGQPELR-------TNKNGRDVTKY 549
P+++ + L++ L S E+F+ +K++HQ +G P LR + K +
Sbjct: 542 NPDRQRFVRLIDRLDAGLISWEDFSSKVKKMHQHRIGAPYLRQPGKAGMSPKLEENFYAN 601
Query: 550 PVPECMCNDSHAQS 563
P+P C+C+ S Q+
Sbjct: 602 PLPGCVCDTSEEQT 615
>At1g22460 Similar to auxin-independent growth promoter
Length = 565
Score = 253 bits (645), Expect = 3e-67
Identities = 154/431 (35%), Positives = 226/431 (51%), Gaps = 23/431 (5%)
Query: 142 EFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQRNQIVDAVVIARI 201
+ WK P + PC+S + Y E R YL+V +GG+NQ R I D V IARI
Sbjct: 134 KLWKHPPNRNFVPCVSPNPSYTSPLES-----RGYLLVHTNGGLNQMRAGICDMVAIARI 188
Query: 202 LGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPSTHIMTRPVEGRPP 261
+ A LVVP L W D S+F D+FD +HF L+ D+RV+ LP V
Sbjct: 189 INATLVVPELDKRSFWQDTSKFSDVFDEDHFINALSKDIRVIKKLPKGIDGLTKVVKHFK 248
Query: 262 LHATPSWIRARYLRRLNREGVLLLRGLDSRLSKD-LPSDLQKLRCKVAFNALRFAEPIEE 320
++ S+ + + V+ DSRL+ + LP D+QKLRC+ + ALRF+ I
Sbjct: 249 SYSGLSYYQNEIASMWDEYKVIRAAKSDSRLANNNLPPDIQKLRCRACYEALRFSTKIRS 308
Query: 321 LGNKIADRMKSKGPYLALHLRMEKDVWVRTGCLPGLTPEYDEVINNERIQRPELLTARSN 380
+G + DRM+S G Y+ALHLR EK++ +GC GL+ E RI++ +
Sbjct: 309 MGELLVDRMRSYGLYIALHLRFEKEMLAFSGCNHGLSAS--EAAELRRIRKNTAYWKVKD 366
Query: 381 MTYHQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFYN 440
+ +++ G CPL EV LL ALG D +Y A G+ GG+ L L F +
Sbjct: 367 IDGRVQRLKGYCPLTPKEVGILLTALGYSSDTPVYIAAGEIYGGESRLADLRSRFSMLMS 426
Query: 441 KEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAFTGHKKYIT 500
KE LA EL+ F+ ++ MAA+DYIVS +SDVF+PS+ GNM RA++GHR F GH+K I+
Sbjct: 427 KEKLATREELKTFMNHSTQMAALDYIVSIESDVFIPSYSGNMARAVEGHRRFLGHRKTIS 486
Query: 501 PNKRHMLPYFLNSSLSEEEFNR----IIKELHQDSLGQPE-----------LRTNKNGRD 545
P+++ ++ E+ NR I E+H+ G P L +++
Sbjct: 487 PDRKAIVRLVDRIGRGAEKDNRKVYERINEIHKTRQGSPRRRKGPASGTKGLERHRSEES 546
Query: 546 VTKYPVPECMC 556
+ P+P+C+C
Sbjct: 547 FYENPLPDCLC 557
>At1g76270 putative auxin-independent growth promoter (At1g76270)
Length = 572
Score = 247 bits (630), Expect = 1e-65
Identities = 152/434 (35%), Positives = 230/434 (52%), Gaps = 21/434 (4%)
Query: 138 KGSSEFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQRNQIVDAVV 197
+ + W+ N + C + S+ + DR YL++ SGG+NQQR IVDAVV
Sbjct: 73 RSDRDIWRSRNAEFFFGCSNASSKFANSKAVTRNDR--YLVIATSGGLNQQRTGIVDAVV 130
Query: 198 IARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPSTHIMTRPVE 257
ARIL A LVVP L W D S+F IFD++ F L+ DVR++ LP T
Sbjct: 131 AARILNATLVVPKLDQKSYWKDASDFSHIFDVDWFISFLSGDVRIIKQLPLKGGRTWSTS 190
Query: 258 G-RPPLHATPSWIRARYLRRLNREGVLLLRGLDSRLSKDLPSDLQKLRCKVAFNALRFAE 316
R P R L L + + L D RLS L DLQKLRC+V ++AL+F +
Sbjct: 191 RMRVPRKCNERCYINRVLPVLLKRHAVQLNKFDYRLSNKLSDDLQKLRCRVNYHALKFTD 250
Query: 317 PIEELGNKIADRMKSKGP-YLALHLRMEKDVWVRTGCLPGLTPEYDEVINNERIQRPELL 375
PI +GN++ RM+ + ++ALHLR E D+ +GC G ++ +R
Sbjct: 251 PILTMGNELVRRMRLRSKHFIALHLRYEPDMLAFSGCYYG---------GGDKERRELAA 301
Query: 376 TARSNMTYH-----QRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLP 430
R T H +++ G CPL EV +L+ALG D IY A G+ GG+E+L P
Sbjct: 302 IRRRWKTLHINNPEKQRRQGRCPLTPEEVGLMLRALGYGSDVHIYVASGEVYGGEESLAP 361
Query: 431 LIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHR 490
L FP+FY+K+ +A EL+PF +S MAA+D++V ++SDVF+ ++ GNM + + G R
Sbjct: 362 LKALFPHFYSKDTIATKEELEPFSSYSSRMAALDFLVCDESDVFVTNNNGNMAKILAGRR 421
Query: 491 AFTGHKKYITPNKRHMLPYFLN-SSLSEEEFNRIIKELHQDSLGQPELRTNKNGRDVTKY 549
+ GHK + PN + + F+N + + EEF+ ++ + +G+P + + GR
Sbjct: 422 RYLGHKPTVRPNAKKLYRLFMNKENTTWEEFSSKVRSFQRGFMGEP--KEVRAGRGEFHE 479
Query: 550 PVPECMCNDSHAQS 563
C+C D+ A++
Sbjct: 480 NPSTCICEDTEAKA 493
>At2g37980 similar to axi 1 protein from Nicotiana tabacum
Length = 638
Score = 245 bits (625), Expect = 5e-65
Identities = 148/444 (33%), Positives = 235/444 (52%), Gaps = 18/444 (4%)
Query: 129 LVSHVQLQAKGSSEFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQ 188
+ +V ++ E W+KP Y+ C S + R+ + YL+V +GG+NQ
Sbjct: 186 MTENVVIEKLPIPEIWQKPESGNYRQCASRP----KNRSRLRRKTNGYLLVHANGGLNQM 241
Query: 189 RNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPS 248
R I D V A+I+ A LV+P+L W D S F DIFD HF VL +DV +V LP
Sbjct: 242 RTGICDMVAAAKIMNATLVLPLLDHESFWTDPSTFKDIFDWRHFMNVLKDDVDIVEYLPP 301
Query: 249 THIMTRPVEGRPPLHATPSWIRARYLRRLNREGVLLLRGLDSRLSKD-LPSDLQKLRCKV 307
+ RP+ P + S+ R+ L L + V+ DSRL+ + LP +Q+LRC+
Sbjct: 302 RYAAMRPLLKAPVSWSKASYYRSEMLPLLKKHKVIKFTHTDSRLANNGLPPSIQRLRCRA 361
Query: 308 AFNALRFAEPIEELGNKIADRMKSKG-PYLALHLRMEKDVWVRTGCLPGLTPEYDEVINN 366
+ AL +++ IE+ G + +R+++ P++ALHLR EKD+ TGC LT E +
Sbjct: 362 NYQALGYSKEIEDFGKVLVNRLRNNSEPFIALHLRYEKDMLAFTGCSHNLTAGEAEEL-- 419
Query: 367 ERIQRPELL-TARSNMTYHQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGK 425
RI R + + +R++ G CP++ E LKA+G P +Y G+ GG
Sbjct: 420 -RIMRYNVKHWKEKEIDSRERRIQGGCPMSPREAAIFLKAMGYPSSTTVYIVAGEIYGG- 477
Query: 426 EALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRA 485
++ +E+PN + LA EL+PF + +AA+DYIV+ +SDVF+ ++ GNM +A
Sbjct: 478 NSMDAFREEYPNVFAHSYLATEEELEPFKPYQNRLAALDYIVALESDVFVYTYDGNMAKA 537
Query: 486 IQGHRAFTGHKKYITPNKRH---MLPYFLNSSLSEEEFNRIIKELHQDSLGQPELRT--- 539
+QGHR F G KK I P++ + ++ + +S +EF+ +K LH + +G P R
Sbjct: 538 VQGHRRFEGFKKTINPDRLNFVRLIDHLDEGVMSWDEFSSEVKRLHNNRIGAPYARLPGE 597
Query: 540 -NKNGRDVTKYPVPECMCNDSHAQ 562
+ + P P+C+CN S +
Sbjct: 598 FPRLEENFYANPQPDCICNKSQPE 621
>At4g16650 growth regulator like protein
Length = 549
Score = 240 bits (612), Expect = 2e-63
Identities = 153/393 (38%), Positives = 220/393 (55%), Gaps = 26/393 (6%)
Query: 176 YLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRV 235
YL++ SGG+NQQR I DAVV+ARIL A LVVP L + W D+S+F DIFD+ F
Sbjct: 125 YLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHHSYWKDDSDFSDIFDVNWFISS 184
Query: 236 LANDVRVVSALPSTHIMTR---PVEGRPPLHATPSWIRARYLRRLNREGVLLLRGLDSRL 292
LA DV +V +P + P R P +T + + L L R VL L D RL
Sbjct: 185 LAKDVTIVKRVPDRVMRAMEKPPYTTRVPRKSTLEYYLDQVLPILTRRHVLQLTKFDYRL 244
Query: 293 SKDLPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSKGP-YLALHLRMEKDVWVRTG 351
+ DL D+QKLRC+V ++ALRF + I+ +G K+ RM+ ++A+HLR E D+ +G
Sbjct: 245 ANDLDEDMQKLRCRVNYHALRFTKRIQSVGMKVVKRMRKMAKRFIAVHLRFEPDMLAFSG 304
Query: 352 CLPGLTPEYDEVINNERIQRPELLTARS------NMTYHQRKMAGLCPLNVMEVMRLLKA 405
C G E+ +R EL R ++ + + G CPL EV +L+A
Sbjct: 305 CDFG---------GGEK-ERAELAEIRKRWDTLPDLDPLEERKRGKCPLTPHEVGLMLRA 354
Query: 406 LGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDY 465
LG D IY A G+ GG++ L PL + FPNFY KE LA EL+P + +S +AAIDY
Sbjct: 355 LGFTNDTYIYVASGEIYGGEKTLKPLRELFPNFYTKEMLA-NDELKPLLPYSSRLAAIDY 413
Query: 466 IVSEKSDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNKRHMLPYFLNSSLSE-EEFNRII 524
IVS++SDVF+ ++ GNM + + G R + GHK+ I PN + + F++ E + F + +
Sbjct: 414 IVSDESDVFITNNNGNMAKILAGRRRYMGHKRTIRPNAKKLSALFMDREKMEWQTFAKKV 473
Query: 525 KELHQDSLGQPELRTNKNGR-DVTKYPVPECMC 556
K + +G P+ K GR + +YP C+C
Sbjct: 474 KSCQRGFMGDPD--EFKPGRGEFHEYP-QSCIC 503
>At4g38390 putative growth regulator protein
Length = 551
Score = 237 bits (605), Expect = 1e-62
Identities = 150/430 (34%), Positives = 221/430 (50%), Gaps = 13/430 (3%)
Query: 138 KGSSEFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQRNQIVDAVV 197
+ + W Y C + ++ ++ R +R YL++ SGG+NQQR I+DAVV
Sbjct: 90 RSDRQLWSSRLSNLYYACSNATDTFQVTDTRSQTNR--YLLIATSGGLNQQRTGIIDAVV 147
Query: 198 IARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPS---THIMTR 254
A IL A LVVP L W D S F DIFD++ F L+ DV+++ LP + I T
Sbjct: 148 AAYILNATLVVPKLDQKSYWKDTSNFEDIFDVDWFISHLSKDVKIIKELPKEEQSRISTS 207
Query: 255 PVEGRPPLHATPSWIRARYLRRLNREGVLLLRGLDSRLSKDLPSDLQKLRCKVAFNALRF 314
R P TPS R L L ++ V+ L D RLS L ++LQKLRC+V ++A+R+
Sbjct: 208 LQSMRVPRKCTPSCYLQRVLPILTKKHVVQLSKFDYRLSNALDTELQKLRCRVNYHAVRY 267
Query: 315 AEPIEELGNKIADRMKSKGP-YLALHLRMEKDVWVRTGCLPGLTPEYDEVINNERIQRPE 373
E I +G + DRM+ K ++ALHLR E D+ +GC G + + R +
Sbjct: 268 TESINRMGQLLVDRMRKKAKHFVALHLRFEPDMLAFSGCYYGGGQKERLELGAMRRRWKT 327
Query: 374 LLTARSNMTYHQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLPLIQ 433
L A + G CPL E+ +L+ LG K+ +Y A G+ GG++ L PL
Sbjct: 328 LHAANPEKV----RRHGRCPLTPEEIGLMLRGLGFGKEVHLYVASGEVYGGEDTLAPLRA 383
Query: 434 EFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAFT 493
FPN + KE L EL PF +S MAA+D+IV +KSD F+ ++ GNM R + G R +
Sbjct: 384 LFPNLHTKETLTSKKELAPFANFSSRMAALDFIVCDKSDAFVTNNNGNMARILAGRRRYL 443
Query: 494 GHKKYITPNKRHMLPYFLN-SSLSEEEFNRIIKELHQDSLGQPELRTNKNGRDVTKYPVP 552
GHK I PN + + F N +++ EF+ ++ +G+P+ K G
Sbjct: 444 GHKVTIRPNAKKLYEIFKNRHNMTWGEFSSKVRRYQTGFMGEPD--EMKPGEGEFHENPA 501
Query: 553 ECMCNDSHAQ 562
C+C S A+
Sbjct: 502 SCICRTSEAR 511
>At5g15740 unknown protein
Length = 508
Score = 235 bits (599), Expect = 6e-62
Identities = 137/371 (36%), Positives = 201/371 (53%), Gaps = 10/371 (2%)
Query: 176 YLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRV 235
YLMV +GG+NQ R I D V +AR + L+VP L W D SEF DIFD++HF
Sbjct: 109 YLMVSCNGGLNQMRAAICDMVTVARYMNVTLIVPELDKTSFWNDPSEFKDIFDVDHFISS 168
Query: 236 LANDVRVVSALP---STHIMTRPVEGRPPLH-ATPSWIRARYLRRLNREGVLLLRGLDSR 291
L ++VR++ LP + PP+ + S+ + + L + + VL L D+R
Sbjct: 169 LRDEVRILKELPPRLKKRVELGVYHEMPPISWSNMSYYQNQILPLVKKHKVLHLNRTDTR 228
Query: 292 LSKD-LPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSKGPYLALHLRMEKDVWVRT 350
L+ + LP ++QKLRC+V FN L+F IEELG ++ ++ KGP+L LHLR E D+ +
Sbjct: 229 LANNGLPVEVQKLRCRVNFNGLKFTPQIEELGRRVVKILREKGPFLVLHLRYEMDMLAFS 288
Query: 351 GCLPGLTPEYDEVINNERIQRPELLTARSNMTYHQRKMAGLCPLNVMEVMRLLKALGAPK 410
GC G PE +E + R P N ++ GLCPL E L ALG +
Sbjct: 289 GCSHGCNPEEEEELTRMRYAYPWWKEKVINSELKRKD--GLCPLTPEETALTLTALGIDR 346
Query: 411 DARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVSEK 470
+ +IY A G+ GG+ + L FPN KE L +L +S MAA+DY+V+ +
Sbjct: 347 NVQIYIAAGEIYGGQRRMKALTDAFPNVVRKETLLESSDLDFCRNHSSQMAALDYLVALE 406
Query: 471 SDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNKR---HMLPYFLNSSLSEEEFNRIIKEL 527
SD+F+P++ GNM R ++GHR F G KK I N+R ++ + LS + F+ +K
Sbjct: 407 SDIFVPTNDGNMARVVEGHRRFLGFKKTIQLNRRFLVKLIDEYTEGLLSWDVFSSTVKAF 466
Query: 528 HQDSLGQPELR 538
H +G P+ R
Sbjct: 467 HSTRMGSPKRR 477
>At5g65470 unknown protein
Length = 504
Score = 231 bits (589), Expect = 8e-61
Identities = 159/473 (33%), Positives = 227/473 (47%), Gaps = 59/473 (12%)
Query: 129 LVSHVQLQAKGSSEFWKKPNGLGYKPCLSFSNDYRRQSERVLKDRRKYLMVVVSGGMNQQ 188
L S +Q + S+ W+ G+ PC+ N S + Y+ V + GG+NQQ
Sbjct: 46 LKSALQRSSGEQSDLWRPLTDQGWSPCIDLGN-----SPSLPDKTAGYVQVFLDGGLNQQ 100
Query: 189 RNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVLANDVRVVSALPS 248
R I DAV +A+IL A LV+P L+VN +W D S F DIFD++HF L +D+RVV LP
Sbjct: 101 RMGICDAVAVAKILNATLVIPYLEVNPVWQDSSSFVDIFDVDHFIDSLKDDIRVVRELPD 160
Query: 249 THI-MTRPVEG---------RPPLHATPSWIRARYLRRLNREGVLLLRGLDSRLSKD-LP 297
+ TR G P+HA+ +W L G+ + RLS D LP
Sbjct: 161 EYSWSTREYYGTAVRETRVKTAPVHASANWYIENVSPVLQSYGIAAISPFSHRLSFDHLP 220
Query: 298 SDLQKLRCKVAFNALRFAEPIEELGNKIADRMKS-------------------------- 331
+++Q+LRCKV F ALRF I LG+ + R+++
Sbjct: 221 AEIQRLRCKVNFQALRFVPHITSLGDALVSRLRNPSWRSNKEQKNVDHLGDMTNPHRRQE 280
Query: 332 KGPYLALHLRMEKDVWVRTGCLPG-------LTPEYDEVINNERIQRPELLTARSNMTYH 384
G + LHLR +KD+ + C G +Y ++I R+ S T
Sbjct: 281 PGKFAVLHLRFDKDMAAHSACDFGGGKAEKLSLAKYRQMIWQGRV-------LNSQFTDE 333
Query: 385 QRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDL 444
+ + G CPL E+ LL A G + R+Y A K GG+ + L Q FP +K L
Sbjct: 334 ELRSQGRCPLTPEEMGLLLAAFGFDNNTRLYLASHKVYGGEARISTLRQVFPRMEDKRSL 393
Query: 445 ALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNKR 504
A E KASL+AA+DY VS SD+F+ + GNM A+ GHR F + K I PN
Sbjct: 394 ASSEERARIKGKASLLAALDYYVSMHSDIFISASPGNMHNALVGHRTFE-NLKTIRPNMA 452
Query: 505 HMLPYFLNSSLSEEEFNRIIKELHQDSLGQPELRTNKNGRDVTKYPVPECMCN 557
+ FLN S++ +F + + E H + GQ LR K + + YP P+CMC+
Sbjct: 453 LIGQLFLNKSITWVDFQQALGEGHVNRQGQIRLRKPK--QSIYTYPAPDCMCH 503
>At4g24530 PsRT17-1 like protein
Length = 431
Score = 230 bits (586), Expect = 2e-60
Identities = 154/429 (35%), Positives = 211/429 (48%), Gaps = 64/429 (14%)
Query: 176 YLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRV 235
+L V + GG+NQQR I DAV +A+I+ LV+P L+VN +W D S F DIFDL+HF V
Sbjct: 16 FLQVFLDGGLNQQRMGICDAVAVAKIMNVTLVIPRLEVNTVWQDSSSFTDIFDLDHFISV 75
Query: 236 LANDVRVVSALP----------------STHIMTRPVEGRPPLHATPSWIRARYLRRLNR 279
L ++VR+V LP +T I T PV HA+ W L +
Sbjct: 76 LKDEVRIVRELPIQYAWSTRDYYATGIRATRIKTAPV------HASAEWYLENVLPIIQS 129
Query: 280 EGVLLLRGLDSRLSKD-LPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSK------ 332
G+ + RL+ D LP +Q+LRCKV F AL F I ELG+ + R+++
Sbjct: 130 YGIAAVAPFSHRLAFDNLPESIQRLRCKVNFEALNFVPHIRELGDALVHRLRNPPSSSQT 189
Query: 333 ------------------GPYLALHLRMEKDVWVRTGC-LPGLTPE------YDEVINNE 367
G + LHLR +KD+ +GC G E Y +VI
Sbjct: 190 SGTMDPTDRINTIVKAGAGKFAVLHLRFDKDMAAHSGCDFEGGKAEKLALAKYRQVIWQG 249
Query: 368 RIQRPELLTARSNMTYHQRKMAGLCPLNVMEVMRLLKALGAPKDARIYWAGGKPLGGKEA 427
R+ S T + + G CPL E+ LL ALG + R+Y A + GG+
Sbjct: 250 RV-------LNSQFTDEELRNKGRCPLTPEEIGLLLSALGFSNNTRLYLASHQVYGGEAR 302
Query: 428 LLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVSEKSDVFMPSHGGNMGRAIQ 487
+ L + FP NK+ LA EL KASLMAA+DY VS KSD+F+ + GNM A+Q
Sbjct: 303 ISTLRKLFPGIENKKSLASAEELADVQGKASLMAAVDYYVSMKSDIFISASPGNMHNALQ 362
Query: 488 GHRAFTGHKKYITPNKRHMLPYFLNSSLSEEEFNRIIKELHQDSLGQPELRTNKNGRDVT 547
HRA+ + K I PN + F+N SL EF + H++ GQ LR K + +
Sbjct: 363 AHRAYL-NLKTIRPNMILLGQVFVNKSLDWSEFEGAVMNGHKNRQGQ--LRLRKQKQSIY 419
Query: 548 KYPVPECMC 556
YP P+CMC
Sbjct: 420 TYPAPDCMC 428
>At2g03280 axi 1 -like protein
Length = 481
Score = 229 bits (584), Expect = 3e-60
Identities = 141/374 (37%), Positives = 207/374 (54%), Gaps = 18/374 (4%)
Query: 177 LMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVL 236
L+V +GG+NQ R I D V +AR+L LVVP L W D S+F DIFD++HF L
Sbjct: 92 LLVSCNGGLNQMRAAICDMVTVARLLNLTLVVPELDKKSFWADTSDFEDIFDIKHFIDSL 151
Query: 237 ANDVRVVSALPSTHIMTRPVE--GRPPLHATPSWIRARY-----LRRLNREGVLLLRGLD 289
++VR++ LP + + PP+ SW +Y L R ++ V+ D
Sbjct: 152 RDEVRIIRRLPKRYSKKYGFKLFEMPPV----SWSNDKYYLQQVLPRFSKRKVIHFVRSD 207
Query: 290 SRLSKD-LPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSKGPYLALHLRMEKDVWV 348
+RL+ + L DLQ+LRC+V F LRF IE LG+K+ ++ +G ++ALHLR E D+
Sbjct: 208 TRLANNGLSLDLQRLRCRVNFQGLRFTPRIEALGSKLVRILQQRGSFVALHLRYEMDMLA 267
Query: 349 RTGCLPGLTPEYDEVINNERIQRPELLTARSNMTYHQRKMAGLCPLNVMEVMRLLKALGA 408
+GC G T E E + R P + +R++ GLCPL E + +LKALG
Sbjct: 268 FSGCTHGCTDEEAEELKKMRYAYP--WWREKEIVSEERRVQGLCPLTPEEAVLVLKALGF 325
Query: 409 PKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVS 468
KD +IY A G+ GG + L L + FP KE L P ELQ F +S MAA+D+IVS
Sbjct: 326 QKDTQIYIAAGEIFGGAKRLALLKESFPRIVKKEMLLDPTELQQFQNHSSQMAALDFIVS 385
Query: 469 EKSDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNKR---HMLPYFLNSSLSEEEFNRIIK 525
S+ F+P++ GNM + ++GHR + G KK I +++ +L N +LS ++F +K
Sbjct: 386 VASNTFIPTYYGNMAKVVEGHRRYLGFKKTILLDRKRLVELLDLHNNKTLSWDQFAVAVK 445
Query: 526 ELHQD-SLGQPELR 538
+ HQ +G+P R
Sbjct: 446 DAHQGRRMGEPTHR 459
>At3g02250 putative auxin-independent growth promoter
Length = 512
Score = 228 bits (581), Expect = 7e-60
Identities = 135/371 (36%), Positives = 198/371 (52%), Gaps = 10/371 (2%)
Query: 176 YLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRV 235
YLMV +GG+NQ R I D V IAR + L+VP L W D SEF DIFD++HF
Sbjct: 109 YLMVSCNGGLNQMRAAICDMVTIARYMNVTLIVPELDKTSFWNDPSEFKDIFDVDHFISS 168
Query: 236 LANDVRVVSALP---STHIMTRPVEGRPPLH-ATPSWIRARYLRRLNREGVLLLRGLDSR 291
L ++VR++ LP + PP+ + S+ + + L + + V+ L D+R
Sbjct: 169 LRDEVRILKELPPRLKRRVRLGLYHTMPPISWSNMSYYQDQILPLVKKYKVVHLNKTDTR 228
Query: 292 LSK-DLPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSKGPYLALHLRMEKDVWVRT 350
L+ +LP ++QKLRC+ FN LRF IEELG ++ ++ KGP+L LHLR E D+ +
Sbjct: 229 LANNELPVEIQKLRCRANFNGLRFTPKIEELGRRVVKILREKGPFLVLHLRYEMDMLAFS 288
Query: 351 GCLPGLTPEYDEVINNERIQRPELLTARSNMTYHQRKMAGLCPLNVMEVMRLLKALGAPK 410
GC G +E + R P + ++ GLCPL E L ALG +
Sbjct: 289 GCSHGCNRYEEEELTRMRYAYP--WWKEKVIDSELKRKEGLCPLTPEETALTLSALGIDR 346
Query: 411 DARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVSEK 470
+ +IY A G+ GGK L L FPN KE L +L +S MAA+DY++S +
Sbjct: 347 NVQIYIAAGEIYGGKRRLKALTDVFPNVVRKETLLDSSDLSFCKNHSSQMAALDYLISLE 406
Query: 471 SDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNKR---HMLPYFLNSSLSEEEFNRIIKEL 527
SD+F+P++ GNM + ++GHR F G KK I N++ ++ + LS E F+ +K
Sbjct: 407 SDIFVPTYYGNMAKVVEGHRRFLGFKKTIELNRKLLVKLIDEYYEGLLSWEVFSTTVKAF 466
Query: 528 HQDSLGQPELR 538
H +G P+ R
Sbjct: 467 HATRMGGPKRR 477
>At1g14020 growth regulator like protein
Length = 499
Score = 223 bits (567), Expect = 3e-58
Identities = 141/399 (35%), Positives = 212/399 (52%), Gaps = 21/399 (5%)
Query: 177 LMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRVL 236
L+V +GG+NQ R+ I D V +AR+L LVVP L W D S F DIFD+ HF L
Sbjct: 95 LLVSCNGGLNQMRSAICDMVTVARLLNLTLVVPELDKTSFWADPSGFEDIFDVRHFIDSL 154
Query: 237 ANDVRVVSALPSTHIMTRPVE--GRPPLHATPSWIRARY-----LRRLNREGVLLLRGLD 289
++VR++ LP + PP+ SW +Y L ++ V+ D
Sbjct: 155 RDEVRILRRLPKRFSRKYGYQMFEMPPV----SWSDEKYYLKQVLPLFSKHKVVHFNRTD 210
Query: 290 SRLSKD-LPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSKGPYLALHLRMEKDVWV 348
+RL+ + LP LQ LRC+V F L+F +E LG+K+ ++ +GP++ALHLR E D+
Sbjct: 211 TRLANNGLPLSLQWLRCRVNFQGLKFTPQLEALGSKLVRILQQRGPFVALHLRYEMDMLA 270
Query: 349 RTGCLPGLTPEYDEVINNERIQRPELLTARSNMTYHQRKMAGLCPLNVMEVMRLLKALGA 408
+GC G T E E + R P + +R+ GLCPL EV +LKALG
Sbjct: 271 FSGCTHGCTEEEAEELKKMRYTYP--WWREKEIVSEERRAQGLCPLTPEEVALVLKALGF 328
Query: 409 PKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAAIDYIVS 468
K+ +IY A G+ G + L L + FP KE L ELQ F +S MAA+D++VS
Sbjct: 329 EKNTQIYIAAGEIYGSEHRLSVLREAFPRIVKKEMLLESAELQQFQNHSSQMAALDFMVS 388
Query: 469 EKSDVFMPSHGGNMGRAIQGHRAFTGHKKYITPNKR---HMLPYFLNSSLSEEEFNRIIK 525
S+ F+P++ GNM + ++GHR + G+KK I +++ +L N +L+ ++F +K
Sbjct: 389 VASNTFIPTYDGNMAKVVEGHRRYLGYKKTILLDRKRLVELLDLHHNKTLTWDQFAVAVK 448
Query: 526 ELHQDSLGQPELR---TNKNGRDVTKYPVP-ECMCNDSH 560
E H+ G P R ++K + Y P EC+C ++
Sbjct: 449 EAHERRAGAPTHRRVISDKPKEEDYFYANPQECLCEGTN 487
>At3g26370 unknown protein
Length = 557
Score = 215 bits (547), Expect = 6e-56
Identities = 138/398 (34%), Positives = 216/398 (53%), Gaps = 20/398 (5%)
Query: 176 YLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRV 235
Y+ + GG+NQQR I +AV +A+I+ A L++PVL+ + IW D ++F DIFD++HF
Sbjct: 161 YVFIHAEGGLNQQRIAICNAVAVAKIMNATLILPVLKQDQIWKDTTKFEDIFDVDHFIDY 220
Query: 236 LANDVRVVSALPSTHI--------MTRPVEGRPPLHATPSWIRARYLRRLNREGVLLLRG 287
L +DVR+V +P + R V+ P A +I L R+ + ++ L+
Sbjct: 221 LKDDVRIVRDIPDWFTDKAELFSSIRRTVKNIPKYAAAQFYID-NVLPRIKEKKIMALKP 279
Query: 288 LDSRLSKD-LPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMKSK----GPYLALHLRM 342
RL D +P ++ +LRC+V ++AL+F IE++ + + RM+++ PY+ALHLR
Sbjct: 280 FVDRLGYDNVPQEINRLRCRVNYHALKFLPEIEQMADSLVSRMRNRTGNPNPYMALHLRF 339
Query: 343 EKDVWVRTGC-LPGLTPEYDEVINNERIQRPELLTARSNMTYH--QRKMAGLCPLNVMEV 399
EK + + C G E ++ + + P S++ Q++ G CPL EV
Sbjct: 340 EKGMVGLSFCDFVGTREEKVKMAEYRQKEWPRRFKNGSHLWQLALQKRKEGRCPLEPGEV 399
Query: 400 MRLLKALGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASL 459
+L+A+G PK+ +IY A G+ GG+ + PL FPN KEDLA EL F + +
Sbjct: 400 AVILRAMGYPKETQIYVASGQVYGGQNRMAPLRNMFPNLVTKEDLAGKEELTTFRKHVTS 459
Query: 460 MAAIDYIVSEKSDVFMPSHGGNMGRAIQGHRAFTGHK-KYITPNKRHMLPYFLNSSLSEE 518
+AA+D++V KSDVF+ +HGGN + I G R + GH+ K I P+K M F + +
Sbjct: 460 LAALDFLVCLKSDVFVMTHGGNFAKLIIGARRYMGHRQKSIKPDKGLMSKSFGDPYMGWA 519
Query: 519 EFNRIIKELHQDSLGQPELRTNKNGRDVTKYPVPECMC 556
F + HQ G PE T N D+ + P+ CMC
Sbjct: 520 TFVEDVVVTHQTRTGLPE-ETFPN-YDLWENPLTPCMC 555
>At1g14970 auxin-independent growth promoter, putative
Length = 458
Score = 212 bits (539), Expect = 5e-55
Identities = 136/398 (34%), Positives = 202/398 (50%), Gaps = 20/398 (5%)
Query: 176 YLMVVVSGGMNQQRNQIVDAVVIARILGAALVVPVLQVNVIWGDESEFGDIFDLEHFKRV 235
++ + +GG+NQQR I +AV +A L A LV+P + IW D S+FGDI+D E+F
Sbjct: 54 FIFIEANGGLNQQRTSICNAVAVAGYLNATLVIPNFHYHSIWKDPSKFGDIYDEEYFIDT 113
Query: 236 LANDVRVVSALPS------THIMTRPVEGRPPLHATPSWIRARYLRRLNREGVLLLRGLD 289
LANDVRVV +P + +T R A S+ R L +L E V+ +
Sbjct: 114 LANDVRVVDTVPEYLMERFDYNLTNVYNFRVKAWAPTSYYRDSVLPKLLEEKVIRISPFA 173
Query: 290 SRLSKDLPSDLQKLRCKVAFNALRFAEPIEELGNKIADRMK-----SKGPYLALHLRMEK 344
+RLS D P +Q+ RC ALRF++PI G + ++MK + G Y+++HLR E+
Sbjct: 174 NRLSFDAPRAVQRFRCLANNVALRFSKPILTQGETLVNKMKGLSANNAGKYVSVHLRFEE 233
Query: 345 DVWVRTGCL--PGLTPEYDEVINNERIQRPELLTARSNMTYHQRKMAGLCPLNVMEVMRL 402
D+ + C+ G + D + ER + + + ++ G CPL +EV +
Sbjct: 234 DMVAFSCCVFDGGDQEKQDMIAARERGWKGKFTKPGRVIRPGANRLNGKCPLTPLEVGLM 293
Query: 403 LKALGAPKDARIYWAGGKPLGGKEALLPLIQEFPNFYNKEDLALPGELQPFVRKASLMAA 462
L+ +G K I+ A G + PL++ FPN KE LA +L PF +S MAA
Sbjct: 294 LRGMGFNKSTYIFLAAGPIYSANRTMAPLLEMFPNLQTKEMLASEEDLAPFKNFSSRMAA 353
Query: 463 IDYIVSEKSDVFMPSHGGNMGRAIQGHR--AFTGHKKYITPNKRHMLPYFLNSSLSEEEF 520
IDY V S+VF+ + GGN + GHR F GH K I P+KR + F N L + F
Sbjct: 354 IDYTVCLHSEVFVTTQGGNFPHFLMGHRRYLFGGHSKTIQPDKRKLAVLFDNPKLGWKSF 413
Query: 521 NRIIKEL--HQDSLGQPELRTNKNGRDVTKYPVPECMC 556
R + + H DS G ++ + +P P+CMC
Sbjct: 414 KRQMLSMRSHSDSKG---FELKRSSDSIYIFPCPDCMC 448
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,967,755
Number of Sequences: 26719
Number of extensions: 568690
Number of successful extensions: 1433
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1275
Number of HSP's gapped (non-prelim): 48
length of query: 563
length of database: 11,318,596
effective HSP length: 104
effective length of query: 459
effective length of database: 8,539,820
effective search space: 3919777380
effective search space used: 3919777380
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC133864.7


