

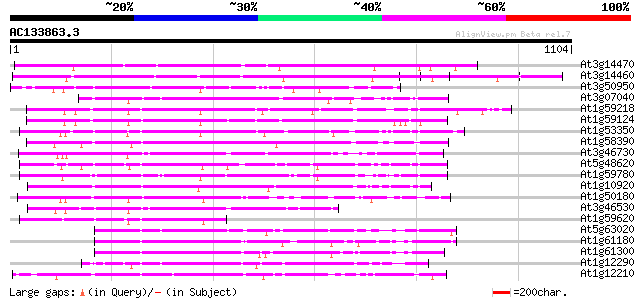
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133863.3 + phase: 0
(1104 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g14470 disease resistance protein, putative 565 e-161
At3g14460 disease resistance protein, putative 490 e-138
At3g50950 putative disease resistance protein 241 1e-63
At3g07040 disease resistance gene (RPM1) 240 4e-63
At1g59218 resistance protein RPP13, putative 231 1e-60
At1g59124 PRM1 homolog (RF45) 208 1e-53
At1g53350 hypothetical protein 204 2e-52
At1g58390 disease resistance protein PRM1 198 1e-50
At3g46730 disease resistance protein RPP8-like protein 198 2e-50
At5g48620 disease resistance protein 194 2e-49
At1g59780 hypothetical protein 189 6e-48
At1g10920 disease resistance protein RPM1 isolog 186 6e-47
At1g50180 disease resistance protein, putative 184 3e-46
At3g46530 disease resistance protein RPP13-like protein 172 1e-42
At1g59620 PRM1 homolog (CW9) 169 1e-41
At5g63020 NBS/LRR disease resistance like protein 168 1e-41
At1g61180 164 2e-40
At1g61300 similar to disease resistance protein 159 6e-39
At1g12290 hypothetical protein 159 1e-38
At1g12210 NBS/LRR disease resistance protein 158 1e-38
>At3g14470 disease resistance protein, putative
Length = 1054
Score = 565 bits (1455), Expect = e-161
Identities = 363/979 (37%), Positives = 553/979 (56%), Gaps = 78/979 (7%)
Query: 9 FLQSSFQVIFEKLASVDIRDYFSSKNVDD-LVKELNIALNSINHVLEEAEIKQYQIIYVK 67
FL + Q +F+ L S R +F + +++ L++ L+ AL +I VL +AE KQ V+
Sbjct: 8 FLAAFLQALFQTLVSEPFRSFFKRRELNENLLERLSTALLTITAVLIDAEEKQITNPVVE 67
Query: 68 KWLDKLKHVVYEADQLLDEISTDAMLNKLKAESEPLTT--NLLGVLGLAEGPSASNEGL- 124
KW+++L+ VVY A+ LD+I+T+A+ + AES L G + L + ++E L
Sbjct: 68 KWVNELRDVVYHAEDALDDIATEALRLNIGAESSSSNRLRQLRGRMSLGDFLDGNSEHLE 127
Query: 125 -------------------------VSWKPSKRLSSTALVDESSIYGRDVDKEELIKFLL 159
+ P +RL +T+LVDES ++GRD DK+E+++FL+
Sbjct: 128 TRLEKVTIRLERLASQRNILGLKELTAMIPKQRLPTTSLVDESEVFGRDDDKDEIMRFLI 187
Query: 160 AGNDSGTQVPIISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAI 219
N + +++IVG+GG+GKTTL++L+YN+ + +F K W +VSE +DV +TK +
Sbjct: 188 PENGKDNGITVVAIVGIGGVGKTTLSQLLYNDQHVRSYFGTKVWAHVSEEFDVFKITKKV 247
Query: 220 LKSFNPSADGEY--LDQLQHQLQHMLMGK--KYLLVLDDIWNGNVEYWEQLLLPFNHGSF 275
+S S E+ LD LQ +L+ L G +LLVLDD+WN N W+ L PF H +
Sbjct: 248 YESVT-SRPCEFTDLDVLQVKLKERLTGTGLPFLLVLDDLWNENFADWDLLRQPFIHAAQ 306
Query: 276 GSKIIVTTREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRK 335
GS+I+VTTR + VA ++ + + +L+ L DCW LF+ F + C ++ + +
Sbjct: 307 GSQILVTTRSQRVA-SIMCAVHVHNLQPLSDGDCWSLFMKTVFGNQEPCLNREIGDLAER 365
Query: 336 IMDKCEGLPLAIISLGQLLRKKFSQDEWMKILETDMWRLSDVDNKINPVLRLSYHNLPSD 395
I+ KC GLPLA+ +LG +LR + EW ++L + +W L + + PVLR+SY+ LP+
Sbjct: 366 IVHKCRGLPLAVKTLGGVLRFEGKVIEWERVLSSRIWDLPADKSNLLPVLRVSYYYLPAH 425
Query: 396 QKRCFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQ 455
KRCFA+CSIFPKG+ FEKD+++ LWMAEG L+ S K+ EE GNE F +LES S Q+
Sbjct: 426 LKRCFAYCSIFPKGHAFEKDKVVLLWMAEGFLQQTRSSKNLEELGNEYFSELESRSLLQK 485
Query: 456 SFDKTYGTYEHYVMYNLVNDLAKSVSGEFCMQIE-GARVEGSLERTRHIRFSLRSNCLNK 514
T Y+M++ +N+LA+ SGEF + E G +++ S ERTR++ + LR N
Sbjct: 486 -------TKTRYIMHDFINELAQFASGEFSSKFEDGCKLQVS-ERTRYLSY-LRDNYAEP 536
Query: 515 L-LETTCELKGLRSL----ILDVHRGTLISNNVQLDLFSRLNFLRTLSFRWCGLSEL-VD 568
+ E E+K LR+ + + R + V L L LR LS ++ L D
Sbjct: 537 MEFEALREVKFLRTFLPLSLTNSSRSCCLDQMVSEKLLPTLTRLRVLSLSHYKIARLPPD 596
Query: 569 EISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGC-ELTELPSNFSKLINLRHLEL 627
NI R+LDLS TE+ LP S+C +YNLQT+LL C L ELP++ S LINLR+L+L
Sbjct: 597 FFKNISHARFLDLSRTELEKLPKSLCYMYNLQTLLLSYCSSLKELPTDISNLINLRYLDL 656
Query: 628 --PYLKKMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPED 685
L++MP+ G+L SLQTL F V +GS + EL L+ LHGK+ I L V D D
Sbjct: 657 IGTKLRQMPRRFGRLKSLQTLTTFFVSASDGSRISELGGLHDLHGKLKIVELQRVVDVAD 716
Query: 686 AVTANLKDKKYLEELYMIFYDRKKEVDDSI----VESNVSVLEALQPNRSLKRLSISQYR 741
A ANL KK+L E+ ++ +++ ++ V E L+P+R +++L+I +Y+
Sbjct: 717 AAEANLNSKKHLREIDFVWRTGSSSSENNTNPHRTQNEAEVFEKLRPHRHIEKLAIERYK 776
Query: 742 GNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKIIGEELYGN 801
G RFP+W+ +V +++R C C+ LP LGQLP L+EL IS ++ IG + Y +
Sbjct: 777 GRRFPDWLSDPSFSRIVCIRLRECQYCTSLPSLGQLPCLKELHISGMVGLQSIGRKFYFS 836
Query: 802 NSKI-----DAFRSLEVLEFQRMENLEEWL-----CHEGFLSLKELTIKDCPKLKRALPQ 851
+ ++ FRSLE L F + + +EWL + F SLK+L I CP+L LP
Sbjct: 837 DQQLRDQDQQPFRSLETLRFDNLPDWQEWLDVRVTRGDLFPSLKKLFILRCPELTGTLPT 896
Query: 852 HLPSLQKLSIINCNKLEASMPEGD----NILELCLK-GCDSILIKELP--TSLKKLVL-- 902
LPSL L I C L+ + N+ L +K CD+++ L +L KL +
Sbjct: 897 FLPSLISLHIYKCGLLDFQPDHHEYSYRNLQTLSIKSSCDTLVKFPLNHFANLDKLEVDQ 956
Query: 903 CENRHT-EFFVEHILGNNA 920
C + ++ E EH+ G NA
Sbjct: 957 CTSLYSLELSNEHLRGPNA 975
Score = 35.0 bits (79), Expect = 0.24
Identities = 32/123 (26%), Positives = 58/123 (47%), Gaps = 8/123 (6%)
Query: 965 LFTNLHSLYLYNCPELVSFPEGGLPSNLSCFSIFDCPKLIASREEWGLFQLNSLKEFRVS 1024
LF +L L++ CPEL LPS +S I+ C L+ + + + +L+ +
Sbjct: 875 LFPSLKKLFILRCPELTGTLPTFLPSLIS-LHIYKC-GLLDFQPDHHEYSYRNLQTLSIK 932
Query: 1025 DEFENVESFPEENLLPPNLRILLLYKCSKLRIMNYKGFLHLL---SLSHLKIYNCPSLER 1081
+ + FP + NL L + +C+ L + HL +L +L+I +C +L+
Sbjct: 933 SSCDTLVKFPLNHF--ANLDKLEVDQCTSLYSLELSN-EHLRGPNALRNLRINDCQNLQL 989
Query: 1082 LPE 1084
LP+
Sbjct: 990 LPK 992
>At3g14460 disease resistance protein, putative
Length = 1424
Score = 490 bits (1262), Expect = e-138
Identities = 320/913 (35%), Positives = 497/913 (54%), Gaps = 67/913 (7%)
Query: 5 VAGAFLQSSFQVIFEKL-ASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQI 63
+A ++L S V+ E++ S ++ + K+ L+K L +AL + N VL +A+ + +
Sbjct: 1 MANSYLSSCANVMVERINTSQELVELCKGKSSSALLKRLKVALVTANPVLADADQRAEHV 60
Query: 64 IYVKKWLDKLKHVVYEADQLLDEISTDAMLNKLKAESEPLT---TNLLG----------- 109
VK WL +K ++A+ +LDE+ T+A+ ++ AE+ L NL+
Sbjct: 61 REVKHWLTGIKDAFFQAEDILDELQTEALRRRVVAEAGGLGGLFQNLMAGREAIQKKIEP 120
Query: 110 -----------------VLGLAEGPSASNEGLVSWKPSKRLSSTALVDESSIYGRDVDKE 152
V+GL E W+ + R L + + GR DK
Sbjct: 121 KMEKVVRLLEHHVKHIEVIGLKEYSETREP---QWRQASRSRPDDL-PQGRLVGRVEDKL 176
Query: 153 ELIKFLLAGNDSGTQVP-IISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESYD 211
L+ LL+ ++ P +IS+VG+ G+GKTTL ++V+N+ ++ EHFE+K W+ +++
Sbjct: 177 ALVNLLLSDDEISIGKPAVISVVGMPGVGKTTLTEIVFNDYRVTEHFEVKMWISAGINFN 236
Query: 212 VVGLTKAILKSFNPSA-DGEYLDQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPF 270
V +TKA+L+ SA + E L LQ QL+ L GK++LLVLDD W+ + WE + F
Sbjct: 237 VFTVTKAVLQDITSSAVNTEDLPSLQIQLKKTLSGKRFLLVLDDFWSESDSEWESFQVAF 296
Query: 271 NHGSFGSKIIVTTREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDY-PKL 329
GSKI++TTR E+ V K+ + ++ + +CW L AF SV +L
Sbjct: 297 TDAEEGSKIVLTTRS-EIVSTVAKAEKIYQMKLMTNEECWELISRFAFGNISVGSINQEL 355
Query: 330 ESIGRKIMDKCEGLPLAIISLGQLLRKKFSQDEWMKILETDMWRLSDVDNKINPVLRLSY 389
E IG++I ++C+GLPLA ++ LR K + D+W + + S N I PVL+LSY
Sbjct: 356 EGIGKRIAEQCKGLPLAARAIASHLRSKPNPDDWYAVSKN----FSSYTNSILPVLKLSY 411
Query: 390 HNLPSDQKRCFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDLES 449
+LP KRCFA CSIFPKG+ F+++EL+ LWMA LL S + E+ GN+ GDL +
Sbjct: 412 DSLPPQLKRCFALCSIFPKGHVFDREELVLLWMAIDLLYQPRSSRRLEDIGNDYLGDLVA 471
Query: 450 ISFFQQSFDKTYGTYEHYVMYNLVNDLAKSVSGEFCMQIEGARVEGSLERTRHIRFSLRS 509
SFFQ + T +VM++L+NDLAK+VSG+FC ++E + TRH FS RS
Sbjct: 472 QSFFQ----RLDITMTSFVMHDLMNDLAKAVSGDFCFRLEDDNIPEIPSTTRHFSFS-RS 526
Query: 510 NC-LNKLLETTCELKGLRSLILDVHRGTL----ISNNVQLDLFSRLNFLRTLSFRWCGLS 564
C + + C + LR+++ +L ++ V L + L+ LR LS ++
Sbjct: 527 QCDASVAFRSICGAEFLRTILPFNSPTSLESLQLTEKVLNPLLNALSGLRILSLSHYQIT 586
Query: 565 ELVDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGC-ELTELPSNFSKLINLR 623
L + +KLLRYLDLS T+I LP+ +C L NLQT+LL C +LT LP + ++LINLR
Sbjct: 587 NLPKSLKGLKLLRYLDLSSTKIKELPEFVCTLCNLQTLLLSNCRDLTSLPKSIAELINLR 646
Query: 624 HLEL--PYLKKMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVF 681
L+L L +MP I KL SLQ L FV+ +G+ L EL++L+HL G + I L V
Sbjct: 647 LLDLVGTPLVEMPPGIKKLRSLQKLSNFVIGRLSGAGLHELKELSHLRGTLRISELQNVA 706
Query: 682 DPEDAVTANLKDKKYLEELYMIFYDRKKEV----DDSIVESNVSVLEALQPNRSLKRLSI 737
+A A LK K +L+ L + + + +++ VL L+P+ LK I
Sbjct: 707 FASEAKDAGLKRKPFLDGLILKWTVKGSGFVPGSFNALACDQKEVLRMLEPHPHLKTFCI 766
Query: 738 SQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKIIGEE 797
Y+G FP W+ + S+ + C LC LPP+GQLPSL+ LSI ++ +G +
Sbjct: 767 ESYQGGAFPKWLGDSSFFGITSVTLSSCNLCISLPPVGQLPSLKYLSIEKFNILQKVGLD 826
Query: 798 LY--GNNSKIDAFRSLEVLEFQRMENLEEWLCHE----GFLSLKELTIKDCPKLKRALPQ 851
+ NNS+ F+SL++L+F M +EW+C E F L++L I+ CP L++ P+
Sbjct: 827 FFFGENNSRGVPFQSLQILKFYGMPRWDEWICPELEDGIFPCLQKLIIQRCPSLRKKFPE 886
Query: 852 HLPSLQKLSIINC 864
LPS +++I +C
Sbjct: 887 GLPSSTEVTISDC 899
Score = 98.2 bits (243), Expect = 2e-20
Identities = 88/292 (30%), Positives = 133/292 (45%), Gaps = 14/292 (4%)
Query: 809 RSLEVLEFQRMENLEEWLCHEGFLSLKELTIKDCPKLKRALPQHLPS-LQKLSIINCNKL 867
+SL + + +L E L E + +L EL I C L+ H P+ L+ L I +C KL
Sbjct: 1094 QSLHIDSCDGLTSLPENLT-ESYPNLHELLIIACHSLESFPGSHPPTTLKTLYIRDCKKL 1152
Query: 868 ---EASMPEGD-NILELCLKGCDSILIKELPTSL-KKLVLCENRHTEFFVEHILGNNAYL 922
E+ P + LE G + P SL KL R E F +
Sbjct: 1153 NFTESLQPTRSYSQLEYLFIGSSCSNLVNFPLSLFPKLRSLSIRDCESFKTFSIHAGLGD 1212
Query: 923 AELCLDLSGFVECPSLDLRCYNSLRTLSIIGWRSSS------LSFSLYLFTNLHSLYLYN 976
+ L+ +CP+L+ L T + S+ L L+ T+L SL++
Sbjct: 1213 DRIALESLEIRDCPNLETFPQGGLPTPKLSSMLLSNCKKLQALPEKLFGLTSLLSLFIIK 1272
Query: 977 CPELVSFPEGGLPSNLSCFSIFDCPKLIASREEWGLFQLNSLKEFRVSDEFENVESFPEE 1036
CPE+ + P GG PSNL I C KL R EWGL L +L+ + E++ESFPEE
Sbjct: 1273 CPEIETIPGGGFPSNLRTLCISLCDKL-TPRIEWGLRDLENLRNLEIDGGNEDIESFPEE 1331
Query: 1037 NLLPPNLRILLLYKCSKLRIMNYKGFLHLLSLSHLKIYNCPSLERLPEKGLP 1088
LLP ++ L + + L+ +N KGF ++ ++I C L+ ++ LP
Sbjct: 1332 GLLPKSVFSLRISRFENLKTLNRKGFHDTKAIETMEISGCDKLQISIDEDLP 1383
Score = 52.0 bits (123), Expect = 2e-06
Identities = 71/247 (28%), Positives = 109/247 (43%), Gaps = 37/247 (14%)
Query: 768 CSHLP--PLGQLPSLRELSISNCKRIKIIGEELYGNNSKIDAFRSLEVLEFQRMENLEEW 825
CS+L PL P LR LSI +C+ K + +I +LE LE + NLE +
Sbjct: 1176 CSNLVNFPLSLFPKLRSLSIRDCESFKTFSIHAGLGDDRI----ALESLEIRDCPNLETF 1231
Query: 826 LCHEGFL---SLKELTIKDCPKLKRALPQH---LPSLQKLSIINCNKLEASMPEG---DN 876
+G L L + + +C KL +ALP+ L SL L II C ++E ++P G N
Sbjct: 1232 --PQGGLPTPKLSSMLLSNCKKL-QALPEKLFGLTSLLSLFIIKCPEIE-TIPGGGFPSN 1287
Query: 877 ILELCLKGCDSILIKELPTSLKKLVLCENRHTEFFVEHILGNNAYLAELCLDLSGFVECP 936
+ LC+ CD L + L+ L N + E I ++ E L S F
Sbjct: 1288 LRTLCISLCDK-LTPRIEWGLRDLENLRNLEIDGGNEDI---ESFPEEGLLPKSVF---- 1339
Query: 937 SLDLRCYNSLRTLSIIGWRSSSLSFSLYLFTNLHSLYLYNCPELVSFPEGGLPSNLSCFS 996
SL + + +L+TL+ G+ + + ++ + C +L + LP LSC
Sbjct: 1340 SLRISRFENLKTLNRKGFHDTKA---------IETMEISGCDKLQISIDEDLPP-LSCLR 1389
Query: 997 IFDCPKL 1003
I C L
Sbjct: 1390 ISSCSLL 1396
Score = 41.2 bits (95), Expect = 0.003
Identities = 39/142 (27%), Positives = 65/142 (45%), Gaps = 23/142 (16%)
Query: 754 LPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRI------------KIIGEELYGN 801
L +L+SL + C +P G +LR L IS C ++ + E+ G
Sbjct: 1262 LTSLLSLFIIKCPEIETIPGGGFPSNLRTLCISLCDKLTPRIEWGLRDLENLRNLEIDGG 1321
Query: 802 NSKIDAF-------RSLEVLEFQRMENLEEWLCHEGF---LSLKELTIKDCPKLKRALPQ 851
N I++F +S+ L R ENL+ L +GF +++ + I C KL+ ++ +
Sbjct: 1322 NEDIESFPEEGLLPKSVFSLRISRFENLKT-LNRKGFHDTKAIETMEISGCDKLQISIDE 1380
Query: 852 HLPSLQKLSIINCNKLEASMPE 873
LP L L I +C+ L + E
Sbjct: 1381 DLPPLSCLRISSCSLLTETFAE 1402
Score = 38.5 bits (88), Expect = 0.021
Identities = 37/122 (30%), Positives = 59/122 (48%), Gaps = 13/122 (10%)
Query: 757 LVSLQMRHCGLCSHLPPLG-QLPSLRELSISNCKRIKIIGEELYGNNSKIDAFRSLEVLE 815
L SL++R C P G P L + +SNCK+++ + E+L+G S + SL +++
Sbjct: 1217 LESLEIRDCPNLETFPQGGLPTPKLSSMLLSNCKKLQALPEKLFGLTSLL----SLFIIK 1272
Query: 816 FQRMENLEEWLCHEGFLS-LKELTIKDCPKLKRALP---QHLPSLQKLSIINCNKLEASM 871
+E + GF S L+ L I C KL + + L +L+ L I N+ S
Sbjct: 1273 CPEIETIP----GGGFPSNLRTLCISLCDKLTPRIEWGLRDLENLRNLEIDGGNEDIESF 1328
Query: 872 PE 873
PE
Sbjct: 1329 PE 1330
>At3g50950 putative disease resistance protein
Length = 852
Score = 241 bits (616), Expect = 1e-63
Identities = 235/815 (28%), Positives = 385/815 (46%), Gaps = 88/815 (10%)
Query: 1 MAELVAGAFLQSSFQVIFEKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQ 60
M + V FL+ + ++ EK +V DY K ++DL EL + L++AE ++
Sbjct: 1 MVDAVVTVFLEKTLNILEEKGRTVS--DY--RKQLEDLQSELKY----MQSFLKDAERQK 52
Query: 61 YQIIYVKKWLDKLKHVVYEADQLL------------DEISTDAMLNKLKAESEPL----- 103
++ + L+ +VYEA+ +L ++ S++A L++L PL
Sbjct: 53 RTNETLRTLVADLRELVYEAEDILVDCQLADGDDGNEQRSSNAWLSRLHPARVPLQYKKS 112
Query: 104 ----------TTNLLGVLGLAEGPSASNEGLVSWKPSKRLSSTALVDESSIYGRDVDKEE 153
T V E + SN G + + R SS + D + + G + DK +
Sbjct: 113 KRLQEINERITKIKSQVEPYFEFITPSNVGRDNG--TDRWSSP-VYDHTQVVGLEGDKRK 169
Query: 154 LIKFLLAGNDSGTQVPIISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESYDVV 213
+ ++L NDS Q+ I++ VG+GG+GKTT+A+ V+N+ +IE FE + WV VS+++
Sbjct: 170 IKEWLFRSNDS--QLLIMAFVGMGGLGKTTIAQEVFNDKEIEHRFERRIWVSVSQTFTEE 227
Query: 214 GLTKAILKSFNPSADGEYLDQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHG 273
+ ++IL++ ++ G+ + L ++Q L+GK+YL+V+DD+W+ N+ +W+++ G
Sbjct: 228 QIMRSILRNLGDASVGDDIGTLLRKIQQYLLGKRYLIVMDDVWDKNLSWWDKIYQGLPRG 287
Query: 274 SFGSKIIVTTREKEVAYHVVKSTMLCDLRQLVKSD-CWRLFVTHAFQGK-SVCDYPKLES 331
GS +IVTTR + VA V +L+ D W LF AF C+ P+LE
Sbjct: 288 QGGS-VIVTTRSESVAKRVQARDDKTHRPELLSPDNSWLLFCNVAFAANDGTCERPELED 346
Query: 332 IGRKIMDKCEGLPLAIISLGQLLR-KKFSQDEWMKILETDMWRL----SDVDNKINPVLR 386
+G++I+ KC+GLPL I ++G LL K EW +I E L S+ DN ++ L+
Sbjct: 347 VGKEIVTKCKGLPLTIKAVGGLLLCKDHVYHEWRRIAEHFQDELRGNTSETDNVMSS-LQ 405
Query: 387 LSYHNLPSDQKRCFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGD 446
LSY LPS K C S++P+ K +L+ W+ EG + +S E G + F
Sbjct: 406 LSYDELPSHLKSCILTLSLYPEDCVIPKQQLVHGWIGEGFVMWRNG-RSATESGEDCFSG 464
Query: 447 LESISFFQQSFDKTY-GTYEHYVMYNLVNDLAKSVSGEFCMQIEGARVEGSLERTRHIRF 505
L + + DKTY GT ++++V DL ++ + + EG RH+
Sbjct: 465 LTNRCLI-EVVDKTYSGTIITCKIHDMVRDLVIDIAKKDSF----SNPEGL--NCRHLGI 517
Query: 506 SLRSNCLNKLLETTCELKGLRSLILDVHRGTLISNNVQLDLFSRLNFLRTL----SFRWC 561
S N K ++ +L+G+ S L N+ F+ +LR L S
Sbjct: 518 S--GNFDEKQIKVNHKLRGVVSTTKTGEVNKL--NSDLAKKFTDCKYLRVLDISKSIFDA 573
Query: 562 GLSELVDEISNIKLLRYLDLSFTE-ITSLPDSICMLYNLQTILLQGC----ELTELPSNF 616
LSE++DEI++++ L L LS T + P S+ L+NLQ + C +L F
Sbjct: 574 PLSEILDEIASLQHLACLSLSNTHPLIQFPRSMEDLHNLQILDASYCQNLKQLQPCIVLF 633
Query: 617 SKLINLRHLELPYLKKMPKHIGKLNSLQTLPYF-VVEEKNGSDLKELEKLNHLHGKICID 675
KL+ L L+ PK IG L L+ L F NG L E++ L +L
Sbjct: 634 KKLLVLDMTNCGSLECFPKGIGSLVKLEVLLGFKPARSNNGCKLSEVKNLTNLR------ 687
Query: 676 GLGYVFDPEDAVTANLKDKKY-LEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKR 734
LG D + D L +L I + DS + ++ ++AL P L
Sbjct: 688 KLGLSLTRGDQIEEEELDSLINLSKLMSISIN----CYDSYGDDLITKIDALTPPHQLHE 743
Query: 735 LSISQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCS 769
LS+ Y G P+W+ LP +R+ +CS
Sbjct: 744 LSLQFYPGKSSPSWLSPHKLP-----MLRYMSICS 773
>At3g07040 disease resistance gene (RPM1)
Length = 926
Score = 240 bits (612), Expect = 4e-63
Identities = 212/772 (27%), Positives = 374/772 (47%), Gaps = 94/772 (12%)
Query: 135 STALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLGGMGKTTLAKLVYNNNKI 194
S+ E+S+ G D K +LI LL+ Q ++++VG+GG GKTTL+ ++ + +
Sbjct: 163 SSLFFSENSLVGIDAPKGKLIGRLLSPEP---QRIVVAVVGMGGSGKTTLSANIFKSQSV 219
Query: 195 EEHFELKAWVYVSESYDVVGLTKAILKSFNPSADGEY--------LDQLQHQLQHMLMGK 246
HFE AWV +S+SY + + + ++K F AD + +L +L L K
Sbjct: 220 RRHFESYAWVTISKSYVIEDVFRTMIKEFYKEADTQIPAELYSLGYRELVEKLVEYLQSK 279
Query: 247 KYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAY--HVVKSTMLCDLRQL 304
+Y++VLDD+W + W ++ + G +GS++++TTR+ VA + + ST ++ L
Sbjct: 280 RYIVVLDDVWTTGL--WREISIALPDGIYGSRVMMTTRDMNVASFPYGIGSTK-HEIELL 336
Query: 305 VKSDCWRLFVTHAFQGK-SVCDYPKLESIGRKIMDKCEGLPLAIISLGQLLRKKFSQDEW 363
+ + W LF AF C LE I RK++++C+GLPLAI SLG ++ K + EW
Sbjct: 337 KEDEAWVLFSNKAFPASLEQCRTQNLEPIARKLVERCQGLPLAIASLGSMMSTKKFESEW 396
Query: 364 MKILETDMWRLSDVDNK--INPVLRLSYHNLPSDQKRCFAFCSIFPKGYTFEKDELIKLW 421
K+ T W L++ + ++ LS+++LP KRCF +CS+FP Y ++ LI++W
Sbjct: 397 KKVYSTLNWELNNNHELKIVRSIMFLSFNDLPYPLKRCFLYCSLFPVNYRMKRKRLIRMW 456
Query: 422 MAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQSFDKTYGTYEHYVMYNLVNDLAKSVS 481
MA+ ++ K+ EE + +L + Q +G + + M++++ ++A SVS
Sbjct: 457 MAQRFVEPIRGVKA-EEVADSYLNELVYRNMLQVILWNPFGRPKAFKMHDVIWEIALSVS 515
Query: 482 --GEFCMQIEGARVEGSLERTRHIRFSLRSNCLNK-LLETTCELKGLRSLILDVHRGTLI 538
FC + +G + R C+ K + + L SL++
Sbjct: 516 KLERFC-DVYNDDSDGDDAAETMENYGSRHLCIQKEMTPDSIRATNLHSLLV------CS 568
Query: 539 SNNVQLDLFSRLNFLRTLSFRWCGLSELVDEISNIKLLRYLDLSFTEITSLPDSICMLYN 598
S +++L LN LR L +S+L D + + L+YL+LS T++ LP + L N
Sbjct: 569 SAKHKMELLPSLNLLRALDLEDSSISKLPDCLVTMFNLKYLNLSKTQVKELPKNFHKLVN 628
Query: 599 LQTILLQGCELTELPSNFSKLINLRHL-----------ELPYL---KKMPKHIGKLNSLQ 644
L+T+ + ++ ELP KL LR+L Y+ + +PK I +L LQ
Sbjct: 629 LETLNTKHSKIEELPLGMWKLKKLRYLITFRRNEGHDSNWNYVLGTRVVPK-IWQLKDLQ 687
Query: 645 TLPYFVVEE---KNGSDLKELEKLNHL-----HGKICIDGLGYVFDPEDAVTANLKDKKY 696
+ F E+ KN + +L +++ + HG+ D L +K ++
Sbjct: 688 VMDCFNAEDELIKNLGCMTQLTRISLVMVRREHGRDLCDSLN-----------KIKRIRF 736
Query: 697 LEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYRGNRFPNWIRGCHLPN 756
L L I + E+DD I + S+++L ++ + R P+W L N
Sbjct: 737 L-SLTSIDEEEPLEIDDLIATA------------SIEKLFLAG-KLERVPSWFN--TLQN 780
Query: 757 LVSLQMRHCGLCSH-LPPLGQLPSLRELSISNCKRIKIIGEELYGNNSKIDAFRSLEVLE 815
L L +R L + + + LP L LS N +G L F++L++LE
Sbjct: 781 LTYLGLRGSQLQENAILSIQTLPRLVWLSFYNA----YMGPRL----RFAQGFQNLKILE 832
Query: 816 FQRMENLEEWLCHEG-FLSLKELTIKDCPKLK---RALPQHLPSLQKLSIIN 863
+M++L E + +G L++L ++ C L+ R + ++L +LQ+L +I+
Sbjct: 833 IVQMKHLTEVVIEDGAMFELQKLYVRACRGLEYVPRGI-ENLINLQELHLIH 883
>At1g59218 resistance protein RPP13, putative
Length = 1049
Score = 231 bits (590), Expect = 1e-60
Identities = 263/1034 (25%), Positives = 452/1034 (43%), Gaps = 128/1034 (12%)
Query: 33 KNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVYEADQLLDEISTDAM 92
+ V+D V EL LN ++ L++A+ K++ VK ++++K ++Y+ + ++ +
Sbjct: 25 QGVEDQVTELKRDLNLLSSFLKDADAKKHTSAVVKNCVEEIKEIIYDGEDTIETFVLEQN 84
Query: 93 LNKLKAESEPLTT----------NLLGVLGLAEGPSASNEGLVSW--------------- 127
L K + + LG+ GL+ S + S+
Sbjct: 85 LGKTSGIKKSIRRLACIIPDRRRYALGIGGLSNRISKVIRDMQSFGVQQAIVDGGYKQPQ 144
Query: 128 --KPSKRLSSTALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLGGMGKTTLA 185
K + + D+S G + + ++L+ +L+ V ++SI G+GG+GKTTLA
Sbjct: 145 GDKQREMRPRFSKDDDSDFVGLEANVKKLVGYLV----DEANVQVVSITGMGGLGKTTLA 200
Query: 186 KLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFNPSADGEYL-----DQLQHQLQ 240
K V+N+ ++ F+ +WV VS+ + + + + IL+ P + + + D LQ +L
Sbjct: 201 KQVFNHEDVKHQFDGLSWVCVSQDFTRMNVWQKILRDLKPKEEEKKIMEMTQDTLQGELI 260
Query: 241 HMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHVVKSTMLCD 300
+L K L+VLDDIW E WE L+ P + G K+++T+R + VA S +
Sbjct: 261 RLLETSKSLIVLDDIW--EKEDWE-LIKPIFPPTKGWKVLLTSRNESVAMRRNTSYINFK 317
Query: 301 LRQLVKSDCWRLFVTHAFQGKSVCDY---PKLESIGRKIMDKCEGLPLAIISLGQLLRKK 357
L D W LF A K ++ + E +G+ ++ C GLPLAI LG +L +K
Sbjct: 318 PECLTTEDSWTLFQRIALPMKDAAEFKIDEEKEELGKLMIKHCGGLPLAIRVLGGMLAEK 377
Query: 358 FSQDEWMKILETDMWRL--------SDVDNKINPVLRLSYHNLPSDQKRCFAFCSIFPKG 409
++ +W ++ E L D +N N VL LS+ LPS K CF + + FP
Sbjct: 378 YTSHDWRRLSENIGSHLVGGRTNFNDDNNNTCNYVLSLSFEELPSYLKHCFLYLAHFPDD 437
Query: 410 YTFEKDELIKLWMAEGLLKCCGSYKSE--EEFGNEIFGDLESISFFQQSFDKTYGTYEHY 467
Y L W AEG+ + Y E + G+ +L + D +E
Sbjct: 438 YEINVKNLSYYWAAEGIFQ-PRHYDGEIIRDVGDVYIEELVRRNMVISERDVKTSRFETC 496
Query: 468 VMYNLVND--LAKSVSGEFCMQIEGARVEG----SLERTRHIRFSLRSNCLNKLLETTCE 521
+++++ + L K+ F +QI +R S+ +R + + + +E
Sbjct: 497 HLHDMMREVCLLKAKEENF-LQITSSRTSTGNSLSIVTSRRLVYQYP---ITLDVEKDIN 552
Query: 522 LKGLRSLILDVHRGTLISNNVQLDL---FSRLNFLRTLSFRWCGL--SELVDEISNIKLL 576
LRSL++ + + L F RL LR L L +L I + L
Sbjct: 553 DPKLRSLVVVANTYMFWGGWSWMLLGSSFIRLELLRVLDIHRAKLKGGKLASSIGQLIHL 612
Query: 577 RYLDLSFTEITSLPDSI----CMLYNLQTILLQGCELTELPSNFSKLINLRHLELP--YL 630
RYL+L E+T +P S+ ++Y IL+ G L +P+ ++ LR+L LP
Sbjct: 613 RYLNLKHAEVTHIPYSLGNLKLLIYLNLVILVSGSTL--VPNVLKEMQQLRYLALPKDMG 670
Query: 631 KKMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPEDAVTAN 690
+K + L L+TL F + + DL+ + +L L ++ + + + A+
Sbjct: 671 RKTKLELSNLVKLETLKNFSTKNCSLEDLRGMVRLRTLTIELRKE------TSLETLAAS 724
Query: 691 LKDKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYRGNRFPNWIR 750
+ KYLE L + + ++ + + L+ L + RLS Q+ FP+
Sbjct: 725 IGGLKYLESLTITDLGSEMRTKEAGIVFDFVYLKTLTLKLYMPRLSKEQH----FPS--- 777
Query: 751 GCHLPNLVSLQMRHCGLCSH-LPPLGQLPSLRELSISNCKRIKIIGEELYGNNSKIDAFR 809
+L +L ++HC L +P L +L L+EL + +R G+E+ ++ F
Sbjct: 778 -----HLTTLYLQHCRLEEDPMPILEKLHQLKELEL---RRKSFSGKEMVCSSG---GFP 826
Query: 810 SLEVLEFQRMENLEEWLCHEGFLS-LKELTIKDCPKLKRALPQHLPS-LQKLSIINCNKL 867
L+ L + +E E+W E + L L I+DC KLK+ +HLPS L +S+ C
Sbjct: 827 QLQKLSIKGLEEWEDWKVEESSMPVLHTLDIRDCRKLKQLPDEHLPSHLTSISLFFCCLE 886
Query: 868 EASMPEGDNILEL--------CLKGCDSILIKELPTSLKKLVLCE-NRHTEFFVEHILGN 918
E MP + ++ L G + L KL L E + E+ VE
Sbjct: 887 EDPMPTLERLVHLKELQLLFRSFSGRIMVCAGSGFPQLHKLKLSELDGLEEWIVEDGSMP 946
Query: 919 NAYLAEL--CLDL----SGFVECPSLDLRCYNSLRTLSIIGWRSSSLSFSLYLFTNLHSL 972
+ E+ C L +GF + +L+L N L W S L LH+L
Sbjct: 947 QLHTLEIRRCPKLKKLPNGFPQLQNLEL---NELEEWE--EWIVEDGSMPL-----LHTL 996
Query: 973 YLYNCPELVSFPEG 986
++NCP+L P+G
Sbjct: 997 RIWNCPKLKQLPDG 1010
>At1g59124 PRM1 homolog (RF45)
Length = 1155
Score = 208 bits (530), Expect = 1e-53
Identities = 236/938 (25%), Positives = 404/938 (42%), Gaps = 130/938 (13%)
Query: 33 KNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVYEADQLLDEISTDAM 92
+ V+D V EL LN ++ L++A K++ VK ++++K ++Y+ + ++ +
Sbjct: 25 QGVEDQVTELKRDLNMLSSFLKDANAKKHTSAVVKNCVEEIKEIIYDGEDTIETFVLEQN 84
Query: 93 LNKLKAESEPLTT----------NLLGVLGLAEGPSASNEGLVSW--------------- 127
L K + + LG+ GL+ S + S+
Sbjct: 85 LGKTSGIKKSIRRLACIIPDRRRYALGIGGLSNRISKVIRDMQSFGVQQAIVDGGYKQPQ 144
Query: 128 --KPSKRLSSTALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLGGMGKTTLA 185
K + + D+S G + + ++L+ +L+ V ++SI G+GG+GKTTLA
Sbjct: 145 GDKQREMRQKFSKDDDSDFVGLEANVKKLVGYLV----DEANVQVVSITGMGGLGKTTLA 200
Query: 186 KLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFNPSADGEYL-----DQLQHQLQ 240
K V+N+ ++ F+ +WV VS+ + + + + IL+ P + + + D LQ +L
Sbjct: 201 KQVFNHEDVKHQFDGLSWVCVSQDFTRMNVWQKILRDLKPKEEEKKIMEMTQDTLQGELI 260
Query: 241 HMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHVVKSTMLCD 300
+L K L+VLDDIW E WE L+ P + G K+++T+R + VA S +
Sbjct: 261 RLLETSKSLIVLDDIWEK--EDWE-LIKPIFPPTKGWKVLLTSRNESVAMRRNTSYINFK 317
Query: 301 LRQLVKSDCWRLFVTHAFQGKSVCDYP---KLESIGRKIMDKCEGLPLAIISLGQLLRKK 357
L D W LF A K ++ + E +G+ ++ C GLPLAI LG +L +K
Sbjct: 318 PECLTTEDSWTLFQRIALPMKDAAEFKIDEEKEELGKLMIKHCGGLPLAIRVLGGMLAEK 377
Query: 358 FSQDEWMKILETDMWRL--------SDVDNKINPVLRLSYHNLPSDQKRCFAFCSIFPKG 409
++ +W ++ E L D +N N VL LS+ LPS K CF + + FP+
Sbjct: 378 YTSHDWRRLSENIGSHLVGGRTNFNDDNNNTCNNVLSLSFEELPSYLKHCFLYLAHFPED 437
Query: 410 YTFEKDELIKLWMAEGLLKCCGSYKSE--EEFGNEIFGDLESISFFQQSFDKTYGTYEHY 467
Y + + L W AEG+ + Y E + G+ +L + D +E
Sbjct: 438 YEIKVENLSYYWAAEGIFQP-RHYDGETIRDVGDVYIEELVRRNMVISERDVKTSRFETC 496
Query: 468 VMYNLVND--LAKSVSGEFCMQIEGARVE-GSLERTRHIRFSLRSNCLNKLLETTCELKG 524
+++++ + L K+ F +QI +R +L+ T R + +E
Sbjct: 497 HLHDMMREVCLLKAKEENF-LQITSSRPSTANLQSTVTSRRFVYQYPTTLHVEKDINNPK 555
Query: 525 LRSLILDVHRGTLISNNVQLDLFSRLNFLRTLSFRWCGLS--ELVDEISNIKLLRYLDLS 582
LR+L++ TL S N+ F+RL LR L + +L I + LRYL L
Sbjct: 556 LRALVVV----TLGSWNLAGSSFTRLELLRVLDLIEVKIKGGKLASCIGKLIHLRYLSLE 611
Query: 583 FTEITSLPDSICMLYNLQTILLQGC-ELTELPSNFSKLINLRHLELP--YLKKMPKHIGK 639
+ E+T +P S+ L L + L T +P+ + LR+L LP +K +
Sbjct: 612 YAEVTHIPYSLGNLKLLIYLNLASFGRSTFVPNVLMGMQELRYLALPSDMGRKTKLELSN 671
Query: 640 LNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPEDAVTANLKDKKYLEE 699
L L+TL F E + DL + +L+ L+ K+ + + + A++ KYLE+
Sbjct: 672 LVKLETLENFSTENSSLEDLCGMVRLSTLNIKLIEE------TSLETLAASIGGLKYLEK 725
Query: 700 LYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYRGNRFPN-WIRGCHLPN-- 756
L + + + ++ + + L+ L + RLS Q+ + ++ C L
Sbjct: 726 LEIYDHGSEMRTKEAGIVFDFVHLKRLWLKLYMPRLSTEQHFPSHLTTLYLESCRLEEDP 785
Query: 757 ------LVSLQMRHCG---------LCSH--LPPLGQL------------------PSLR 781
L+ L+ G +CS P L +L P LR
Sbjct: 786 MPILEKLLQLKELELGFESFSGKKMVCSSGGFPQLQRLSLLKLEEWEDWKVEESSMPLLR 845
Query: 782 ELSISNCKRIKIIGEELYGNNSKID--------------AFRSLEVLEFQRMENLEEWLC 827
L I + +G +Y ++ F L+ L R+E EEW+
Sbjct: 846 TLDIQK-DPLPTLGRLVYLKELQLGFRTFSGRIMVCSGGGFPQLQKLSIYRLEEWEEWIV 904
Query: 828 HEGFLS-LKELTIKDCPKLKRALP---QHLPSLQKLSI 861
+G + L L I DCPKLK+ LP Q + SL+ L I
Sbjct: 905 EQGSMPFLHTLYIDDCPKLKK-LPDGLQFIYSLKNLKI 941
>At1g53350 hypothetical protein
Length = 906
Score = 204 bits (520), Expect = 2e-52
Identities = 230/936 (24%), Positives = 409/936 (43%), Gaps = 107/936 (11%)
Query: 19 EKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVY 78
EKL + R+ +D+ V L L + +L++A+ K+ + V+ +L+ +K +VY
Sbjct: 11 EKLWELLSRESARLNGIDEQVDGLKRQLGRLQSLLKDADAKKNETERVRNFLEDVKDIVY 70
Query: 79 EADQLLDEISTDAMLNKLK--------------------AESEPLTTNL---------LG 109
+AD +++ + + K K ++ E +T + LG
Sbjct: 71 DADDIIESFLLNELRGKEKGIKKQVRTLACFLVDRRKFASDIEGITKRISEVIVGMQSLG 130
Query: 110 VLGLAEGPSASNEGLVSWKPSKRLSST-ALVDESSIYGRDVDKEELIKFLLAGNDSGTQV 168
+ +A+G S L + + + T + ES + G D EEL+ L+ NDS V
Sbjct: 131 IQHIADGGGRS---LSLQERQREIRQTFSRNSESDLVGLDQSVEELVDHLVE-NDS---V 183
Query: 169 PIISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFNPSAD 228
++S+ G+GG+GKTTLA+ V++++ + HF+ +WV VS+ + + + IL+ P +
Sbjct: 184 QVVSVSGMGGIGKTTLARQVFHHDIVRRHFDGFSWVCVSQQFTRKDVWQRILQDLRPYDE 243
Query: 229 G-----EYLDQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTT 283
G EY LQ +L +L +YLLVLDD+W E W+++ F H G K+++T+
Sbjct: 244 GIIQMDEY--TLQGELFELLESGRYLLVLDDVW--KEEDWDRIKAVFPH-KRGWKMLLTS 298
Query: 284 REKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCEGL 343
R + + H + R L W+LF + ++ E++G++++ C GL
Sbjct: 299 RNEGLGLHADPTCFAFRPRILTPEQSWKLFERIVSSRRDKTEFKVDEAMGKEMVTYCGGL 358
Query: 344 PLAIISLGQLLRKKFSQDEWMKILETDMWRL-------SDVDNKINPVLRLSYHNLPSDQ 396
PLA+ LG LL KK + EW ++ + + D N + VL LSY +LP
Sbjct: 359 PLAVKVLGGLLAKKHTVLEWKRVHSNIVTHIVGKSGLSDDNSNSVYRVLSLSYEDLPMQL 418
Query: 397 KRCFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQS 456
K CF + + FP+ Y + L W+AEG++ + ++ G +L +
Sbjct: 419 KHCFFYLAHFPEDYKIDVKILFNYWVAEGIITPFHDGSTIQDTGESYLEELVRRNMVVVE 478
Query: 457 FDKTYGTYEHYVMYNLVNDLAKSVSGEFCMQIEGARVEGSLERT-------RHIRFSLRS 509
E+ M++++ ++ S + E I +V + T R R L S
Sbjct: 479 ESYLTSRIEYCQMHDMMREVCLSKAKEENF-IRVVKVPTTTSTTINAQSPCRSRRLVLHS 537
Query: 510 NCLNKLLETTCELKGLRSLILDVHRGTLISNNVQLDLFSRLNFLRTLSFRWCGL--SELV 567
+L K LI V F L LR L + +L
Sbjct: 538 GNALHMLGHKDNKKARSVLIFGVEEKFWKPRG-----FQCLPLLRVLDLSYVQFEGGKLP 592
Query: 568 DEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCE--LTELPSNFSKLINLRHL 625
I ++ LR+L L ++ LP S+ L L + L + L +P+ ++ LR+L
Sbjct: 593 SSIGDLIHLRFLSLYEAGVSHLPSSLGNLKLLLCLNLGVADRLLVHVPNVLKEMQELRYL 652
Query: 626 ELPYLKKMPK----HIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVF 681
LP + MP +G L +L++L F + + +DL + KL+ L+ +F
Sbjct: 653 RLP--RSMPAKTKLELGDLVNLESLTNFSTKHGSVTDLLRMTKLSVLN---------VIF 701
Query: 682 DPEDAVTANLKDKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYR 741
E L + L L + + ++V + + VL+ + LK L++S +
Sbjct: 702 SGECTFETLLLSLRELRNLETLSFHDFQKVSVANHGGELLVLDFIH----LKDLTLSMHL 757
Query: 742 GNRFPNWIRGCHLPNLVSLQMRHCGLCSH-LPPLGQLPSLRELSISNCKRIKIIGEELYG 800
RFP+ R P+L + + C + +P L +L L+ + +S+ +G +
Sbjct: 758 -PRFPDQYR--FPPHLAHIWLIGCRMEEDPMPILEKLLHLKSVYLSSG---AFLGRRMVC 811
Query: 801 NNSKIDAFRSLEVLEFQRMENLEEWLCHEGFL-SLKELTIKDCPKLKRALPQHLPSLQKL 859
+ F L L+ + L EW EG + L+ LTI +C KLK+ LP L + L
Sbjct: 812 SKG---GFPQLLALKMSYKKELVEWRVEEGSMPCLRTLTIDNCKKLKQ-LPDGLKYVTCL 867
Query: 860 SIINCNKLEASMPEGDNILELCLKGCDSILIKELPT 895
+ +++ E L + G D ++ +P+
Sbjct: 868 KELKIERMKREWTE-----RLVIGGEDYYKVQHIPS 898
>At1g58390 disease resistance protein PRM1
Length = 907
Score = 198 bits (504), Expect = 1e-50
Identities = 217/888 (24%), Positives = 395/888 (44%), Gaps = 97/888 (10%)
Query: 33 KNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVYEADQLLDEI----- 87
+ V+D V L LN ++ L++A+ K++ V+ ++++K +VY+A+ +++
Sbjct: 25 QGVEDQVTGLKRDLNLLSSFLKDADAKKHTTAVVRNVVEEIKEIVYDAEDIIETYLLKEK 84
Query: 88 --STDAMLNKLKAESEPLTT---NLLGVLGLAEGPSASNEGLVSWKPSKRLSSTALVD-- 140
T + +++ + ++ N L V G+ S + S+ + + +
Sbjct: 85 LWKTSGIKMRIRRHACIISDRRRNALDVGGIRTRISDVIRDMQSFGVQQAIVDGGYMQPQ 144
Query: 141 ---------------ESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLGGMGKTTLA 185
ES G +V+ ++L+ +L+ + V ++SI G+GG+GKTTLA
Sbjct: 145 GDRQREMRQTFSKDYESDFVGLEVNVKKLVGYLVDEEN----VQVVSITGMGGLGKTTLA 200
Query: 186 KLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFNPSADGEYLDQLQH-----QLQ 240
+ V+N+ ++ F+ AWV VS+ + + + IL++ + + Q++ +L
Sbjct: 201 RQVFNHEDVKHQFDRLAWVCVSQEFTRKNVWQMILQNLTSREKKDEILQMEEAELHDKLF 260
Query: 241 HMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHVVKSTMLCD 300
+L K L+V DDIW E W+ L+ P + G K+++T++ + VA +
Sbjct: 261 QLLETSKSLIVFDDIWKD--EDWD-LIKPIFPPNKGWKVLLTSQNESVAVRGDIKYLNFK 317
Query: 301 LRQLVKSDCWRLFVTHAFQGKSVCDY---PKLESIGRKIMDKCEGLPLAIISLGQLLRKK 357
L D W LF AF K + ++E +G++++ C GLPLAI LG LL K
Sbjct: 318 PECLAIEDSWTLFQRIAFPKKDASESKVDEEMEDMGKQMLKHCGGLPLAIKVLGGLLAAK 377
Query: 358 FSQDEWMKI---LETD-MWRLSDVDNKINPVLRLSYHNLPSDQKRCFAFCSIFPKGYTFE 413
++ +W ++ + +D + R S ++ I VL +S+ LPS K CF + + FP+ +
Sbjct: 378 YTMHDWERLSVNIGSDIVGRTSSNNSSIYHVLSMSFEELPSYLKHCFLYLAHFPEDHKIN 437
Query: 414 KDELIKLWMAEGLLKCCGSYKSE--EEFGNEIFGDLESISFFQQSFDKTYGTYEHYVMYN 471
++L W AEG+ + E ++ G +L + D T + +++
Sbjct: 438 VEKLSYCWAAEGISTAEDYHNGETIQDVGQSYLEELVRRNMIIWERDATASRFGTCHLHD 497
Query: 472 LVNDLAK-SVSGEFCMQIEGARVEGSLERTRHIRFSLRSNCLNKLLETTCELK------G 524
++ ++ E +QI V + T + + RS L TT ++
Sbjct: 498 MMREVCLFKAKEENFLQIAVKSVGVTSSSTGNSQSPCRSRRLVYQCPTTLHVERDINNPK 557
Query: 525 LRSLILDVHRGTLISNNVQLDLFSRLNFLRTLSFRWCGLS--ELVDEISNIKLLRYLDLS 582
LRSL++ H + + + F+RL LR L + +L I N+ LRYL L
Sbjct: 558 LRSLVVLWHDLWVENWKLLGTSFTRLKLLRVLDLFYVDFEGMKLPFGIGNLIHLRYLSLQ 617
Query: 583 FTEITSLPDSICMLYNLQTILLQ-GCELTELPSNFSKLINLRHLELP--YLKKMPKHIGK 639
+++ LP S+ L L + L E +P F ++ LR+L+LP KK +
Sbjct: 618 DAKVSHLPSSLGNLMLLIYLNLDVDTEFIFVPDVFMRMHELRYLKLPLHMHKKTRLSLRN 677
Query: 640 LNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPEDAVTANLKDKKYLEE 699
L L+TL YF + DL + +L L ++ + ++A++ + LE
Sbjct: 678 LVKLETLVYFSTWHSSSKDLCGMTRLMTLAIRLT------RVTSTETLSASISGLRNLEY 731
Query: 700 LYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYRGNRFPNWIRGCHLPNLVS 759
LY++ KK ++ IV + + L L + R FP+ L
Sbjct: 732 LYIVGTHSKKMREEGIVLDFIHLKHLL--------LDLYMPRQQHFPS--------RLTF 775
Query: 760 LQMRHCGLCSH-LPPLGQLPSLRELSI---SNCKRIKIIGEELYGNNSKIDAFRSLEVLE 815
+++ CGL +P L +L L+ + + S C R + F L+ LE
Sbjct: 776 VKLSECGLEEDPMPILEKLLHLKGVILLKGSYCGRRMVCSG---------GGFPQLKKLE 826
Query: 816 FQRMENLEEWLCHEGFLSLKE-LTIKDCPKLKRALPQHLPSLQKLSII 862
+ EEWL EG + L E L+I DC +LK +P L + L ++
Sbjct: 827 IVGLNKWEEWLVEEGSMPLLETLSILDCEELKE-IPDGLRFIYSLELV 873
>At3g46730 disease resistance protein RPP8-like protein
Length = 847
Score = 198 bits (503), Expect = 2e-50
Identities = 226/889 (25%), Positives = 390/889 (43%), Gaps = 109/889 (12%)
Query: 17 IFEKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHV 76
+ K+ I + + V D ++EL L I+ L++ E ++ + K+W + +
Sbjct: 9 VLNKIGGYLINEVLALMGVKDDLEELKTELTCIHGYLKDVEAREREDEVSKEWTKLVLDI 68
Query: 77 VYEADQLLDEISTDA-----------MLNKLKAESEP---------LTTNLLGV------ 110
Y+ + +LD + NK+ + + L +L +
Sbjct: 69 AYDIEDVLDTYFLKLEERSLRRGLLRLTNKIGKKRDAYNIVEDIRTLKRRILDITRKRET 128
Query: 111 LGLAEGPSASNEGLVSWKPSKRLSSTALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPI 170
G+ E + + + + + + E + G + D + L+ LL+ N+ I
Sbjct: 129 FGIGSFNEPRGENITNVRVRQLRRAPPVDQEELVVGLEDDVKILLVKLLSDNEKDKSY-I 187
Query: 171 ISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFN-PSADG 229
ISI G+GG+GKT LA+ +YN+ ++ F+ +AW YVS+ Y + I++S SA+
Sbjct: 188 ISIFGMGGLGKTALARKLYNSGDVKRRFDCRAWTYVSQEYKTRDILIRIIRSLGIVSAEE 247
Query: 230 -------EYLDQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQL--LLPFNHGSFGSKII 280
E ++L+ L +L GK Y++V+DD+W+ + WE L LP +H GSK+I
Sbjct: 248 MEKIKMFEEDEELEVYLYGLLEGKNYMVVVDDVWDPDA--WESLKRALPCDHR--GSKVI 303
Query: 281 VTTREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKC 340
+TTR + +A V + LR L + W LF AF D L+ G++++ KC
Sbjct: 304 ITTRIRAIAEGVEGTVYAHKLRFLTFEESWTLFERKAFSNIEKVD-EDLQRTGKEMVKKC 362
Query: 341 EGLPLAIISLGQLLRKKFSQDEWMKILETDMWRLSDVDNKINPVLRLSYHNLPSDQKRCF 400
GLPLAI+ L LL +K +EW ++ + RL D I+ V LS+ + + K CF
Sbjct: 363 GGLPLAIVVLSGLLSRK-RTNEWHEVCASLWRRLKDNSIHISTVFDLSFKEMRHELKLCF 421
Query: 401 AFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQSFDKT 460
+ S+FP+ Y + ++LI L +AEG + + +EE E +S K
Sbjct: 422 LYFSVFPEDYEIKVEKLIHLLVAEGFI------QEDEEMMMEDVARCYIDELVDRSLVKA 475
Query: 461 YGTYEHYVMYNLVNDLAKSVSGEFCMQIEGARVEGSLERTRHIRFSLRSNCLNKLLET-- 518
VM ++DL + ++ + ++ V + + I R ++ L+
Sbjct: 476 ERIERGKVMSCRIHDLLRDLAIKKAKELNFVNVYNEKQHSSDI---CRREVVHHLMNDYY 532
Query: 519 TCEL---KGLRSLILDVHR---GTLISNNVQLDLFSRLNFLRTLSFRWCGLSELVDEISN 572
C+ K +RS + R G + + N++L L LN L + L D I
Sbjct: 533 LCDRRVNKRMRSFLFIGERRGFGYVNTTNLKLKLLRVLNMEGLLFVSKNISNTLPDVIGE 592
Query: 573 IKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHLELPYLKK 632
+ LRYL ++ T ++ LP SI L LQT+ G + + ++ SKL +LRH+ ++ +
Sbjct: 593 LIHLRYLGIADTYVSILPASISNLRFLQTLDASGNDPFQYTTDLSKLTSLRHVIGKFVGE 652
Query: 633 MPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPEDAVTANLK 692
IG+ +LQTL KLNH + NL+
Sbjct: 653 C--LIGEGVNLQTL--------RSISSYSWSKLNH------------------ELLRNLQ 684
Query: 693 DKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYR---GNRFPNWI 749
D + YD K VD V N V + N + +L + ++ +R +
Sbjct: 685 D--------LEIYDHSKWVDQRRVPLNF-VSFSKPKNLRVLKLEMRNFKLSSESRTTIGL 735
Query: 750 RGCHLPNLVSLQMRHCGLCSH-LPPLGQLPSLRELSISNC--KRIKIIGEELYGNNSKID 806
+ P+L SL + L + +P L +LP L +L + +C +KI+ G
Sbjct: 736 VDVNFPSLESLTLVGTTLEENSMPALQKLPRLEDLVLKDCNYSGVKIMSISAQG----FG 791
Query: 807 AFRSLEVLEFQRMENLEEW-LCHEGFLSLKELTIKDCPKL-KRALPQHL 853
++LE+ +R L+E + E SL +LT+K +L K +P L
Sbjct: 792 RLKNLEMSMERRGHGLDELRIEEEAMPSLIKLTVKGRLELTKLMIPDRL 840
>At5g48620 disease resistance protein
Length = 908
Score = 194 bits (493), Expect = 2e-49
Identities = 232/917 (25%), Positives = 414/917 (44%), Gaps = 127/917 (13%)
Query: 19 EKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVY 78
EKL + R+ + +D+ + L L S+ +L++A+ K++ V+ +L+ +K +V+
Sbjct: 11 EKLWDLLSRESERLQGIDEQLDGLKRQLRSLQSLLKDADAKKHGSDRVRNFLEDVKDLVF 70
Query: 79 EADQLLDEISTDAMLNKLKAES------------------------EPLTTNLLGVLGLA 114
+A+ +++ +LNKL+ E E +T + V+G
Sbjct: 71 DAEDIIESY----VLNKLRGEGKGVKKHVRRLARFLTDRHKVASDIEGITKRISDVIG-- 124
Query: 115 EGPSASNEGLVSWKPSKRLSSTALVD-----------ESSIYGRDVDKEELIKFLLAGND 163
E S + ++ S L V ES + G + EEL+ L+ ND
Sbjct: 125 EMQSFGIQQIIDGVRSLSLQERQRVQREIRQTYPDSSESDLVGVEQSVEELVGHLVE-ND 183
Query: 164 SGTQVPIISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSF 223
++SI G+GG+GKTTLA+ V++++ + HF+ AWV VS+ + + + + IL+
Sbjct: 184 I---YQVVSIAGMGGIGKTTLARQVFHHDLVRRHFDGFAWVCVSQQFTLKHVWQRILQEL 240
Query: 224 NPSADGEYLDQ----LQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKI 279
P DG L LQ +L +L +YLLVLDD+W E W+++ F G K+
Sbjct: 241 QPH-DGNILQMDESALQPKLFQLLETGRYLLVLDDVW--KKEDWDRIKAVFPR-KRGWKM 296
Query: 280 IVTTREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDY---PKLESIGRKI 336
++T+R + V H + + L + W+L F + + ++E++G+++
Sbjct: 297 LLTSRNEGVGIHADPTCLTFRASILNPEESWKLCERIVFPRRDETEVRLDEEMEAMGKEM 356
Query: 337 MDKCEGLPLAIISLGQLLRKKFSQDEWMKILETDMWRL---SDVD----NKINPVLRLSY 389
+ C GLPLA+ +LG LL K + EW ++ + ++ S +D N +N +L LSY
Sbjct: 357 VTHCGGLPLAVKALGGLLANKHTVPEWKRVSDNIGSQIVGGSCLDDNSLNSVNRILSLSY 416
Query: 390 HNLPSDQKRCFAFCSIFPKGYTFEKDELIKLWMAEGL-----LKCCGSYKSEEEF-GNEI 443
+LP+ K F + + FP+ +L W AEG+ ++ G Y EE N +
Sbjct: 417 EDLPTHLKHRFLYLAHFPEDSKIYTQDLFNYWAAEGIYDGSTIQDSGEYYLEELVRRNLV 476
Query: 444 FGDLESISFFQQSFDKTYGTYEHYVMYNLVND--LAKSVSGEFCMQIEGARVEGSLER-- 499
D +S + M++++ + L+K+ F I+ ++
Sbjct: 477 IADNRYLSL----------EFNFCQMHDMMREVCLSKAKEENFLQIIKDPTSTSTINAQS 526
Query: 500 -TRHIRFSLRSNCLNKLLETTCELKGLRSLILDVHRGTLISNNVQLDLFSRLNFLRTLSF 558
+R RFS+ S +L K +RSLI+ + +F L LR L
Sbjct: 527 PSRSRRFSIHSGKAFHILGHRNNPK-VRSLIVSRFEEDFWIRSA--SVFHNLTLLRVLDL 583
Query: 559 RWCGL--SELVDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTIL-----LQGCELTE 611
+L I + LRYL L ++ LP + + NL+ +L + E
Sbjct: 584 SRVKFEGGKLPSSIGGLIHLRYLSLYGAVVSHLPST---MRNLKLLLFLNLRVDNKEPIH 640
Query: 612 LPSNFSKLINLRHLELPYL--KKMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLH 669
+P+ +++ LR+L LP K +G L +L+ L YF + + +DL + KL +L
Sbjct: 641 VPNVLKEMLELRYLSLPQEMDDKTKLELGDLVNLEYLWYFSTQHSSVTDLLRMTKLRNL- 699
Query: 670 GKICIDGLGYVFDPEDAVTANLKDKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPN 729
G + + + ++++L++ + LE L ++F VD VL+
Sbjct: 700 GVSLSERCNF-----ETLSSSLRELRNLEMLNVLFSPEIVMVD----HMGEFVLDHF--- 747
Query: 730 RSLKRLSISQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCSH-LPPLGQLPSLRELSISNC 788
LK+L ++ R ++ P+ + P+L + + HC + +P L +L L+ +++S
Sbjct: 748 IHLKQLGLA-VRMSKIPD--QHQFPPHLAHIHLVHCVMKEDPMPILEKLLHLKSVALSYG 804
Query: 789 KRIKIIGEELYGNNSKIDAFRSLEVLEFQRMENLEEWLCHEGFL-SLKELTIKDCPKLKR 847
IG + + F L L LEEW+ EG + L+ LTI DC KLK
Sbjct: 805 ---AFIGRRVVCSKG---GFPQLCALGISGESELEEWIVEEGSMPCLRTLTIHDCEKLKE 858
Query: 848 ALP---QHLPSLQKLSI 861
LP +++ SL++L I
Sbjct: 859 -LPDGLKYITSLKELKI 874
>At1g59780 hypothetical protein
Length = 906
Score = 189 bits (481), Expect = 6e-48
Identities = 220/897 (24%), Positives = 399/897 (43%), Gaps = 109/897 (12%)
Query: 19 EKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVY 78
EKL + ++Y + V++ + EL L + L +A+ K+ + L+++K + Y
Sbjct: 16 EKLWKLLSQEYERFQGVEEQITELRDDLKMLMAFLSDADAKKQTRALARNCLEEIKEITY 75
Query: 79 EADQLLDEISTDAMLNKLKAESEP------------------LTTNLLGVLGLAEGPSAS 120
+A+ +++ +N P ++ LG+
Sbjct: 76 DAEDIIEIFLLKGSVNMRSLACFPGGRREIALQITSISKRISKVIQVMQNLGIKSDIMDG 135
Query: 121 NEGLVSWKPSKRLSST-ALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLGGM 179
+ + + L T + ES++ G + + E+L++ L+ GNDS V SI GLGG+
Sbjct: 136 VDSHAQLERKRELRHTFSSESESNLVGLEKNVEKLVEELV-GNDSSHGV---SITGLGGL 191
Query: 180 GKTTLAKLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFNPSADGEYL--DQLQH 237
GKTTLA+ +++++K++ HF+ AWV VS+ + + K IL + +P L D +Q
Sbjct: 192 GKTTLARQIFDHDKVKSHFDGLAWVCVSQEFTRKDVWKTILGNLSPKYKDSDLPEDDIQK 251
Query: 238 QLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHVVKSTM 297
+L +L KK L+V DD+W E W ++ F G K+++T+R + H V T
Sbjct: 252 KLFQLLETKKALIVFDDLWKR--EDWYRIAPMFPERKAGWKVLLTSRNDAIHPHCV--TF 307
Query: 298 LCDLRQLVKSDCWRLFVTHAF-QGKSVCDY---PKLESIGRKIMDKCEGLPLAIISLGQL 353
+L L +CW+L AF + K++ Y ++ + +++ C+ LPLA+ LG L
Sbjct: 308 KPEL--LTHDECWKLLQRIAFSKQKTITGYIIDKEMVKMAKEMTKHCKRLPLAVKLLGGL 365
Query: 354 LRKKFSQDEWMKILETDMWRL--------SDVDNKINPVLRLSYHNLPSDQKRCFAFCSI 405
L K + +W I E + + + + +N VL LS+ LP K C + +
Sbjct: 366 LDAKHTLRQWKLISENIISHIVVGGTSSNENDSSSVNHVLSLSFEGLPGYLKHCLLYLAS 425
Query: 406 FPKGYTFEKDELIKLWMAEGLLKCCGSYKSE--EEFGNEIFGDLESISFFQQSFDKTYGT 463
+P+ + E + L +W AEG + G+Y+ + + +L + D
Sbjct: 426 YPEDHEIEIERLSYVWAAEG-ITYPGNYEGATIRDVADLYIEELVKRNMVISERDALTSR 484
Query: 464 YEHYVMYNLVND--LAKSVSGEFCMQIEGARVEGSLERTRHIRFSLRSNCLNKLL---ET 518
+E +++L+ + L K+ F + S+ R S R N + E
Sbjct: 485 FEKCQLHDLMREICLLKAKEENFLQIVTDPTSSSSVHSLASSR-SRRLVVYNTSIFSGEN 543
Query: 519 TCELKGLRSLI---LDVHRGTLISNNVQLDLFSRLNFLRTLSFRWCGLSELVDEISNIKL 575
+ LRSL+ + R ++ SN ++L L L+ L F+ +L I +
Sbjct: 544 DMKNSKLRSLLFIPVGYSRFSMGSNFIELPLLRVLD-LDGAKFKG---GKLPSSIGKLIH 599
Query: 576 LRYLDLSFTEITSLPDSICMLYNLQTIL-----LQGCELTELPSNFSKLINLRHLELPYL 630
L+YL L +T LP S L NL+++L + +L +P+ F +++ LR+L LP+
Sbjct: 600 LKYLSLYQASVTYLPSS---LRNLKSLLYLNLRINSGQLINVPNVFKEMLELRYLSLPWE 656
Query: 631 KK--MPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPEDAVT 688
+ +G L L+TL F ++ + +DL + KL L I +GL + ++
Sbjct: 657 RSSLTKLELGNLLKLETLINFSTKDSSVTDLHRMTKLRTLQILISGEGLHM-----ETLS 711
Query: 689 ANLKDKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYRGNRFPNW 748
+ L +LE+L ++ S SV + + + R + + FP+
Sbjct: 712 SALSMLGHLEDL-------------TVTPSENSV--QFKHPKLIYRPMLPDVQ--HFPS- 753
Query: 749 IRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKIIGEELYGNNSKIDAF 808
+L ++ + +C L P+ L L +L + + +G + F
Sbjct: 754 -------HLTTISLVYCFL--EEDPMPTLEKLLQLKVVSLWYNAYVGRRMVCTGG---GF 801
Query: 809 RSLEVLEFQRMENLEEWLCHEGFLS-LKELTIKDCPKLKRALP---QHLPSLQKLSI 861
L LE ++ LEEW+ EG + L L I DC KLK +P + + SL++L+I
Sbjct: 802 PPLHRLEIWGLDALEEWIVEEGSMPLLHTLHIVDCKKLKE-IPDGLRFISSLKELAI 857
>At1g10920 disease resistance protein RPM1 isolog
Length = 821
Score = 186 bits (472), Expect = 6e-47
Identities = 200/821 (24%), Positives = 368/821 (44%), Gaps = 58/821 (7%)
Query: 35 VDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVYEADQLLDEISTDAMLN 94
+ + V L L + +L++A+ K+++ V+ +L+ ++ +VY+A+ +++ +
Sbjct: 27 IGEQVDGLKRQLGRLQSLLKDADAKKHESERVRNFLEDVRDIVYDAEDIIESFLLNEFRT 86
Query: 95 K---LKAESEPLTTNL-LGVLGLAEGPSASNEGLVSWKPSKRLSSTALVDESSIYGRDVD 150
K +K + L L LG+ + +G S+ + + + + A ES + G +
Sbjct: 87 KEKGIKKHARRLACFLSLGIQEIIDGASSMSLQERQREQKEIRQTFANSSESDLVGVEQS 146
Query: 151 KEELIKFLLAGNDSGTQVPIISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESY 210
E L L+ ++ + ++SI G+GG+GKTTLA+ V++++ ++ HF+ AWV+VS+ +
Sbjct: 147 VEALAGHLVENDN----IQVVSISGMGGIGKTTLARQVFHHDMVQRHFDGFAWVFVSQQF 202
Query: 211 DVVGLTKAILKSFNP-SADGEYLDQ--LQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLL 267
+ + I + P + D ++D+ LQ +L +L +YL+VLDD+W E W+++
Sbjct: 203 TQKHVWQRIWQELQPQNGDISHMDEHILQGKLFKLLETGRYLVVLDDVWKE--EDWDRIK 260
Query: 268 LPFNHGSFGSKIIVTTREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCD-Y 326
F G K+++T+R + V H + R L + W+L F +
Sbjct: 261 AVFPRKR-GWKMLLTSRNEGVGIHADPKSFGFKTRILTPEESWKLCEKIVFHRRDETGTL 319
Query: 327 PKLESIGRKIMDKCEGLPLAIISLGQLLRKKFSQDEWMKILET------DMWRLSDVDNK 380
+E++G++++ C GLPLA+ LG LL K + EW ++ + L D N
Sbjct: 320 SDMEAMGKEMVTCCGGLPLAVKVLGGLLATKHTVPEWKRVYDNIGPHLAGRSSLDDNLNS 379
Query: 381 INPVLRLSYHNLPSDQKRCFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFG 440
I VL LSY NLP K CF + + FP+ Y L AEG++ + ++ G
Sbjct: 380 IYRVLSLSYENLPMCLKHCFLYLAHFPEYYEIHVKRLFNYLAAEGIITSSDDGTTIQDKG 439
Query: 441 NEIFGDLESISFFQQSFDKTYGTYEHYVMYNLVNDLAKS-VSGEFCMQIEGARVEGSLER 499
+ +L + + + +H M++++ ++ S E ++I S
Sbjct: 440 EDYLEELARRNMITIDKNYMFLRKKHCQMHDMMREVCLSKAKEENFLEIFKVSTATSAIN 499
Query: 500 TRHIRFSLR-----SNCLNKLLETTCELKGLRSLILDVHRGTLISNNVQLDLFSRLNFLR 554
R + S R N L L +T K +RSL+ F L LR
Sbjct: 500 ARSLSKSRRLSVHGGNALPSLGQTIN--KKVRSLLYFAFEDEFCILESTTPCFRSLPLLR 557
Query: 555 TLSFRWCGLS--ELVDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCELTEL 612
L +L I ++ LR+L L I+ LP S+ L L + L + +
Sbjct: 558 VLDLSRVKFEGGKLPSSIGDLIHLRFLSLHRAWISHLPSSLRNLKLLLYLNLGFNGMVHV 617
Query: 613 PSNFSKLINLRHLELPYLK--KMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHG 670
P+ ++ LR+L+LP K + L +L++L F + + DL + KL L
Sbjct: 618 PNVLKEMQELRYLQLPMSMHDKTKLELSDLVNLESLMNFSTKYASVMDLLHMTKLREL-S 676
Query: 671 KICIDGLGYVFDPEDAVTANLKDKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPNR 730
DG D ++++L + LE L++ YDR++ VL +
Sbjct: 677 LFITDG------SSDTLSSSLGQLRSLEVLHL--YDRQEP--RVAYHGGEIVLNCIH--- 723
Query: 731 SLKRLSISQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCSH-LPPLGQLPSLRELSISNCK 789
LK L ++ + RFP+ + P+L + + C + +P L +L L+ + ++
Sbjct: 724 -LKELELAIHMP-RFPD--QYLFHPHLSHIYLWCCSMEEDPIPILERLLHLKSVILTFGA 779
Query: 790 RIKIIGEELYGNNSKIDAFRSLEVLEFQRMENLEEWLCHEG 830
+G + + F L L+ + +E LEEW+ EG
Sbjct: 780 ---FVGRRMVCSKG---GFPQLCFLKLEELEELEEWIVEEG 814
>At1g50180 disease resistance protein, putative
Length = 857
Score = 184 bits (466), Expect = 3e-46
Identities = 233/909 (25%), Positives = 395/909 (42%), Gaps = 146/909 (16%)
Query: 16 VIFEKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKH 75
V +KL + + + + D VK+L L +N L++A+ KQ++ V+ W+ ++
Sbjct: 8 VTVQKLGQLLLEEPLFLFGIGDQVKQLQDELKRLNCFLKDADEKQHESERVRNWVAGIRE 67
Query: 76 VVYEADQLLDEISTDAMLNKLK-----------------------AESEPLTTNL----L 108
Y+A+ +L+ A K K +E +T+ L
Sbjct: 68 ASYDAEDILEAFFLKAESRKQKGMKRVLRRLACILNEAVSLHSVGSEIREITSRLSKIAA 127
Query: 109 GVLGLAEGPSASNEGL-VSWKPSKRLSSTALVDESSIYGRDVDKEELIKFLLAGNDSGTQ 167
+L S EGL +S ++ S V E ++ G + E+L+ L++G G +
Sbjct: 128 SMLDFGIKESMGREGLSLSDSLREQRQSFPYVVEHNLVGLEQSLEKLVNDLVSG---GEK 184
Query: 168 VPIISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFNPSA 227
+ + SI G+GG+GKTTLAK +++++K+ HF+ AWVYVS+ + + I + +
Sbjct: 185 LRVTSICGMGGLGKTTLAKQIFHHHKVRRHFDRFAWVYVSQDCRRRHVWQDIFLNLSYKD 244
Query: 228 DGEYL-----DQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVT 282
+ + + +QL +L L K L+VLDDIW + W+ L F H + GS+II+T
Sbjct: 245 ENQRILSLRDEQLGEELHRFLKRNKCLIVLDDIWGKDA--WDCLKHVFPHET-GSEIILT 301
Query: 283 TREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCD---YPKLESIGRKIMDK 339
TR KEVA + +L + + L + W L + G+ + K+E IG++I+ +
Sbjct: 302 TRNKEVALYADPRGVLHEPQLLTCEESWELLEKISLSGRENIEPMLVKKMEEIGKQIVVR 361
Query: 340 CEGLPLAIISLGQLLRKKFSQDEWMKILETDMWRLSDVDNK-------INPVLRLSYHNL 392
C GLPLAI LG LL K + +EW ++ E +S+ + + VL LSY L
Sbjct: 362 CGGLPLAITVLGGLLATKSTWNEWQRVCENIKSYVSNGGSSNGSKNMLVADVLCLSYEYL 421
Query: 393 PSDQKRCFAFCSIFPKGYTFEKDELIKLWMAEGL---LKCCGSYKSEEEFGNEIFGDLES 449
P K+CF + + +P+ Y L+ +AEG+ +K + + E+ G + +L
Sbjct: 422 PPHVKQCFLYFAHYPEDYEVHVGTLVSYCIAEGMVMPVKHTEAGTTVEDVGQDYLEELVK 481
Query: 450 ISFFQQSFDKTYGTYEHYVMYNLVNDLAKSVSGEFCMQIEGARVEGSLERTRHIRF-SLR 508
S + T E VM ++DL + V + Q +V S ++ F SL
Sbjct: 482 RSMVMVG-RRDIVTSE--VMTCRMHDLMREVCLQKAKQESFVQVIDSRDQDEAEAFISLS 538
Query: 509 SNCLNKLLETTCELKGLRSLILDVHRGTLISNNVQLDLFSRLNFLRTLSFRWCGLSELVD 568
+N R + + +H G + + L +SFR
Sbjct: 539 TNT-------------SRRISVQLHGGAEEHH---------IKSLSQVSFR--------- 567
Query: 569 EISNIKLLRYLDLSFTEIT--SLPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHLE 626
+KLLR LDL +I LPD + L +L+ + ++ + EL S+ L + L+
Sbjct: 568 ---KMKLLRVLDLEGAQIEGGKLPDDVGDLIHLRNLSVRLTNVKELTSSIGNLKLMITLD 624
Query: 627 LPYLKKMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPEDA 686
L ++K G+L L F V + N DL + L L + +V
Sbjct: 625 L-FVK------GQLYIPNQLWDFPVGKCNPRDLLAMTSLRRLSINLSSQNTDFV------ 671
Query: 687 VTANLKDKKYLEELYMIFYDRKKE-----VDDSIVESNVSVLEALQPNRSLKRLSISQYR 741
V ++L K L+ L + + E VD + + S + L L+ L++L Q
Sbjct: 672 VVSSL--SKVLKRLRGLTINVPCEPMLPPVDVTQLVSAFTNLCELELFLKLEKLPGEQSF 729
Query: 742 GNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKIIGEELYGN 801
+ +L +L++ CGL P L L L I +G +L +
Sbjct: 730 SS------------DLGALRLWQCGLVD--DPFMVLEKLPNLKILQLFEGSFVGSKLCCS 775
Query: 802 NSKIDAFRSLEVLEFQRMENLEEWLCHEG-FLSLKELTIKDCPKLKRALPQ---HLPSLQ 857
+ +ENLEEW +G + L + +K C KLK ++P+ L +LQ
Sbjct: 776 ---------------KNLENLEEWTVEDGAMMRLVTVELKCCNKLK-SVPEGTRFLKNLQ 819
Query: 858 KLSIINCNK 866
++ I N K
Sbjct: 820 EVEIGNRTK 828
>At3g46530 disease resistance protein RPP13-like protein
Length = 835
Score = 172 bits (435), Expect = 1e-42
Identities = 179/654 (27%), Positives = 287/654 (43%), Gaps = 65/654 (9%)
Query: 35 VDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVYEADQLLDEIS------ 88
V + ++EL L I+ L++ E ++ + K+W + Y+ + +LD
Sbjct: 27 VKEDLEELKTELTCIHGYLKDVEAREREDEVSKEWSKLVLDFAYDVEDVLDTYHLKLEER 86
Query: 89 ---------TDAMLNKLKAESEPLTTNLL--------------GVLGLAEGPSASNEGLV 125
T+ + K+ A S +L G+ GL E N +
Sbjct: 87 SQRRGLRRLTNKIGRKMDAYSIVDDIRILKRRILDITRKRETYGIGGLKEPQGGGNTSSL 146
Query: 126 SWKPSKRLSSTALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLGGMGKTTLA 185
+ +R S E + G + D + L++ LL + IISI G+GG+GKT LA
Sbjct: 147 RVRQLRRARSVD--QEEVVVGLEDDAKILLEKLLDYEEKNRF--IISIFGMGGLGKTALA 202
Query: 186 KLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFNPSADGEYL--------DQLQH 237
+ +YN+ ++E FE +AW YVS+ Y + I++S ++ GE L ++L+
Sbjct: 203 RKLYNSRDVKERFEYRAWTYVSQEYKTGDILMRIIRSLGMTS-GEELEKIRKFAEEELEV 261
Query: 238 QLQHMLMGKKYLLVLDDIWNGNVEYWEQL--LLPFNHGSFGSKIIVTTREKEVAYHVVKS 295
L +L GKKYL+V+DDIW E W+ L LP NH GS++I+TTR K VA V
Sbjct: 262 YLYGLLEGKKYLVVVDDIWER--EAWDSLKRALPCNHE--GSRVIITTRIKAVAEGVDGR 317
Query: 296 TMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCEGLPLAIISLGQLLR 355
LR L + W LF AF+ D L++ G++++ KC GLPL I+ L LL
Sbjct: 318 FYAHKLRFLTFEESWELFEQRAFRNIQRKDEDLLKT-GKEMVQKCRGLPLCIVVLAGLLS 376
Query: 356 KKFSQDEWMKILETDMWRLSDVDNKINP-VLRLSYHNLPSDQKRCFAFCSIFPKGYTFEK 414
+K + EW + + RL D + P V LS+ L + K CF + SIFP+ Y +
Sbjct: 377 RK-TPSEWNDVCNSLWRRLKDDSIHVAPIVFDLSFKELRHESKLCFLYLSIFPEDYEIDL 435
Query: 415 DELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQSFDKTYGTYEHYVMYNLVN 474
++LI L +AEG + + +EE E +S + VM ++
Sbjct: 436 EKLIHLLVAEGFI------QGDEEMMMEDVARYYIEELIDRSLLEAVRRERGKVMSCRIH 489
Query: 475 DLAKSVSGEFCMQIEGARVEGSLERTRHIRFSLRSNCLNKLLETTCELKGLRSLILDVHR 534
DL + V+ + ++ V +H + R ++ + K +
Sbjct: 490 DLLRDVAIKKSKELNFVNVYND-HVAQHSSTTCRREVVHHQFKRYSSEKRKNKRMRSFLY 548
Query: 535 GTLISNNVQLDLFSRLNFLRTLSFRWCGLSELVDEISNIKLLRYLDLSFTEIT--SLPDS 592
+ V LD F L LR L F L ++ ++ LRYL + I +
Sbjct: 549 FGEFDHLVGLD-FETLKLLRVLDFGSLWLPFKIN--GDLIHLRYLGIDGNSINDFDIAAI 605
Query: 593 ICMLYNLQTILLQGCELTELPSNFSKLINLRHLELPYLKKMPKHIGKLNSLQTL 646
I L LQT+ + E + KL +LRH+ + + IG + +LQTL
Sbjct: 606 ISKLRFLQTLFVSDNYFIEETIDLRKLTSLRHVIGNFFGGL--LIGDVANLQTL 657
>At1g59620 PRM1 homolog (CW9)
Length = 628
Score = 169 bits (427), Expect = 1e-41
Identities = 122/428 (28%), Positives = 219/428 (50%), Gaps = 31/428 (7%)
Query: 19 EKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVY 78
EKL + +R+ + V EL LN + LE+A+ K++Q V + ++K +VY
Sbjct: 11 EKLWDLLVRESDRFQGVKKQFNELRSDLNKLRCFLEDADAKKHQSAMVSNTVKEVKEIVY 70
Query: 79 EADQLLDEISTDAMLNKLKAESEPLTTNLLGVLGLAEGPSASNEGL---VSWKPSKRLSS 135
+ + +++ L + + + + VL + EGL ++ K + +
Sbjct: 71 DTEDIIETFLRKKQLGRTRGMKKRIK-EFACVLPDRRKIAIDMEGLSKRIAKKDKRNMRQ 129
Query: 136 T-ALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLGGMGKTTLAKLVYNNNKI 194
T + +ES + G + + ++L+ L+ DS ++SI G+GG+GKTTLA+ V+N+ +
Sbjct: 130 TFSNNNESVLVGLEENVKKLVGHLVEVEDSSQ---VVSITGMGGIGKTTLARQVFNHETV 186
Query: 195 EEHFELKAWVYVSESYDVVGLTKAILKSFNPSADGEYL------DQLQHQLQHMLMGKKY 248
+ HF AWV VS+ + + + IL+ P EY+ D+LQ +L +L +K
Sbjct: 187 KSHFAQLAWVCVSQQFTRKYVWQTILRKVGP----EYIKLEMTEDELQEKLFRLLGTRKA 242
Query: 249 LLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHVVKSTMLCDLRQLVKSD 308
L+VLDDIW E W+ + F G G K+++T+R + VA + + L +
Sbjct: 243 LIVLDDIW--REEDWDMIEPIFPLGK-GWKVLLTSRNEGVALRANPNGFIFKPDCLTPEE 299
Query: 309 CWRLFVTHAFQGKSVCDY---PKLESIGRKIMDKCEGLPLAIISLGQLLRKKFSQDEWMK 365
W +F F G++ +Y K+E +G++++ C GLPLA+ LG LL F+ DEW +
Sbjct: 300 SWTIFRRIVFPGENTTEYKVDEKMEELGKQMIKHCGGLPLALKVLGGLLVVHFTLDEWKR 359
Query: 366 I---LETDMWRLSDVDNK----INPVLRLSYHNLPSDQKRCFAFCSIFPKGYTFEKDELI 418
I +++ + + ++K + +L LS+ LP K CF + + FP+ +T + ++L
Sbjct: 360 IYGNIKSHIVGGTSFNDKNMSSVYHILHLSFEELPIYLKHCFLYLAQFPEDFTIDLEKLS 419
Query: 419 KLWMAEGL 426
W AEG+
Sbjct: 420 YYWAAEGM 427
Score = 34.3 bits (77), Expect = 0.40
Identities = 37/126 (29%), Positives = 61/126 (48%), Gaps = 17/126 (13%)
Query: 571 SNIKLLRYLDLSFTEITS-LPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHLEL-- 627
+ ++L+R LDL E LP SI +L +L+ + L + + LPS+ L L +L L
Sbjct: 506 TRLQLMRVLDLHGVEFGGELPSSIGLLIHLRYLSLYRAKASHLPSSMQNLKMLLYLNLCV 565
Query: 628 -----PYLKKMPKHIGKLNSLQTLPYFV-----VEEKNGSDLKELEKLNHLHGKICIDGL 677
Y+ K + +L L +LP + +E N +L++LE + HG + G
Sbjct: 566 QESCYIYIPNFLKEMLELKYL-SLPLRMDDKVKLELGNLVNLEKLENFSTEHGGV---GG 621
Query: 678 GYVFDP 683
V+DP
Sbjct: 622 SSVYDP 627
>At5g63020 NBS/LRR disease resistance like protein
Length = 888
Score = 168 bits (426), Expect = 1e-41
Identities = 186/749 (24%), Positives = 332/749 (43%), Gaps = 109/749 (14%)
Query: 167 QVPIISIVGLGGMGKTTLAKLVYNN-NKIEEHFELKAWVYVSESYDVVGLTKAI---LKS 222
++ I+ + G+GG+GKTTL + N +++ F++ W+ VS+ + + I L+S
Sbjct: 173 EIGILGLHGMGGVGKTTLLSHINNRFSRVGGEFDIVIWIVVSKELQIQRIQDEIWEKLRS 232
Query: 223 FNPSADGEYLDQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVT 282
N + D + ++L K+++L+LDDIW+ V+ E + +PF G KI+ T
Sbjct: 233 DNEKWKQKTEDIKASNIYNVLKHKRFVLLLDDIWS-KVDLTE-VGVPFPSRENGCKIVFT 290
Query: 283 TREKEVAYHV-VKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCE 341
TR KE+ + V S M ++R L D W LF T ++ +P++ ++ R + KC
Sbjct: 291 TRLKEICGRMGVDSDM--EVRCLAPDDAWDLF-TKKVGEITLGSHPEIPTVARTVAKKCR 347
Query: 342 GLPLAIISLGQLLRKKFSQDEW---MKILETDMWRLSDVDNKINPVLRLSYHNLPSDQ-K 397
GLPLA+ +G+ + K + EW + +L + S ++++I P+L+ SY NL S+Q K
Sbjct: 348 GLPLALNVIGETMAYKRTVQEWRSAIDVLTSSAAEFSGMEDEILPILKYSYDNLKSEQLK 407
Query: 398 RCFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQSF 457
CF +C++FP+ + EK++L+ W+ EG + + E G EI G L +
Sbjct: 408 LCFQYCALFPEDHNIEKNDLVDYWIGEGFID--RNKGKAENQGYEIIGILVRSCLLMEEN 465
Query: 458 DKTYGTYEHYVMYNLVNDLAKSVSGEFCMQIEGARVEGSLERTRHI----------RFSL 507
+T M+++V ++A ++ +F Q E V+ L+ +R+I R SL
Sbjct: 466 QET------VKMHDVVREMALWIASDFGKQKENFIVQAGLQ-SRNIPEIEKWKVARRVSL 518
Query: 508 RSNCLNKLLETTCELKGLRSLILDVHRGTLISNNVQLDLFSRLNFLRTLSFRW-CGLSEL 566
N + + + E L +L+L + ++ F + L L L L
Sbjct: 519 MFNNIESIRDAP-ESPQLITLLLRKN----FLGHISSSFFRLMPMLVVLDLSMNRDLRHL 573
Query: 567 VDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHLE 626
+EIS L+YL LS T I P + L L + L+ + E S L +L+ L
Sbjct: 574 PNEISECVSLQYLSLSRTRIRIWPAGLVELRKLLYLNLEYTRMVESICGISGLTSLKVLR 633
Query: 627 LPYLKKMPKH---IGKLNSLQTLPYFVVEEKNGSDLKEL---EKLNHLHGKICIDGLGYV 680
L ++ P+ + +L L+ L + S L++ ++L + I+ L
Sbjct: 634 L-FVSGFPEDPCVLNELQLLENLQTLTITLGLASILEQFLSNQRLASCTRALRIENL--- 689
Query: 681 FDPEDAVTANLKDKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQY 740
+P+ +V + + L+EL+ D I E V E + P
Sbjct: 690 -NPQSSVISFVATMDSLQELHF--------ADSDIWEIKVKRNETVLP------------ 728
Query: 741 RGNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKIIGEELYG 800
H+P + P+L ++S+ C R++ + ++
Sbjct: 729 -----------LHIPTTTTF----------------FPNLSQVSLEFCTRLRDLTWLIFA 761
Query: 801 NNSKIDAFRSLEVLEFQRMENLE--EWLCHEGFLSLKELTIKDCPKLKRAL--PQHLPSL 856
N + R + + + + N E E F LKEL +++ LK P P L
Sbjct: 762 PN--LTVLRVISASDLKEVINKEKAEQQNLIPFQELKELRLENVQMLKHIHRGPLPFPCL 819
Query: 857 QKLSIINCNKLE------ASMPEGDNILE 879
QK+ + C++L S+P GD ++E
Sbjct: 820 QKILVNGCSELRKLPLNFTSVPRGDLVIE 848
>At1g61180
Length = 889
Score = 164 bits (416), Expect = 2e-40
Identities = 197/746 (26%), Positives = 321/746 (42%), Gaps = 92/746 (12%)
Query: 168 VPIISIVGLGGMGKTTLAKLVYNN-NKIEEHFELKAWVYVSESYDVVGLTKAILKSFNPS 226
V I+ + G+GG+GKTTL K ++N +I F++ W+ VS+ + L + I + +
Sbjct: 172 VGIMGLHGMGGVGKTTLFKKIHNKFAEIGGTFDIVIWIVVSKGVMISKLQEDIAEKLHLC 231
Query: 227 AD---GEYLDQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTT 283
D + + +L GK+++L+LDDIW V+ E + +P+ K+ TT
Sbjct: 232 DDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWE-KVDL-EAIGIPYPSEVNKCKVAFTT 289
Query: 284 REKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCEGL 343
R +EV + + + L D W LF D P + + R++ KC GL
Sbjct: 290 RSREVCGEMGDHKPM-QVNCLEPEDAWELFKNKVGDNTLSSD-PVIVELAREVAQKCRGL 347
Query: 344 PLAIISLGQLLRKKFSQDEW---MKILETDMWRLSDVDNKINPVLRLSYHNLPSDQ-KRC 399
PLA+ +G+ + K EW + + T SD+ NKI P+L+ SY +L + K C
Sbjct: 348 PLALNVIGETMSSKTMVQEWEHAIHVFNTSAAEFSDMQNKILPILKYSYDSLGDEHIKSC 407
Query: 400 FAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQSFDK 459
F +C++FP+ ++LI W+ EG + K G + G L + +
Sbjct: 408 FLYCALFPEDGEIYNEKLIDYWICEGFIGEDQVIKRARNKGYAMLGTLTRANLLTK---- 463
Query: 460 TYGTYEHYVMYNLVNDLAKSVSGEFCMQIEGARVEG--SLERTRHIR--FSLRSNCL--N 513
GTY + VM+++V ++A ++ +F Q E V+ L ++ ++R L N
Sbjct: 464 -VGTY-YCVMHDVVREMALWIASDFGKQKENFVVQAGVGLHEIPKVKDWGAVRKMSLMDN 521
Query: 514 KLLETTCELK--GLRSLILDVHR-----GTLI---SNNVQLDLFSRLNFLRTLSFRWCGL 563
+ E TCE K L +L L ++ G I V LDL +F
Sbjct: 522 DIEEITCESKCSELTTLFLQSNKLKNLPGAFIRYMQKLVVLDLSYNRDF----------- 570
Query: 564 SELVDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCELTELPSNFSKLINLR 623
++L ++IS + L++LDLS T I +P + L L + L + S S+L++LR
Sbjct: 571 NKLPEQISGLVSLQFLDLSNTSIEHMPIGLKELKKLTFLDLTYTDRLCSISGISRLLSLR 630
Query: 624 HLELPYLK-----KMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLG 678
L L K + K + +L +LQ L V E D ++L L +CI+ G
Sbjct: 631 LLRLLGSKVHGDASVLKELQQLQNLQELAITVSAELISLD----QRLAKLISNLCIE--G 684
Query: 679 YVFDPED----AVTANLKDKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKR 734
++ P D A NL + + R+ E + S + N + +L R
Sbjct: 685 FLQKPFDLSFLASMENLSSLRVENSYFSEIKCRESETESSYLRINPKI----PCFTNLSR 740
Query: 735 LSISQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKII 794
L I + + WI PNLV L + RE+ +II
Sbjct: 741 LEIMKCHSMKDLTWI--LFAPNLVVL---------------LIEDSREVG-------EII 776
Query: 795 GEELYGNNSKIDAFRSLEVLEFQRMENLEE-WLCHEGFLSLKELTIKDCPKLKRALPQHL 853
+E N + I F LE L + LE + F L + + +CPKL R LP +
Sbjct: 777 NKEKATNLTSITPFLKLEWLILYNLPKLESIYWSPLPFPVLLTMDVSNCPKL-RKLPLNA 835
Query: 854 PSLQKLSIINCNKLEASMPEGDNILE 879
S+ K+ + PE +N LE
Sbjct: 836 TSVSKVEEFEIHMYPP--PEQENELE 859
Score = 36.6 bits (83), Expect = 0.081
Identities = 46/168 (27%), Positives = 71/168 (41%), Gaps = 18/168 (10%)
Query: 922 LAELCLDLSGFVECPSLDLRCYNSLRTLSIIGWRSSSLSFSLYLFTNLHSLYLYNCPELV 981
++ LC++ GF++ P DL S+ LS + +S S + S YL P++
Sbjct: 677 ISNLCIE--GFLQKP-FDLSFLASMENLSSLRVENSYFSEIKCRESETESSYLRINPKIP 733
Query: 982 SFPEGGLPSNLSCFSIFDCPKLIASREEWGLFQLNS----LKEFRVSDEFENVESFPEEN 1037
F +NLS I C + W LF N +++ R E N E
Sbjct: 734 CF------TNLSRLEIMKCHSM--KDLTWILFAPNLVVLLIEDSREVGEIINKEKATNLT 785
Query: 1038 LLPPNLRI--LLLYKCSKLRIMNYKGFLHLLSLSHLKIYNCPSLERLP 1083
+ P L++ L+LY KL + Y L L + + NCP L +LP
Sbjct: 786 SITPFLKLEWLILYNLPKLESI-YWSPLPFPVLLTMDVSNCPKLRKLP 832
>At1g61300 similar to disease resistance protein
Length = 763
Score = 159 bits (403), Expect = 6e-39
Identities = 188/716 (26%), Positives = 317/716 (44%), Gaps = 75/716 (10%)
Query: 167 QVPIISIVGLGGMGKTTLAKLVYNN-NKIEEHFELKAWVYVSESYDVVGLTKAILKSFNP 225
+V I+ + G+GG+GKTTL K ++N K+ F++ W+ VS+ + L + I + +
Sbjct: 60 RVGIMGLHGMGGVGKTTLFKKIHNKFAKMSSRFDIVIWIVVSKGAKLSKLQEDIAEKLHL 119
Query: 226 SAD---GEYLDQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVT 282
D + + +L GK+++L+LDDIW V+ E + +P+ K+ T
Sbjct: 120 CDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWE-KVDL-EAIGVPYPSEVNKCKVAFT 177
Query: 283 TREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCEG 342
TR+++V + + ++ L D W LF D P + + R++ KC G
Sbjct: 178 TRDQKVCGEMGDHKPM-QVKCLEPEDAWELFKNKVGDNTLRSD-PVIVELAREVAQKCRG 235
Query: 343 LPLAIISLGQLLRKKFSQDEW---MKILETDMWRLSDVDNKINPVLRLSYHNLPSDQ-KR 398
LPLA+ +G+ + K EW + +L S++ NKI P+L+ SY +L + K
Sbjct: 236 LPLALSVIGETMASKTMVQEWEHAIDVLTRSAAEFSNMGNKILPILKYSYDSLGDEHIKS 295
Query: 399 CFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQSFD 458
CF +C++FP+ ++LI W+ EG + K G E+ G L + +
Sbjct: 296 CFLYCALFPEDDEIYNEKLIDYWICEGFIGEDQVIKRARNKGYEMLGTLTLANLLTK--- 352
Query: 459 KTYGTYEHYVMYNLVNDLAKSVSGEFCMQIEG----ARVEGSLERTRH------IRFSLR 508
GT EH VM+++V ++A ++ +F Q E ARV G ER R SL
Sbjct: 353 --VGT-EHVVMHDVVREMALWIASDFGKQKENFVVRARV-GLHERPEAKDWGAVRRMSLM 408
Query: 509 SNCLNKLLETTCELKGLRSLILDVHRGTLISNNVQLDLFSRLNFLRTLSFRWCGLSELVD 568
N + E TCE K L + L + + + + + + LS+ ++L +
Sbjct: 409 DNHIE---EITCESKCSELTTLFLQSNQLKNLSGEFIRYMQKLVVLDLSYN-RDFNKLPE 464
Query: 569 EISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHLELP 628
+IS + L++LDLS T I LP + L L + L S S+L++LR L L
Sbjct: 465 QISGLVSLQFLDLSNTSIKQLPVGLKKLKKLTFLNLAYTVRLCSISGISRLLSLRLLRLL 524
Query: 629 YLK-----KMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDP 683
K + K + KL +LQ L + ++L ++L +L + I+ G++ P
Sbjct: 525 GSKVHGDASVLKELQKLQNLQHLAITL-----SAELSLNQRLANLISILGIE--GFLQKP 577
Query: 684 EDAVTANLKDKKYLEELYM--IFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYR 741
D + L + L L++ ++ K + S + + + +L RL +S+
Sbjct: 578 FD--LSFLASMENLSSLWVKNSYFSEIKCRESETASSYLRINPKIPCFTNLSRLGLSKCH 635
Query: 742 GNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKIIGEELYGN 801
+ WI PNLV L + RE+ +II +E N
Sbjct: 636 SIKDLTWI--LFAPNLVYL---------------YIEDSREVG-------EIINKEKATN 671
Query: 802 NSKIDAFRSLEVLEFQRMENLEE-WLCHEGFLSLKELTIKDCPKLKRALPQHLPSL 856
+ I F LE L + LE + F L + + DCPKL R LP + S+
Sbjct: 672 LTSITPFLKLERLILYNLPKLESIYWSPLHFPRLLIIHVLDCPKL-RKLPLNATSV 726
Score = 42.7 bits (99), Expect = 0.001
Identities = 72/280 (25%), Positives = 112/280 (39%), Gaps = 28/280 (10%)
Query: 815 EFQRMENLEEWLCHEGFLSLKELTIKDCPKLKRALPQHLPSLQKLSIINCNKLE--ASMP 872
+F ++ L FL L +IK LP L L+KL+ +N S+
Sbjct: 458 DFNKLPEQISGLVSLQFLDLSNTSIKQ-------LPVGLKKLKKLTFLNLAYTVRLCSIS 510
Query: 873 EGDNILELCLKGCDSILIKELPTSLKKLVLCEN-RHTEFFVEHILGNNAYLAELC--LDL 929
+L L L + + LK+L +N +H + L N LA L L +
Sbjct: 511 GISRLLSLRLLRLLGSKVHGDASVLKELQKLQNLQHLAITLSAELSLNQRLANLISILGI 570
Query: 930 SGFVECPSLDLRCYNSLRTLSIIGWRSSSLSFSLYLFTNLHSLYLYNCPELVSFPEGGLP 989
GF++ P DL S+ LS + ++S S + S YL P++ F
Sbjct: 571 EGFLQKP-FDLSFLASMENLSSLWVKNSYFSEIKCRESETASSYLRINPKIPCF------ 623
Query: 990 SNLSCFSIFDCPKLIASREEWGLFQLNS----LKEFRVSDEFENVESFPEENLLPPNLRI 1045
+NLS + C + W LF N +++ R E N E + P L++
Sbjct: 624 TNLSRLGLSKCHSI--KDLTWILFAPNLVYLYIEDSREVGEIINKEKATNLTSITPFLKL 681
Query: 1046 --LLLYKCSKLRIMNYKGFLHLLSLSHLKIYNCPSLERLP 1083
L+LY KL + Y LH L + + +CP L +LP
Sbjct: 682 ERLILYNLPKLESI-YWSPLHFPRLLIIHVLDCPKLRKLP 720
>At1g12290 hypothetical protein
Length = 884
Score = 159 bits (401), Expect = 1e-38
Identities = 183/710 (25%), Positives = 312/710 (43%), Gaps = 81/710 (11%)
Query: 141 ESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLGGMGKTTLAKLVYNNNKI---EEH 197
+ +I G++ E+ L+ D GT+ I+ + G+GG+GKTTL L NN+ ++
Sbjct: 153 QPTIVGQETILEKAWDHLM---DDGTK--IMGLYGMGGVGKTTL--LTQINNRFCDTDDG 205
Query: 198 FELKAWVYVSESYDVVGLTKAILKSFNPSADGEYLDQLQHQ----LQHMLMGKKYLLVLD 253
E+ WV VS + + K I + E+ + ++Q + + L K+++L+LD
Sbjct: 206 VEIVIWVVVSGDLQIHKIQKEIGEKIG-FIGVEWNQKSENQKAVDILNFLSKKRFVLLLD 264
Query: 254 DIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHV-VKSTMLCDLRQLVKSDCWRL 312
DIW VE E + +P G KI TTR + V + V M ++R L D W L
Sbjct: 265 DIWK-RVELTE-IGIPNPTSENGCKIAFTTRCQSVCASMGVHDPM--EVRCLGADDAWDL 320
Query: 313 FVTHAFQGKSVCDYPKLESIGRKIMDKCEGLPLAIISLGQLLRKKFSQDEWMKILETDMW 372
F ++ +P + I RK+ C GLPLA+ +G+ + K + EW + ++
Sbjct: 321 FKKKVGD-ITLSSHPDIPEIARKVAQACCGLPLALNVIGETMACKKTTQEWDRAVDVSTT 379
Query: 373 RLSD---VDNKINPVLRLSYHNLPSDQ-KRCFAFCSIFPKGYTFEKDELIKLWMAEGLLK 428
++ V +I P+L+ SY NL S+ K CF +CS+FP+ EK+ LI W+ EG +
Sbjct: 380 YAANFGAVKERILPILKYSYDNLESESVKTCFLYCSLFPEDDLIEKERLIDYWICEGFID 439
Query: 429 CCGSYKSEEEFGNEIFGDLESISFFQQSFDKTYGTYEHYVMYNLVNDLAKSVSGEF---- 484
+ K G EI G L S + + + M+++V ++A ++ +
Sbjct: 440 GDENKKGAVGEGYEILGTLVCASLLVEG--GKFNNKSYVKMHDVVREMALWIASDLRKHK 497
Query: 485 --CMQIEGARVEGSLERTRHIRFSLRSNCLN---KLLETTCELKGLRSLILDVHRGTLIS 539
C+ G R+ + + + + R + +N K + + E L +L L +R +
Sbjct: 498 DNCIVRAGFRL-NEIPKVKDWKVVSRMSLVNNRIKEIHGSPECPKLTTLFLQDNRHLV-- 554
Query: 540 NNVQLDLFSRLNFLRTLSFRW-CGLSELVDEISNIKLLRYLDLSFTEITSLPDSICMLYN 598
N+ + F + L L W LS L D+IS + LRYLDLS++ I LP + L
Sbjct: 555 -NISGEFFRSMPRLVVLDLSWNVNLSGLPDQISELVSLRYLDLSYSSIGRLPVGLLKLKK 613
Query: 599 LQTILLQGCELTELPSNFSKLINLRHLELPYLKK--MPKHIGKLNSLQTLPYFVVEEKNG 656
L + L+ E S L NL+ + L L+ + +L L+ L +E +
Sbjct: 614 LMHLNLESMLCLESVSGIDHLSNLKTVRLLNLRMWLTISLLEELERLENLEVLTIEIISS 673
Query: 657 SDLKEL---EKLNHLHGKICIDGLGYVFDPEDAVTANLKDKKYLEELYMIFYDRKKEVDD 713
S L++L +L K+ + L D E L L E+++ + D
Sbjct: 674 SALEQLLCSHRLVRCLQKVSVKYL----DEESVRILTLPSIGDLREVFIGGCGMR----D 725
Query: 714 SIVESNVSVLEALQPNRSLKRLSISQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPP 773
I+E N S+ PN L ++ I+ G + W+
Sbjct: 726 IIIERNTSLTSPCFPN--LSKVLITGCNGLKDLTWLLFA--------------------- 762
Query: 774 LGQLPSLRELSISNCKRIKIIGEELYGNNSKIDAFRSLEVLEFQRMENLE 823
P+L L++ N ++I+ I + + + I FR LE L + L+
Sbjct: 763 ----PNLTHLNVWNSRQIEEIISQEKASTADIVPFRKLEYLHLWDLPELK 808
>At1g12210 NBS/LRR disease resistance protein
Length = 885
Score = 158 bits (400), Expect = 1e-38
Identities = 225/896 (25%), Positives = 395/896 (43%), Gaps = 109/896 (12%)
Query: 5 VAGAFLQSSFQVIFEKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQII 64
V+G+++Q+ + E LAS+ + DD+ +N EE + ++
Sbjct: 24 VSGSYIQN----LSENLASLQKAMGVLNAKRDDVQGRIN---------REEFTGHRRRLA 70
Query: 65 YVKKWLDKLKHVVYEADQLLDEISTD-------AMLNKLKAESEPLTTNLLGVLGLAEGP 117
V+ WL +++ + + + LL + + +K S ++ +L EG
Sbjct: 71 QVQVWLTRIQTIENQFNDLLSTCNAEIQRLCLCGFCSKNVKMSYLYGKRVIVLLREVEGL 130
Query: 118 SASNEGLVSWKPSKRLSSTALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPIISIVGLG 177
S+ + + + L +S+I G+D +++ L+ +V I+ + G+G
Sbjct: 131 SSQGVFDIVTEAAPIAEVEELPIQSTIVGQDSMLDKVWNCLMED-----KVWIVGLYGMG 185
Query: 178 GMGKTTLAKLVYNN-NKIEEHFELKAWVYVSESYDVVGLTKAI---LKSFNPSADGEYLD 233
G+GKTTL + N +K+ F++ WV VS++ V + K+I L + D + +
Sbjct: 186 GVGKTTLLTQINNKFSKLGGGFDVVIWVVVSKNATVHKIQKSIGEKLGLVGKNWDEKNKN 245
Query: 234 QLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHVV 293
Q + ++L KK++L+LDDIW VE + + +P+ G G K+ TT KEV +
Sbjct: 246 QRALDIHNVLRRKKFVLLLDDIWE-KVEL-KVIGVPYPSGENGCKVAFTTHSKEVCGRMG 303
Query: 294 KSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCEGLPLAIISLGQL 353
+ ++ L + W L + ++ +P + + RK+ +KC GLPLA+ +G+
Sbjct: 304 VDNPM-EISCLDTGNAWDLLKKKVGEN-TLGSHPDIPQLARKVSEKCCGLPLALNVIGET 361
Query: 354 LRKKFSQDEWMKILE--TDMWRLSDVDNKINPVLRLSYHNLPS-DQKRCFAFCSIFPKGY 410
+ K + EW E T S ++++I P+L+ SY +L D K CF +CS+FP+ +
Sbjct: 362 MSFKRTIQEWRHATEVLTSATDFSGMEDEILPILKYSYDSLNGEDAKSCFLYCSLFPEDF 421
Query: 411 TFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIFGDL-ESISFFQQSFDKTYGTYEHYVM 469
K+ LI+ W+ EG +K + G +I G L S + + DK + M
Sbjct: 422 EIRKEMLIEYWICEGFIKEKQGREKAFNQGYDILGTLVRSSLLLEGAKDK-----DVVSM 476
Query: 470 YNLVNDLAKSVSGEFCMQIEGARVEGSL--------ERTRHI-RFSLRSNCLNKLLETTC 520
+++V ++A + + E V+ + E R + R SL +N K+L +
Sbjct: 477 HDMVREMALWIFSDLGKHKERCIVQAGIGLDELPEVENWRAVKRMSLMNNNFEKILGSP- 535
Query: 521 ELKGLRSLILDVHRGTLISNNVQLDLFSRLNFLRTLSF-RWCGLSELVDEISNIKLLRYL 579
E L +L L + + ++ ++ F + L L LSEL +EIS + L+YL
Sbjct: 536 ECVELITLFLQNNYKLV---DISMEFFRCMPSLAVLDLSENHSLSELPEEISELVSLQYL 592
Query: 580 DLSFTEITSLPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHLELPYLKKMPKHIGK 639
DLS T I LP + L L + L+ E S S L +LR L L
Sbjct: 593 DLSGTYIERLPHGLHELRKLVHLKLERTRRLESISGISYLSSLRTLRL------------ 640
Query: 640 LNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPEDAVTANLKDKKYLEE 699
+S TL +++E L+ LE L + I +G +F + + ++
Sbjct: 641 RDSKTTLDTGLMKE-----LQLLEHLELITTDISSGLVGELF-------CYPRVGRCIQH 688
Query: 700 LYMIFYDRKKEVDDSIVESNVSVLEALQP--NRSLKRLSISQYRGNRFPNWIRGCHLPNL 757
+Y+ D + ++S+ V VL A+ S+ + + + P W + PN
Sbjct: 689 IYI--RDHWERPEESV---GVLVLPAIHNLCYISIWNCWMWEIMIEKTP-WKKNLTNPNF 742
Query: 758 VSLQMRHCGLCSHLPPLGQL---PSLRELSISNCKRIK-IIGEELYGNNSKIDAFRSLEV 813
+L C L L L P+L L + CK ++ II +E K + E+
Sbjct: 743 SNLSNVRIEGCDGLKDLTWLLFAPNLINLRVWGCKHLEDIISKE------KAASVLEKEI 796
Query: 814 LEFQRMENLEEWLCHE---------GFLSLKELTI-KDCPKLKRALPQHLPSLQKL 859
L FQ++E L + E F L+ L I +CPKL R LP S+ K+
Sbjct: 797 LPFQKLECLNLYQLSELKSIYWNALPFQRLRCLDILNNCPKL-RKLPLDSKSVVKV 851
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,538,982
Number of Sequences: 26719
Number of extensions: 1173419
Number of successful extensions: 5132
Number of sequences better than 10.0: 312
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 201
Number of HSP's that attempted gapping in prelim test: 3365
Number of HSP's gapped (non-prelim): 1112
length of query: 1104
length of database: 11,318,596
effective HSP length: 110
effective length of query: 994
effective length of database: 8,379,506
effective search space: 8329228964
effective search space used: 8329228964
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC133863.3


