

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.9 - phase: 0 /pseudo
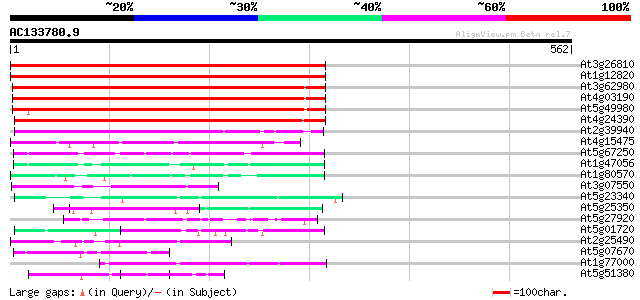
(562 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g26810 transport inhibitor response 1-like protein 505 e-143
At1g12820 transport inhibitor response 1, putative 494 e-140
At3g62980 transport inhibitor response 1 (TIR1) 386 e-107
At4g03190 putative homolog of transport inhibitor response 1 370 e-103
At5g49980 transport inhibitor response 1 protein 307 9e-84
At4g24390 transport inhibitor response-like protein 300 1e-81
At2g39940 unknown protein 167 1e-41
At4g15475 unknown protein 79 5e-15
At5g67250 unknown protein 65 1e-10
At1g47056 hypothetical protein 56 6e-08
At1g80570 unknown protein 54 3e-07
At3g07550 unknown protein 52 7e-07
At5g23340 unknown protein 51 1e-06
At5g25350 leucine-rich repeats containing protein 50 3e-06
At5g27920 unknown protein 49 1e-05
At5g01720 putative F-box protein family, AtFBL3 47 4e-05
At2g25490 putative glucose regulated repressor protein 46 5e-05
At5g07670 unknown protein 45 1e-04
At1g77000 unknown protein 45 1e-04
At5g51380 putative protein 44 2e-04
>At3g26810 transport inhibitor response 1-like protein
Length = 575
Score = 505 bits (1300), Expect = e-143
Identities = 239/316 (75%), Positives = 284/316 (89%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
MNYFPDEVIEHVFD+V SH DRN++SLVCKSWY+IER++RQ+VFIGNCY+I+PERL+ RF
Sbjct: 1 MNYFPDEVIEHVFDFVTSHKDRNAISLVCKSWYKIERYSRQKVFIGNCYAINPERLLRRF 60
Query: 61 PDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELL 120
P LKSLTLKGKPHFADF+LVPH WGGFV PWIEALA+++VGLEELRLKRMVV+DESLELL
Sbjct: 61 PCLKSLTLKGKPHFADFNLVPHEWGGFVLPWIEALARSRVGLEELRLKRMVVTDESLELL 120
Query: 121 SRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
SRSFVNFKSLVLVSCEGFTTDGLA++AANCR LR+LDLQENE++DH+GQWLSCFP++CT+
Sbjct: 121 SRSFVNFKSLVLVSCEGFTTDGLASIAANCRHLRDLDLQENEIDDHRGQWLSCFPDTCTT 180
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
LV+LNFACL+G+ NL ALERLV+RSPNLKSL+LNR+VP+DAL R++ APQ++DLG+GS+
Sbjct: 181 LVTLNFACLEGETNLVALERLVARSPNLKSLKLNRAVPLDALARLMACAPQIVDLGVGSY 240
Query: 241 FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGI 300
+D +S++Y A I KC S+ SLSGFLE AP L+A +PIC NLTSLNLSYAA I G
Sbjct: 241 ENDPDSESYLKLMAVIKKCTSLRSLSGFLEAAPHCLSAFHPICHNLTSLNLSYAAEIHGS 300
Query: 301 ELIKLIRHCGKLQRLW 316
LIKLI+HC KLQRLW
Sbjct: 301 HLIKLIQHCKKLQRLW 316
>At1g12820 transport inhibitor response 1, putative
Length = 577
Score = 494 bits (1273), Expect = e-140
Identities = 235/316 (74%), Positives = 281/316 (88%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
MNYFPDEVIEHVFD+V SH DRNS+SLVCKSW++IERF+R+ VFIGNCY+I+PERL+ RF
Sbjct: 1 MNYFPDEVIEHVFDFVASHKDRNSISLVCKSWHKIERFSRKEVFIGNCYAINPERLIRRF 60
Query: 61 PDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELL 120
P LKSLTLKGKPHFADF+LVPH WGGFV+PWIEALA+++VGLEELRLKRMVV+DESL+LL
Sbjct: 61 PCLKSLTLKGKPHFADFNLVPHEWGGFVHPWIEALARSRVGLEELRLKRMVVTDESLDLL 120
Query: 121 SRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
SRSF NFKSLVLVSCEGFTTDGLA++AANCR LRELDLQENE++DH+GQWL+CFP+SCT+
Sbjct: 121 SRSFANFKSLVLVSCEGFTTDGLASIAANCRHLRELDLQENEIDDHRGQWLNCFPDSCTT 180
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
L+SLNFACLKG+ N+ ALERLV+RSPNLKSL+LNR+VP+DAL R+++ APQL+DLG+GS+
Sbjct: 181 LMSLNFACLKGETNVAALERLVARSPNLKSLKLNRAVPLDALARLMSCAPQLVDLGVGSY 240
Query: 241 FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGI 300
++ + +++A I K S+ SLSGFLEVAP L A YPICQNL SLNLSYAA I G
Sbjct: 241 ENEPDPESFAKLMTAIKKYTSLRSLSGFLEVAPLCLPAFYPICQNLISLNLSYAAEIQGN 300
Query: 301 ELIKLIRHCGKLQRLW 316
LIKLI+ C +LQRLW
Sbjct: 301 HLIKLIQLCKRLQRLW 316
Score = 30.4 bits (67), Expect = 2.7
Identities = 40/176 (22%), Positives = 74/176 (41%), Gaps = 19/176 (10%)
Query: 142 GLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERL 201
GLA VAA C+ L+EL + ++V + S + LV+++ C K L ++
Sbjct: 325 GLAVVAATCKELQELRVFPSDVHGEEDNNASV---TEVGLVAISAGCPK----LHSILYF 377
Query: 202 VSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKS 261
+ N + + ++ P R+ P D H F A + CK
Sbjct: 378 CKQMTNAALIAVAKNCPNFIRFRLCILEPHKPD-------HITFQSLDEGFGAIVQACKG 430
Query: 262 I--TSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
+ S+SG L F +Y + L L++++ AG ++ ++ C K+++L
Sbjct: 431 LRRLSVSGLLTDQVFLYIGMY--AEQLEMLSIAF-AGDTDKGMLYVLNGCKKMRKL 483
>At3g62980 transport inhibitor response 1 (TIR1)
Length = 594
Score = 386 bits (992), Expect = e-107
Identities = 180/313 (57%), Positives = 241/313 (76%), Gaps = 1/313 (0%)
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP+EV+EHVF ++ DRNS+SLVCKSWY IER+ R++VFIGNCY++SP ++ RFP +
Sbjct: 9 FPEEVLEHVFSFIQLDKDRNSVSLVCKSWYEIERWCRRKVFIGNCYAVSPATVIRRFPKV 68
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRS 123
+S+ LKGKPHFADF+LVP GWGG+VYPWIEA++ + LEE+RLKRMVV+D+ LEL+++S
Sbjct: 69 RSVELKGKPHFADFNLVPDGWGGYVYPWIEAMSSSYTWLEEIRLKRMVVTDDCLELIAKS 128
Query: 124 FVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVS 183
F NFK LVL SCEGF+TDGLAA+AA CR+L+ELDL+E++V+D G WLS FP++ TSLVS
Sbjct: 129 FKNFKVLVLSSCEGFSTDGLAAIAATCRNLKELDLRESDVDDVSGHWLSHFPDTYTSLVS 188
Query: 184 LNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
LN +CL +++ ALERLV+R PNLKSL+LNR+VP++ L +L RAPQL +LG G + +
Sbjct: 189 LNISCLASEVSFSALERLVTRCPNLKSLKLNRAVPLEKLATLLQRAPQLEELGTGGYTAE 248
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
+ D Y+ + CK + LSGF + P L A+Y +C LT+LNLSYA + +L+
Sbjct: 249 VRPDVYSGLSVALSGCKELRCLSGFWDAVPAYLPAVYSVCSRLTTLNLSYAT-VQSYDLV 307
Query: 304 KLIRHCGKLQRLW 316
KL+ C KLQRLW
Sbjct: 308 KLLCQCPKLQRLW 320
Score = 28.9 bits (63), Expect = 7.9
Identities = 45/182 (24%), Positives = 74/182 (39%), Gaps = 32/182 (17%)
Query: 142 GLAAVAANCRSLRELDLQENE---VEDHKG---QWLSCFPESCTSLVSLNFACLKGDINL 195
GL +A+ C+ LREL + +E +E + Q L C L S+ + C + +
Sbjct: 329 GLEVLASTCKDLRELRVFPSEPFVMEPNVALTEQGLVSVSMGCPKLESVLYFCRQ--MTN 386
Query: 196 GALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKAT 255
AL + PN+ RL P LT P +D+G G A
Sbjct: 387 AALITIARNRPNMTRFRLCIIEP--KAPDYLTLEP--LDIGFG---------------AI 427
Query: 256 ILKCKSI--TSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQ 313
+ CK + SLSG L F Y + + L++++ AG + + ++ C L+
Sbjct: 428 VEHCKDLRRLSLSGLLTDKVFEYIGTY--AKKMEMLSVAF-AGDSDLGMHHVLSGCDSLR 484
Query: 314 RL 315
+L
Sbjct: 485 KL 486
>At4g03190 putative homolog of transport inhibitor response 1
Length = 585
Score = 370 bits (951), Expect = e-103
Identities = 178/313 (56%), Positives = 234/313 (73%), Gaps = 1/313 (0%)
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP +V+EH+ ++ S+ DRNS+SLVCKSW+ ER TR+RVF+GNCY++SP + RFP++
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRS 123
+SLTLKGKPHFAD++LVP GWGG+ +PWIEA+A LEE+R+KRMVV+DE LE ++ S
Sbjct: 65 RSLTLKGKPHFADYNLVPDGWGGYAWPWIEAMAAKSSSLEEIRMKRMVVTDECLEKIAAS 124
Query: 124 FVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVS 183
F +FK LVL SCEGF+TDG+AA+AA CR+LR L+L+E VED G WLS FPES TSLVS
Sbjct: 125 FKDFKVLVLTSCEGFSTDGIAAIAATCRNLRVLELRECIVEDLGGDWLSYFPESSTSLVS 184
Query: 184 LNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
L+F+CL ++ + LERLVSRSPNLKSL+LN +V +D L +L APQL +LG GSF
Sbjct: 185 LDFSCLDSEVKISDLERLVSRSPNLKSLKLNPAVTLDGLVSLLRCAPQLTELGTGSFAAQ 244
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
L +A++ CK + SLSG +V P L A+Y +C LTSLNLSYA + +L+
Sbjct: 245 LKPEAFSKLSEAFSNCKQLQSLSGLWDVLPEYLPALYSVCPGLTSLNLSYAT-VRMPDLV 303
Query: 304 KLIRHCGKLQRLW 316
+L+R C KLQ+LW
Sbjct: 304 ELLRRCSKLQKLW 316
Score = 33.1 bits (74), Expect = 0.42
Identities = 36/129 (27%), Positives = 53/129 (40%), Gaps = 10/129 (7%)
Query: 91 WIEALAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANC 150
++ AL GL L L V L L R + L ++ GL AVA+ C
Sbjct: 276 YLPALYSVCPGLTSLNLSYATVRMPDLVELLRRCSKLQKLWVMDL--IEDKGLEAVASYC 333
Query: 151 RSLRELDLQENEVE------DHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSR 204
+ LREL + +E + Q L + C L S+ + C++ AL + +
Sbjct: 334 KELRELRVFPSEPDLDATNIPLTEQGLVFVSKGCRKLESVLYFCVQ--FTNAALFTIARK 391
Query: 205 SPNLKSLRL 213
PNLK RL
Sbjct: 392 RPNLKCFRL 400
>At5g49980 transport inhibitor response 1 protein
Length = 619
Score = 307 bits (787), Expect = 9e-84
Identities = 165/320 (51%), Positives = 210/320 (65%), Gaps = 9/320 (2%)
Query: 4 FPDEVIEHVFDYVV----SHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVER 59
FPD V+E+V + V+ S DRN+ SLVCKSW+R+E TR VFIGNCY++SP RL +R
Sbjct: 50 FPDHVLENVLENVLQFLDSRCDRNAASLVCKSWWRVEALTRSEVFIGNCYALSPARLTQR 109
Query: 60 FPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLEL 119
F ++SL LKGKP FADF+L+P WG PW+ +A+ LE++ LKRM V+D+ L L
Sbjct: 110 FKRVRSLVLKGKPRFADFNLMPPDWGANFAPWVSTMAQAYPCLEKVDLKRMFVTDDDLAL 169
Query: 120 LSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCT 179
L+ SF FK L+LV CEGF T G++ VA CR L+ LDL E+EV D + W+SCFPE T
Sbjct: 170 LADSFPGFKELILVCCEGFGTSGISIVANKCRKLKVLDLIESEVTDDEVDWISCFPEDVT 229
Query: 180 SLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGS 239
L SL F C++ IN ALE LV+RSP LK LRLNR V + L R+L APQL LG GS
Sbjct: 230 CLESLAFDCVEAPINFKALEGLVARSPFLKKLRLNRFVSLVELHRLLLGAPQLTSLGTGS 289
Query: 240 FFHD--LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGI 297
F HD S+ + A CKS+ LSGF E+ P L AI+P+C NLTSLN SYA
Sbjct: 290 FSHDEEPQSEQEPDYAAAFRACKSVVCLSGFRELMPEYLPAIFPVCANLTSLNFSYAN-- 347
Query: 298 LGIELIK-LIRHCGKLQRLW 316
+ ++ K +I +C KLQ W
Sbjct: 348 ISPDMFKPIILNCHKLQVFW 367
Score = 34.7 bits (78), Expect = 0.14
Identities = 46/195 (23%), Positives = 74/195 (37%), Gaps = 31/195 (15%)
Query: 128 KSLVLVSCEGFTTDGLAAVAANCRSLREL-----DLQENEVEDHKGQWLSCFPESCTSLV 182
K V + + +GL AVAA C+ LREL D +E+ L E C L
Sbjct: 362 KLQVFWALDSICDEGLQAVAATCKELRELRIFPFDPREDSEGPVSELGLQAISEGCRKLE 421
Query: 183 SLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFH 242
S+ + C R N + ++ + P + R+ D H
Sbjct: 422 SILYFC--------------QRMTNAAVIAMSENCPELTVFRLCIMGRHRPD-------H 460
Query: 243 DLNSDAYAMFKATILKCKSITSL--SGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGI 300
F A + CK +T L SG L F Y + + +L++++ AG +
Sbjct: 461 VTGKPMDEGFGAIVKNCKKLTRLAVSGLLTDQAFRYMGEYG--KLVRTLSVAF-AGDSDM 517
Query: 301 ELIKLIRHCGKLQRL 315
L ++ C +LQ+L
Sbjct: 518 ALRHVLEGCPRLQKL 532
>At4g24390 transport inhibitor response-like protein
Length = 623
Score = 300 bits (769), Expect = 1e-81
Identities = 160/313 (51%), Positives = 200/313 (63%), Gaps = 3/313 (0%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
+ V+E+V ++ S DRN++SLVC+SWYR+E TR VFIGNCYS+SP RL+ RF ++S
Sbjct: 56 ENVLENVLQFLTSRCDRNAVSLVCRSWYRVEAQTRLEVFIGNCYSLSPARLIHRFKRVRS 115
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFV 125
L LKGKP FADF+L+P WG PW+ A AK LE++ LKRM V+D+ L LL+ SF
Sbjct: 116 LVLKGKPRFADFNLMPPNWGAQFSPWVAATAKAYPWLEKVHLKRMFVTDDDLALLAESFP 175
Query: 126 NFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLN 185
FK L LV CEGF T G+A VA CR L+ LDL E+EV D + W+SCFPE T L SL+
Sbjct: 176 GFKELTLVCCEGFGTSGIAIVANKCRQLKVLDLMESEVTDDELDWISCFPEGETHLESLS 235
Query: 186 FACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD-- 243
F C++ IN ALE LV RSP LK LR NR V ++ L R++ RAPQL LG GSF D
Sbjct: 236 FDCVESPINFKALEELVVRSPFLKKLRTNRFVSLEELHRLMVRAPQLTSLGTGSFSPDNV 295
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
+ + A CKSI LSGF E P L AI +C NLTSLN SY A I L
Sbjct: 296 PQGEQQPDYAAAFRACKSIVCLSGFREFRPEYLLAISSVCANLTSLNFSY-ANISPHMLK 354
Query: 304 KLIRHCGKLQRLW 316
+I +C ++ W
Sbjct: 355 PIISNCHNIRVFW 367
Score = 41.6 bits (96), Expect = 0.001
Identities = 63/243 (25%), Positives = 98/243 (39%), Gaps = 39/243 (16%)
Query: 83 GWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDG 142
G+ F ++ A++ L L +S L+ + + N + V + + +G
Sbjct: 319 GFREFRPEYLLAISSVCANLTSLNFSYANISPHMLKPIISNCHNIR--VFWALDSIRDEG 376
Query: 143 LAAVAANCRSLREL-----DLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGA 197
L AVAA C+ LREL D +E+ G L E C L S+ + C ++ GA
Sbjct: 377 LQAVAATCKELRELRIFPFDPREDSEGPVSGVGLQAISEGCRKLESILYFC--QNMTNGA 434
Query: 198 LERLVSRSPNLKSLRL---NRSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKA 254
+ + P L RL R P +T P MD G G A
Sbjct: 435 VTAMSENCPQLTVFRLCIMGRHRPDH-----VTGKP--MDDGFG---------------A 472
Query: 255 TILKCKSITSL--SGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKL 312
+ CK +T L SG L FS Y + + +L++++ AG L ++ C KL
Sbjct: 473 IVKNCKKLTRLAVSGLLTDEAFSYIGEYG--KLIRTLSVAF-AGNSDKALRYVLEGCPKL 529
Query: 313 QRL 315
Q+L
Sbjct: 530 QKL 532
>At2g39940 unknown protein
Length = 592
Score = 167 bits (424), Expect = 1e-41
Identities = 100/311 (32%), Positives = 166/311 (53%), Gaps = 14/311 (4%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
D+VIE V Y+ DR+S SLVC+ W++I+ TR+ V + CY+ +P+RL RFP+L+S
Sbjct: 18 DDVIEQVMTYITDPKDRDSASLVCRRWFKIDSETREHVTMALCYTATPDRLSRRFPNLRS 77
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFV 125
L LKGKP A F+L+P WGG+V PW+ ++ N L+ + +RM+VSD L+ L+++
Sbjct: 78 LKLKGKPRAAMFNLIPENWGGYVTPWVTEISNNLRQLKSVHFRRMIVSDLDLDRLAKARA 137
Query: 126 -NFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSL 184
+ ++L L C GFTTDGL ++ +CR ++ L ++E+ + G+WL + TSL L
Sbjct: 138 DDLETLKLDKCSGFTTDGLLSIVTHCRKIKTLLMEESSFSEKDGKWLHELAQHNTSLEVL 197
Query: 185 NFACLK-GDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
NF + I+ LE + +L S+++ + L A L + GS D
Sbjct: 198 NFYMTEFAKISPKDLETIARNCRSLVSVKVG-DFEILELVGFFKAAANLEEFCGGSLNED 256
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
+ M ++ + + L G + P + ++P + L+L YA L+
Sbjct: 257 IGMPEKYM---NLVFPRKLCRL-GLSYMGPNEMPILFPFAAQIRKLDLLYA-------LL 305
Query: 304 KLIRHCGKLQR 314
+ HC +Q+
Sbjct: 306 ETEDHCTLIQK 316
>At4g15475 unknown protein
Length = 610
Score = 79.3 bits (194), Expect = 5e-15
Identities = 83/309 (26%), Positives = 138/309 (43%), Gaps = 35/309 (11%)
Query: 2 NYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVE--- 58
N P+E+I +F + S +R++ SLVCK W +ERF+R + IG S SP+ +
Sbjct: 9 NCLPEELILEIFRRLESKPNRDACSLVCKRWLSLERFSRTTLRIGA--SFSPDDFISLLS 66
Query: 59 -RFPDLKSLTLKGKPHFADFSLVPH--------GWGGFVYPWIEALAKNKVGLEELRLKR 109
RF + S+ + + + SL P + K G E +
Sbjct: 67 RRFLYITSIHVDERISVSLPSLSPSPKRKRGRDSSSPSSSKRKKLTDKTHSGAENVESSS 126
Query: 110 MVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQ 169
+ +D L L+ F ++L L+ C ++ GL ++A C SL+ LDLQ V D Q
Sbjct: 127 L--TDTGLTALANGFPRIENLSLIWCPNVSSVGLCSLAQKCTSLKSLDLQGCYVGD---Q 181
Query: 170 WLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVD--ALQRILT 227
L+ + C L LN +G ++G ++ +V S +LKS+ + S + +L+ + +
Sbjct: 182 GLAAVGKFCKQLEELNLRFCEGLTDVGVIDLVVGCSKSLKSIGVAASAKITDLSLEAVGS 241
Query: 228 RAPQLMDLGIGS-FFHDLNSDAYAM----FKATILKCKSITSLSGFLEVAPFSLAAIYPI 282
L L + S + HD A A K L+C S+T + + AA+ +
Sbjct: 242 HCKLLEVLYLDSEYIHDKGLIAVAQGCHRLKNLKLQCVSVTDV---------AFAAVGEL 292
Query: 283 CQNLTSLNL 291
C +L L L
Sbjct: 293 CTSLERLAL 301
Score = 38.9 bits (89), Expect = 0.008
Identities = 50/229 (21%), Positives = 98/229 (41%), Gaps = 13/229 (5%)
Query: 92 IEALAKNKVGLEELRLKRMV-VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANC 150
IEA+ K+ L+EL L + + +L+ + + + + L LV C G + ++A C
Sbjct: 364 IEAIGKSCPRLKELALLYCQRIGNSALQEIGKGCKSLEILHLVDCSGIGDIAMCSIAKGC 423
Query: 151 RSLRELDLQENEVEDHKGQWLSCFPESCTSL--VSLNFACLKGDINLGALERLVSRSPNL 208
R+L++L ++ +KG + + C SL +SL F G+ L A + + +L
Sbjct: 424 RNLKKLHIRRCYEIGNKG--IISIGKHCKSLTELSLRFCDKVGNKALIA----IGKGCSL 477
Query: 209 KSLRLN--RSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLS 266
+ L ++ + + I PQL L I S ++ A K + LS
Sbjct: 478 QQLNVSGCNQISDAGITAIARGCPQLTHLDI-SVLQNIGDMPLAELGEGCPMLKDLV-LS 535
Query: 267 GFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
+ L + C+ L + ++ Y GI + ++ C ++++
Sbjct: 536 HCHHITDNGLNHLVQKCKLLETCHMVYCPGITSAGVATVVSSCPHIKKV 584
Score = 32.3 bits (72), Expect = 0.72
Identities = 19/55 (34%), Positives = 27/55 (48%), Gaps = 1/55 (1%)
Query: 258 KCKSITSLS-GFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGK 311
KC S+ SL V LAA+ C+ L LNL + G+ + +I L+ C K
Sbjct: 164 KCTSLKSLDLQGCYVGDQGLAAVGKFCKQLEELNLRFCEGLTDVGVIDLVVGCSK 218
>At5g67250 unknown protein
Length = 527
Score = 64.7 bits (156), Expect = 1e-10
Identities = 81/319 (25%), Positives = 136/319 (42%), Gaps = 27/319 (8%)
Query: 5 PDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISP--ERLVERFPD 62
PDE + HVF ++ + DR SLVCK W ++ +R R+ + IS + RF
Sbjct: 47 PDECLAHVFQFLGA-GDRKRCSLVCKRWLLVDGQSRHRLSLDAKDEISSFLTSMFNRFDS 105
Query: 63 LKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSR 122
+ L L+ D V + L +V L R ++D +E ++
Sbjct: 106 VTKLALR-----CDRKSVSLSDEALAMISVRCLNLTRVKLRGCR----EITDLGMEDFAK 156
Query: 123 SFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLV 182
+ N K L + SC F G+ A+ +C+ L EL ++ H+ L P+ +S
Sbjct: 157 NCKNLKKLSVGSC-NFGAKGVNAMLEHCKLLEELSVKRLR-GIHEAAELIHLPDDASS-S 213
Query: 183 SLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVP--VDALQRILTRAPQLMDLGIGSF 240
SL CLK +N E L++ + LK+L++ R + LQ I L ++
Sbjct: 214 SLRSICLKELVNGQVFEPLLATTRTLKTLKIIRCLGDWDKVLQMIANGKSSLSEI----- 268
Query: 241 FHDLNSDAYAMFKATILKCKSITSLS--GFLEVAPFSLAAIYPICQNLTSLNL-SYAAGI 297
H + + I KC ++ +L E + F L + C+ L L++ +
Sbjct: 269 -HLERLQVSDIGLSAISKCSNVETLHIVKTPECSNFGLIYVAERCKLLRKLHIDGWRTNR 327
Query: 298 LGIE-LIKLIRHCGKLQRL 315
+G E L+ + +HC LQ L
Sbjct: 328 IGDEGLLSVAKHCLNLQEL 346
>At1g47056 hypothetical protein
Length = 518
Score = 55.8 bits (133), Expect = 6e-08
Identities = 85/327 (25%), Positives = 133/327 (39%), Gaps = 44/327 (13%)
Query: 5 PDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSI--SPERLVERFPD 62
PDE + VF ++ S +R +LVC+ W +E R R+ + + S L RF
Sbjct: 44 PDECLALVFQFLNS-GNRKRCALVCRRWMIVEGQNRYRLSLHARSDLITSIPSLFSRFDS 102
Query: 63 LKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSR 122
+ L+LK D V G EAL K + L+ ++ E ++
Sbjct: 103 VTKLSLK-----CDRRSVSIG--------DEALVKISLRCRNLKRLKLRACRELTDVGMA 149
Query: 123 SFV-NFKSLVLVSCEG--FTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCT 179
+F N K L + SC F G+ AV +C +L EL ++ L F +
Sbjct: 150 AFAENCKDLKIFSCGSCDFGAKGVKAVLDHCSNLEELSIKR----------LRGFTDIAP 199
Query: 180 SLV-------SLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQL 232
++ SL CLK N ++ + NLKSL+L R +L +
Sbjct: 200 EMIGPGVAASSLKSICLKELYNGQCFGPVIVGAKNLKSLKLFR---CSGDWDLLLQEMSG 256
Query: 233 MDLGIGSFFHDLNSDAYAMFKATILKCKSITSLS--GFLEVAPFSLAAIYPICQNLTSLN 290
D G+ H + + I C S+ SL E F LAAI C+ L L+
Sbjct: 257 KDHGVVE-IHLERMQVSDVALSAISYCSSLESLHLVKTPECTNFGLAAIAEKCKRLRKLH 315
Query: 291 L-SYAAGILGIE-LIKLIRHCGKLQRL 315
+ + A ++G E L+ + + C +LQ L
Sbjct: 316 IDGWKANLIGDEGLVAVAKFCSQLQEL 342
Score = 41.6 bits (96), Expect = 0.001
Identities = 45/174 (25%), Positives = 77/174 (43%), Gaps = 19/174 (10%)
Query: 101 GLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQE 160
G+ E+ L+RM VSD +L +S + +SL LV T GLAA+A C+ LR+L +
Sbjct: 260 GVVEIHLERMQVSDVALSAISYC-SSLESLHLVKTPECTNFGLAAIAEKCKRLRKLHIDG 318
Query: 161 NEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNR----- 215
+ + L + C+ L L + + +L L ++ NL+ L L
Sbjct: 319 WKANLIGDEGLVAVAKFCSQLQEL--VLIGVNPTTLSLGMLAAKCLNLERLALCGCDTFG 376
Query: 216 -------SVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSI 262
+ AL+++ + + D+GI +L + + K I KCK +
Sbjct: 377 DPELSCIAAKCPALRKLCIKNCPISDVGI----ENLANGCPGLTKVKIKKCKGV 426
Score = 37.0 bits (84), Expect = 0.029
Identities = 23/72 (31%), Positives = 39/72 (53%)
Query: 94 ALAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSL 153
A+AK L+EL L + + SL +L+ +N + L L C+ F L+ +AA C +L
Sbjct: 331 AVAKFCSQLQELVLIGVNPTTLSLGMLAAKCLNLERLALCGCDTFGDPELSCIAAKCPAL 390
Query: 154 RELDLQENEVED 165
R+L ++ + D
Sbjct: 391 RKLCIKNCPISD 402
>At1g80570 unknown protein
Length = 467
Score = 53.5 bits (127), Expect = 3e-07
Identities = 81/329 (24%), Positives = 132/329 (39%), Gaps = 51/329 (15%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPE----RL 56
M+ PD ++ + + + DRNSLSL CK ++ ++ R + IG C + L
Sbjct: 1 MDELPDHLVWDILSKLHTTDDRNSLSLSCKRFFSLDNEQRYSLRIG-CGLVPASDALLSL 59
Query: 57 VERFPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIE-----ALAKNKVGLEELRLKRMV 111
RFP+L + ++ GW + ++ L N L +L L
Sbjct: 60 CRRFPNLSKV-----------EIIYSGWMSKLGKQVDDQGLLVLTTNCHSLTDLTLSFCT 108
Query: 112 -VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQW 170
++D + LS S SL L T G+ ++A C+ LR L L + +W
Sbjct: 109 FITDVGIGHLS-SCPELSSLKLNFAPRITGCGVLSLAVGCKKLRRLHLIRC-LNVASVEW 166
Query: 171 LSCFPESCTSLVSLNFACLKG--DINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTR 228
L F + L +L C+K I G L +L + L SL+ VDA R +
Sbjct: 167 LEYFGK----LETLEELCIKNCRAIGEGDLIKLRNSWRKLTSLQFE----VDANYRYMKV 218
Query: 229 APQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLS-GFLEVAP-FSLAAIYPICQNL 286
QL + ++ C S+ LS G +AP LA + C+NL
Sbjct: 219 YDQL---------------DVERWPKQLVPCDSLVELSLGNCIIAPGRGLACVLRNCKNL 263
Query: 287 TSLNLSYAAGILGIELIKLIRHCGKLQRL 315
L+L G+ ++I L++ L+ +
Sbjct: 264 EKLHLDMCTGVSDSDIIALVQKASHLRSI 292
>At3g07550 unknown protein
Length = 395
Score = 52.4 bits (124), Expect = 7e-07
Identities = 49/209 (23%), Positives = 87/209 (41%), Gaps = 27/209 (12%)
Query: 3 YFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSI-SPERLVERFP 61
+ PD+ + +F + S +D +S L C W I+ +R+ + +S+ +P L + P
Sbjct: 17 HLPDDCLSFIFQRLDSVADHDSFGLTCHRWLNIQNISRRSLQFQCSFSVLNPSSLSQTNP 76
Query: 62 DLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRM-VVSDESLELL 120
D+ S H L W LE L L V++D SL+ L
Sbjct: 77 DVSS-------HHLHRLLTRFQW-----------------LEHLSLSGCTVLNDSSLDSL 112
Query: 121 SRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
+L L C G + DG++ +A+ C +L + L + D + L+ S
Sbjct: 113 RYPGARLHTLYLDCCFGISDDGISTIASFCPNLSVVSLYRCNISDIGLETLARASLS-LK 171
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLK 209
V+L++ L D + AL + + ++K
Sbjct: 172 CVNLSYCPLVSDFGIKALSQACLQLESVK 200
>At5g23340 unknown protein
Length = 405
Score = 51.2 bits (121), Expect = 1e-06
Identities = 76/344 (22%), Positives = 133/344 (38%), Gaps = 60/344 (17%)
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
D+ + V + S D+ LVCK W ++ D K
Sbjct: 12 DDELRWVLSRLDSDKDKEVFGLVCKRWLNLQS-----------------------TDRKK 48
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMV-------VSDESLE 118
L + PH + LA + EL L + + V+D L
Sbjct: 49 LAARAGPHM-----------------LRRLASRFTQIVELDLSQSISRSFYPGVTDSDLA 91
Query: 119 LLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESC 178
++S F + L L +C+G T GLA++ L+ LD+ KG LS E C
Sbjct: 92 VISEGFKFLRVLNLHNCKGITDTGLASIGRCLSLLQFLDVSYCRKLSDKG--LSAVAEGC 149
Query: 179 TSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLN--RSVPVDALQRILTRAPQLMDLG 236
L +L+ A + I +L+ L R +L++L L ++ L ++ ++ L
Sbjct: 150 HDLRALHLAGCR-FITDESLKSLSERCRDLEALGLQGCTNITDSGLADLVKGCRKIKSLD 208
Query: 237 IGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAG 296
I + ++ ++ KA K++ L + +V S++++ C+NL +L +
Sbjct: 209 INKCSNVGDAGVSSVAKACASSLKTLKLLDCY-KVGNESISSLAQFCKNLETLIIGGCRD 267
Query: 297 ILGIELIKLIRHC-GKLQRLWWD*VL*LAH------VKNCKN*E 333
I ++ L C L+ L D L ++ +K CKN E
Sbjct: 268 ISDESIMLLADSCKDSLKNLRMDWCLNISDSSLSCILKQCKNLE 311
Score = 39.3 bits (90), Expect = 0.006
Identities = 44/216 (20%), Positives = 92/216 (42%), Gaps = 45/216 (20%)
Query: 112 VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWL 171
++DESL+ LS + ++L L C T GLA + CR ++ LD+ +
Sbjct: 163 ITDESLKSLSERCRDLEALGLQGCTNITDSGLADLVKGCRKIKSLDINK----------- 211
Query: 172 SCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPV--DALQRILTRA 229
C+++ GD + ++ + + S LK+L+L V +++ +
Sbjct: 212 ------CSNV---------GDAGVSSVAKACASS--LKTLKLLDCYKVGNESISSLAQFC 254
Query: 230 PQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSL 289
L L IG D++ ++ + + + L ++ SL+ I C+NL +L
Sbjct: 255 KNLETLIIGG-CRDISDESIMLLADSCKDSLKNLRMDWCLNISDSSLSCILKQCKNLEAL 313
Query: 290 NLSY-------------AAGILGIELIKLIRHCGKL 312
++ + +LG++++K + +C K+
Sbjct: 314 DIGCCEEVTDTAFRDLGSDDVLGLKVLK-VSNCTKI 348
>At5g25350 leucine-rich repeats containing protein
Length = 623
Score = 50.1 bits (118), Expect = 3e-06
Identities = 49/177 (27%), Positives = 73/177 (40%), Gaps = 31/177 (17%)
Query: 45 IGNCYSISPERLVERFPD-----LKSLTLKGKPHFADFSLV-------------PHGWGG 86
+ NC IS P L+SL+++ P F D SL G G
Sbjct: 411 LANCLGISDFNSESSLPSPSCSSLRSLSIRCCPGFGDASLAFLGKFCHQLQDVELCGLNG 470
Query: 87 FVYPWI-EALAKNKVGLEELRLKRMV-VSDESLELLSRSFVN-FKSLVLVSCEGFTTDGL 143
+ E L N VGL ++ L + VSD ++ +S +SL L C+ T L
Sbjct: 471 VTDAGVRELLQSNNVGLVKVNLSECINVSDNTVSAISVCHGRTLESLNLDGCKNITNASL 530
Query: 144 AAVAANCRSLRELDLQENEVEDHKGQWLSCFPE----------SCTSLVSLNFACLK 190
AVA NC S+ +LD+ V DH + L+ P C+S+ + AC++
Sbjct: 531 VAVAKNCYSVNDLDISNTLVSDHGIKALASSPNHLNLQVLSIGGCSSITDKSKACIQ 587
Score = 44.3 bits (103), Expect = 2e-04
Identities = 68/284 (23%), Positives = 110/284 (37%), Gaps = 46/284 (16%)
Query: 61 PDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMV-VSDESLEL 119
P ++ L L P D LV A+A+N V L +L + V +E L
Sbjct: 193 PMIEKLDLSRCPGITDSGLV-------------AIAENCVNLSDLTIDSCSGVGNEGLRA 239
Query: 120 LSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRS-LRELDLQENEVE-------------- 164
++R VN +S+ + SC G+A + A S L ++ LQ V
Sbjct: 240 IARRCVNLRSISIRSCPRIGDQGVAFLLAQAGSYLTKVKLQMLNVSGLSLAVIGHYGAAV 299
Query: 165 -----------DHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRL 213
+ KG W+ + L SL+ +G ++G LE + + P+LK + L
Sbjct: 300 TDLVLHGLQGVNEKGFWVMGNAKGLKKLKSLSVMSCRGMTDVG-LEAVGNGCPDLKHVSL 358
Query: 214 NRSVPVD--ALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEV 271
N+ + V L + A L L + H +N F SL+ L +
Sbjct: 359 NKCLLVSGKGLVALAKSALSLESLKLEE-CHRINQFGLMGFLMNCGSKLKAFSLANCLGI 417
Query: 272 APFSLAAIY--PICQNLTSLNLSYAAGILGIELIKLIRHCGKLQ 313
+ F+ + P C +L SL++ G L L + C +LQ
Sbjct: 418 SDFNSESSLPSPSCSSLRSLSIRCCPGFGDASLAFLGKFCHQLQ 461
Score = 39.7 bits (91), Expect = 0.004
Identities = 26/100 (26%), Positives = 47/100 (47%), Gaps = 14/100 (14%)
Query: 61 PDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMV-VSDESLEL 119
P L+ ++L P +D L +A++ +E+L L R ++D L
Sbjct: 167 PSLRIVSLWNLPAVSDLGL-------------SEIARSCPMIEKLDLSRCPGITDSGLVA 213
Query: 120 LSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQ 159
++ + VN L + SC G +GL A+A C +LR + ++
Sbjct: 214 IAENCVNLSDLTIDSCSGVGNEGLRAIARRCVNLRSISIR 253
Score = 33.9 bits (76), Expect = 0.25
Identities = 50/223 (22%), Positives = 91/223 (40%), Gaps = 26/223 (11%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRI-ERFTRQRVFIGNCYSISPERLVER 59
++ P+E + + + S +R++ + V K W + +R V + S + VE
Sbjct: 55 IDVLPEECLFEILRRLPSGQERSACACVSKHWLNLLSSISRSEV------NESSVQDVEE 108
Query: 60 FPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLK----RMVVSDE 115
S +L+GK D L G ++ GL +L+++ V+D
Sbjct: 109 GEGFLSRSLEGKKA-TDLRLAAIAVG----------TSSRGGLGKLQIRGSGFESKVTDV 157
Query: 116 SLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFP 175
L ++ + + + L + + GL+ +A +C + +LDL G L
Sbjct: 158 GLGAVAHGCPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSRCPGITDSG--LVAIA 215
Query: 176 ESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVP 218
E+C +L L G N G L + R NL+S+ + RS P
Sbjct: 216 ENCVNLSDLTIDSCSGVGNEG-LRAIARRCVNLRSISI-RSCP 256
>At5g27920 unknown protein
Length = 642
Score = 48.5 bits (114), Expect = 1e-05
Identities = 72/262 (27%), Positives = 117/262 (44%), Gaps = 54/262 (20%)
Query: 55 RLVERFPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMV-VS 113
+ ++ LK++ + G H +D SLV +L+ + L E+ L R V V+
Sbjct: 294 KYIKGLKHLKTIWIDGA-HVSDSSLV-------------SLSSSCRSLMEIGLSRCVDVT 339
Query: 114 DESLELLSRSFVNFKSLVLVSCEGFTTD-GLAAVAANCRSLRELDLQENEVEDHKG-QWL 171
D + L+R+ +N K+L L C GF TD ++AVA +CR+L L L+ + KG Q L
Sbjct: 340 DIGMISLARNCLNLKTLNLACC-GFVTDVAISAVAQSCRNLGTLKLESCHLITEKGLQSL 398
Query: 172 SCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQ 231
C+ + L+ G +N LE +S+ NL+ L+L
Sbjct: 399 GCY---SMLVQELDLTDCYG-VNDRGLE-YISKCSNLQRLKLG-------------LCTN 440
Query: 232 LMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNL 291
+ D GI FH + S + + + +C +GF LAA+ C++L L L
Sbjct: 441 ISDKGI---FH-IGSKCSKLLELDLYRC------AGF---GDDGLAALSRGCKSLNRLIL 487
Query: 292 SYA-----AGILGIELIKLIRH 308
SY G+ I ++L+ H
Sbjct: 488 SYCCELTDTGVEQIRQLELLSH 509
Score = 42.0 bits (97), Expect = 0.001
Identities = 79/337 (23%), Positives = 132/337 (38%), Gaps = 51/337 (15%)
Query: 22 RNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKSLTLKGKPHFADFSLVP 81
R + L+ K + R++ TR + I + L+ ++P+L SL L P D ++
Sbjct: 28 RKTWRLISKDFLRVDSLTRTTIRILRVEFLPT--LLFKYPNLSSLDLSVCPKLDDDVVLR 85
Query: 82 HGWGGFVYPW-IEAL------AKNKVGLEELR-----LKRMVVS------DESLELLSRS 123
G + I++L A GLE L L+R+ VS D LS S
Sbjct: 86 LALDGAISTLGIKSLNLSRSTAVRARGLETLARMCHALERVDVSHCWGFGDREAAALS-S 144
Query: 124 FVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVS 183
+ L + C + GLA + C +L ++ L+ G L C + C L S
Sbjct: 145 ATGLRELKMDKCLSLSDVGLARIVVGCSNLNKISLKWCMEISDLGIDLLC--KICKGLKS 202
Query: 184 LNFACLKGD-----------------------INLGALERLVSRSPNLKSLRLNRS--VP 218
L+ + LK I+ G L+ L + SP+L+ + + R V
Sbjct: 203 LDVSYLKITNDSIRSIALLVKLEVLDMVSCPLIDDGGLQFLENGSPSLQEVDVTRCDRVS 262
Query: 219 VDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAA 278
+ L I+ P + L +++ K LK + G V+ SL +
Sbjct: 263 LSGLISIVRGHPDIQLLKASHCVSEVSGSFLKYIKG--LKHLKTIWIDG-AHVSDSSLVS 319
Query: 279 IYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
+ C++L + LS + I +I L R+C L+ L
Sbjct: 320 LSSSCRSLMEIGLSRCVDVTDIGMISLARNCLNLKTL 356
Score = 33.9 bits (76), Expect = 0.25
Identities = 35/116 (30%), Positives = 53/116 (45%), Gaps = 19/116 (16%)
Query: 101 GLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQE 160
G+E++R LELLS L L + T GLAA+A+ C+ L LD++
Sbjct: 497 GVEQIR---------QLELLSH-------LELRGLKNITGVGLAAIASGCKKLGYLDVKL 540
Query: 161 NEVEDHKGQW-LSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNR 215
E D G W L+ F ++ + N C D L L +SR ++ + L+R
Sbjct: 541 CENIDDSGFWALAYFSKNLRQINLCN--CSVSDTALCMLMSNLSRVQDVDLVHLSR 594
>At5g01720 putative F-box protein family, AtFBL3
Length = 665
Score = 46.6 bits (109), Expect = 4e-05
Identities = 54/209 (25%), Positives = 88/209 (41%), Gaps = 16/209 (7%)
Query: 112 VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWL 171
+S S+ ++ S SL + SC + + + CR L ELDL +NE++D + +
Sbjct: 368 LSRVSITQIANSCPLLVSLKMESCSLVSREAFWLIGQKCRLLEELDLTDNEIDDEGLKSI 427
Query: 172 SCFPESCTSLVSLNFA-CLKGDINLGALERLVSRSPNLKSLRLNRSVPVD--ALQRILTR 228
S SC SL SL CL +I L + NL+ L L RSV + + I
Sbjct: 428 S----SCLSLSSLKLGICL--NITDKGLSYIGMGCSNLRELDLYRSVGITDVGISTIAQG 481
Query: 229 APQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLS--GFLEVAPFSLAAIYPICQNL 286
L + I S+ D+ + ++ KC + + G + LAAI C+ L
Sbjct: 482 CIHLETINI-SYCQDITDKSL----VSLSKCSLLQTFESRGCPNITSQGLAAIAVRCKRL 536
Query: 287 TSLNLSYAAGILGIELIKLIRHCGKLQRL 315
++L I L+ L L+++
Sbjct: 537 AKVDLKKCPSINDAGLLALAHFSQNLKQI 565
Score = 45.8 bits (107), Expect = 6e-05
Identities = 75/347 (21%), Positives = 137/347 (38%), Gaps = 41/347 (11%)
Query: 6 DEVIEHVFDYVVSH-SDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLK 64
+E++ + D + + SD S SL CKS+Y++E ++ R + S R++ R+ +
Sbjct: 18 EELVFIILDLISPNPSDLKSFSLTCKSFYQLE--SKHRGSLKPLRSDYLPRILTRYRNTT 75
Query: 65 SLTLKGKPHFADFSLVPHGW--------------GGFVYPWIEALAKNKVGLEELRLKRM 110
L L P D++L G G F + LA V L E+ L
Sbjct: 76 DLDLTFCPRVTDYALSVVGCLSGPTLRSLDLSRSGSFSAAGLLRLALKCVNLVEIDLSNA 135
Query: 111 VVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQW 170
++ + + + L L C+ T G+ +A C+ L + L+ G
Sbjct: 136 TEMRDADAAVVAEARSLERLKLGRCKMLTDMGIGCIAVGCKKLNTVSLKWCVGVGDLG-- 193
Query: 171 LSCFPESCTSLVSLNFA-------CLKGDINLGALERLVSR------SPNLKSLRLN--- 214
+ C + +L+ + CL + L LE L+ +LKSLR +
Sbjct: 194 VGLLAVKCKDIRTLDLSYLPITGKCLHDILKLQHLEELLLEGCFGVDDDSLKSLRHDCKS 253
Query: 215 -RSVPVDALQRILTRAPQLMDLGIGSFFH-DLNSDAYAM---FKATILKCKSITSLS-GF 268
+ + + Q + R + G G DL+ + + F +++ K ++ S+
Sbjct: 254 LKKLDASSCQNLTHRGLTSLLSGAGYLQRLDLSHCSSVISLDFASSLKKVSALQSIRLDG 313
Query: 269 LEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
V P L AI +C +L ++LS + L L+ L++L
Sbjct: 314 CSVTPDGLKAIGTLCNSLKEVSLSKCVSVTDEGLSSLVMKLKDLRKL 360
>At2g25490 putative glucose regulated repressor protein
Length = 628
Score = 46.2 bits (108), Expect = 5e-05
Identities = 60/232 (25%), Positives = 96/232 (40%), Gaps = 29/232 (12%)
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
++ PDE + +F + +R++ + V K W + RQ+ P ++ E
Sbjct: 64 IDVLPDECLFEIFRRLSGPQERSACAFVSKQWLTLVSSIRQKEID------VPSKITEDG 117
Query: 61 PDLK---SLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLK---RMVVSD 114
D + S +L GK D L G + GL +L ++ VSD
Sbjct: 118 DDCEGCLSRSLDGKKA-TDVRLAAIAVG----------TAGRGGLGKLSIRGSNSAKVSD 166
Query: 115 ESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCF 174
L + RS + SL L + T +GL +A C L +L+L KG L
Sbjct: 167 LGLRSIGRSCPSLGSLSLWNVSTITDNGLLEIAEGCAQLEKLELNRCSTITDKG--LVAI 224
Query: 175 PESCTSLVSLNF-ACLK-GDINLGALERLVS--RSPNLKSLRLNRSVPVDAL 222
+SC +L L AC + GD L A+ R S +S ++K+ L R + +L
Sbjct: 225 AKSCPNLTELTLEACSRIGDEGLLAIARSCSKLKSVSIKNCPLVRDQGIASL 276
Score = 38.1 bits (87), Expect = 0.013
Identities = 45/174 (25%), Positives = 71/174 (39%), Gaps = 41/174 (23%)
Query: 94 ALAKNKVGLEELRLKRMV-VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCR- 151
A+AK+ L EL L+ + DE L ++RS KS+ + +C G+A++ +N
Sbjct: 223 AIAKSCPNLTELTLEACSRIGDEGLLAIARSCSKLKSVSIKNCPLVRDQGIASLLSNTTC 282
Query: 152 SLRELDLQENEVED-------------------------HKGQWLSCFPESCTSLVSLNF 186
SL +L LQ V D KG W+ L SL
Sbjct: 283 SLAKLKLQMLNVTDVSLAVVGHYGLSITDLVLAGLSHVSEKGFWVMGNGVGLQKLNSLTI 342
Query: 187 ACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
+G ++G LE + PN+K + I++++P L D G+ SF
Sbjct: 343 TACQGVTDMG-LESVGKGCPNMK-------------KAIISKSPLLSDNGLVSF 382
Score = 32.0 bits (71), Expect = 0.94
Identities = 27/106 (25%), Positives = 49/106 (45%), Gaps = 2/106 (1%)
Query: 119 LLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESC 178
+ +R+ + L + C T L ++AANC+ L +LD+ + + D Q L+ +
Sbjct: 512 ITARNGWTLEVLNIDGCSNITDASLVSIAANCQILSDLDISKCAISDSGIQALASSDKLK 571
Query: 179 TSLVSLNFACLKGDINLGALERLVSR--SPNLKSLRLNRSVPVDAL 222
++S+ + D +L A+ L S NL+ R + VD L
Sbjct: 572 LQILSVAGCSMVTDKSLPAIVGLGSTLLGLNLQQCRSISNSTVDFL 617
Score = 29.6 bits (65), Expect = 4.7
Identities = 51/178 (28%), Positives = 76/178 (42%), Gaps = 20/178 (11%)
Query: 128 KSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESC-TSLVSLNF 186
+SL + +C GF LAA+ C L ++DL KG S F +SLV +NF
Sbjct: 444 RSLSIRNCPGFGDANLAAIGKLCPQLEDIDLC-----GLKGITESGFLHLIQSSLVKINF 498
Query: 187 ACLKGDINLGALERLVS-----RSPNLKSLRLN--RSVPVDALQRILTRAPQLMDLGIGS 239
+ G NL +R++S L+ L ++ ++ +L I L DL I
Sbjct: 499 S---GCSNL--TDRVISAITARNGWTLEVLNIDGCSNITDASLVSIAANCQILSDLDISK 553
Query: 240 FFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGI 297
+S A+ + LK + I S++G V SL AI + L LNL I
Sbjct: 554 CAIS-DSGIQALASSDKLKLQ-ILSVAGCSMVTDKSLPAIVGLGSTLLGLNLQQCRSI 609
>At5g07670 unknown protein
Length = 476
Score = 45.1 bits (105), Expect = 1e-04
Identities = 42/161 (26%), Positives = 72/161 (44%), Gaps = 14/161 (8%)
Query: 5 PDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLK 64
PD ++ V + +S R +LSLVCK W+R+ + + + +S RL+ RFP+L+
Sbjct: 66 PDLILIRVIQKI-PNSQRKNLSLVCKRWFRLHGRLVRSFKVSDWEFLSSGRLISRFPNLE 124
Query: 65 SLTLK-----GKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLEL 119
++ L P+ LV H F + + E L + + + L+
Sbjct: 125 TVDLVSGCLISPPNLG--ILVNHRIVSFT---VGVGSYQSWSFFEENLLSVELVERGLKA 179
Query: 120 LSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQE 160
L+ N + LV+ + GL VA C L+EL+L +
Sbjct: 180 LAGGCSNLRKLVVTNTSEL---GLLNVAEECSRLQELELHK 217
Score = 36.6 bits (83), Expect = 0.038
Identities = 45/169 (26%), Positives = 74/169 (43%), Gaps = 19/169 (11%)
Query: 107 LKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDH 166
L +VSD L +L++ L LV CEG DG+ + C+ L EL + +N++E
Sbjct: 244 LYNSLVSDIGLMILAQGCKRLVKLELVGCEG-GFDGIKEIGECCQMLEELTVCDNKME-- 300
Query: 167 KGQWLSCFPESCTSLVSLNF-ACLKGDIN--------LGALERLVSRSPNLKSLRLNRSV 217
WL C +L +L +C K D + ALERL L+ +L
Sbjct: 301 -SGWLGGL-RYCENLKTLKLVSCKKIDNDPDESLSCCCPALERL-----QLEKCQLRDKN 353
Query: 218 PVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLS 266
V AL ++ A +++ +D+ S A A + +L + + L+
Sbjct: 354 TVKALFKMCEAAREIVFQDCWGLDNDIFSLAMAFGRVKLLYLEGCSLLT 402
>At1g77000 unknown protein
Length = 360
Score = 44.7 bits (104), Expect = 1e-04
Identities = 60/232 (25%), Positives = 98/232 (41%), Gaps = 13/232 (5%)
Query: 91 WIEALAKNKVGLEELRLKRMVVSDESLEL-LSRSFVNFKSLVLVSCEGFTTDG-LAAVAA 148
W +A++ +GL L L + SL L L+ FV ++LVL + D + A+A
Sbjct: 58 WRDAVS---LGLTRLSLSWCKKNMNSLVLSLAPKFVKLQTLVLRQDKPQLEDNAVEAIAN 114
Query: 149 NCRSLRELDL-QENEVEDHKGQWLSCFPESCTSLVSLNFA-CLK-GDINLGALERLVSRS 205
+C L++LDL + +++ DH L CT+L LN + C D L L R +
Sbjct: 115 HCHELQDLDLSKSSKITDHS---LYSLARGCTNLTKLNLSGCTSFSDTALAHLTRFCRKL 171
Query: 206 PNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSL 265
L +V + LQ I QL L +G + +++ D +++
Sbjct: 172 KILNLCGCVEAVSDNTLQAIGENCNQLQSLNLG-WCENISDDGVMSLAYGCPDLRTLDLC 230
Query: 266 SGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRLWW 317
S L + S+ A+ C +L SL L Y I + L + K + W
Sbjct: 231 SCVL-ITDESVVALANRCIHLRSLGLYYCRNITDRAMYSLAQSGVKNKHEMW 281
>At5g51380 putative protein
Length = 479
Score = 43.9 bits (102), Expect = 2e-04
Identities = 35/104 (33%), Positives = 53/104 (50%), Gaps = 5/104 (4%)
Query: 112 VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWL 171
VSD L +L++ L L CEG + DG+ A+ C L EL + ++ ++D L
Sbjct: 251 VSDIGLTILAQGCKRLVKLELSGCEG-SFDGIKAIGQCCEVLEELSICDHRMDDGWIAAL 309
Query: 172 SCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNR 215
S F T L+S +C K D + G +L+ P L+SL+L R
Sbjct: 310 SYFESLKTLLIS---SCRKIDSSPGP-GKLLGSCPALESLQLRR 349
Score = 42.7 bits (99), Expect = 5e-04
Identities = 42/144 (29%), Positives = 66/144 (45%), Gaps = 9/144 (6%)
Query: 20 SDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKSLTLKG---KPHFAD 76
S +SLVCK W ++ + + + + + RLV RFP L S+ L P
Sbjct: 82 SQNEDVSLVCKRWLSVQGRRLRSMKVFDWEFLLSGRLVSRFPKLTSVDLVNACFNPSSNS 141
Query: 77 FSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCE 136
L+ H F +L N V EE L +V D+ L +L R + LV+++
Sbjct: 142 GILLCHTSISFHVSTDSSLNLNFV--EESLLDNEMV-DKGLRVLGRGSFDLIKLVVINA- 197
Query: 137 GFTTDGLAAVAANCRSLRELDLQE 160
T GL ++A +C L+EL+L +
Sbjct: 198 --TELGLLSLAEDCSDLQELELHK 219
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.340 0.146 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,460,286
Number of Sequences: 26719
Number of extensions: 441186
Number of successful extensions: 2113
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 1881
Number of HSP's gapped (non-prelim): 202
length of query: 562
length of database: 11,318,596
effective HSP length: 104
effective length of query: 458
effective length of database: 8,539,820
effective search space: 3911237560
effective search space used: 3911237560
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC133780.9


