

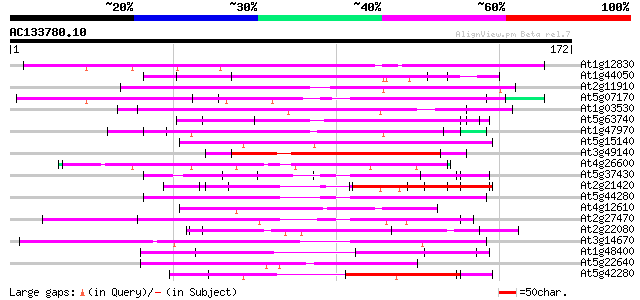
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.10 - phase: 0
(172 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g12830 unknown protein 139 6e-34
At1g44050 unknown protein 64 5e-11
At2g11910 unknown protein 59 1e-09
At5g07170 putative protein 57 6e-09
At1g03530 unknown protein 57 6e-09
At5g63740 unknown protein 56 8e-09
At1g47970 Unknown protein (T2J15.12) 56 1e-08
At5g15140 putative aldose 1-epimerase - like protein 55 2e-08
At3g49140 putative protein 54 3e-08
At4g26600 unknown protein 54 4e-08
At5g37430 contains similarity to unknown protein (pir||C71432) 54 5e-08
At2g21420 Mutator-like transposase 52 2e-07
At5g44280 putative protein 51 3e-07
At4g12610 putative protein 51 3e-07
At2g27470 putative CCAAT-binding transcription factor subunit 51 3e-07
At2g22080 En/Spm-like transposon protein 51 3e-07
At3g14670 hypothetical protein 51 4e-07
At1g48400 50 5e-07
At5g22640 unknown protein 50 6e-07
At5g42280 putative protein 49 1e-06
>At1g12830 unknown protein
Length = 213
Score = 139 bits (351), Expect = 6e-34
Identities = 86/193 (44%), Positives = 114/193 (58%), Gaps = 36/193 (18%)
Query: 5 ETNNHEEPTFPTKRKPDQ--EETHHLPNKNPKLT-STTTVDQETELPTT--------VNS 53
+ N ++ +FP KRK D ++ ++ NK KL S + D E++ T +NS
Sbjct: 3 DVENQQDSSFPVKRKSDLFCQDQDNVTNKAQKLNPSLNSADSESKDGETNGSGQIENLNS 62
Query: 54 SSDAVPNNEP----------------------ENNTDDDDEDDEDEENSDEEPVVDRKGK 91
S++ E E D++DE+++DEE +EE VVDRKGK
Sbjct: 63 STEETIGAESSVLSEKAAEEVESGVIGDVGAEEEQEDEEDEEEDDEEEEEEEEVVDRKGK 122
Query: 92 GIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPS 151
GI R+DKGKGKMIE E+ DD+ DDSDD D+D +G +SDFSDDPLAEVDLDNILPS
Sbjct: 123 GISREDKGKGKMIEVEESDDE--DDSDD-DEDDEGFDEDDESDFSDDPLAEVDLDNILPS 179
Query: 152 RTRRRTAQSGVRI 164
RTRRR++Q GV I
Sbjct: 180 RTRRRSSQPGVFI 192
Score = 29.3 bits (64), Expect = 1.1
Identities = 26/123 (21%), Positives = 50/123 (40%), Gaps = 9/123 (7%)
Query: 2 AEPETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNN 61
AE E + E+ + + ++EE + ++ K S + + + D ++
Sbjct: 93 AEEEQEDEEDEEEDDEEEEEEEE---VVDRKGKGISREDKGKGKMIEVEESDDEDDSDDD 149
Query: 62 EPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEE------DEDDDDSD 115
E + D+DDE D ++ E + + R G I E +++DD+S
Sbjct: 150 EDDEGFDEDDESDFSDDPLAEVDLDNILPSRTRRRSSQPGVFIPNEFSNRHVNDEDDESS 209
Query: 116 DSD 118
DSD
Sbjct: 210 DSD 212
>At1g44050 unknown protein
Length = 734
Score = 63.5 bits (153), Expect = 5e-11
Identities = 34/85 (40%), Positives = 47/85 (55%), Gaps = 8/85 (9%)
Query: 69 DDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDS---DDSDDSDDDSD 125
DDD+D ED++N D++ D KG DD + +++++DDDD DD DD DDD D
Sbjct: 70 DDDDDAEDDDNGDDKEDGDDDNKGDDGDDDNEDDNEDDDNDDDDDDDDDDDDDDDDDDDD 129
Query: 126 GNVSGSDSDFSDDPLAEVDLDNILP 150
G+ D D DD D D +LP
Sbjct: 130 GDDDNEDGDCDDD-----DGDLLLP 149
Score = 55.5 bits (132), Expect = 1e-08
Identities = 27/93 (29%), Positives = 47/93 (50%), Gaps = 19/93 (20%)
Query: 42 DQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKG 101
D + E + D +N+ ++ DD+++D+ED++N D++ DD
Sbjct: 72 DDDAEDDDNGDDKEDGDDDNKGDDGDDDNEDDNEDDDNDDDD------------DD---- 115
Query: 102 KMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSD 134
+++D+DDDD DD D DD+ DG+ D D
Sbjct: 116 ---DDDDDDDDDDDDDDGDDDNEDGDCDDDDGD 145
Score = 47.4 bits (111), Expect = 4e-06
Identities = 25/79 (31%), Positives = 41/79 (51%), Gaps = 25/79 (31%)
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDD 111
N D +NE +N DD+D+DD+D+++ D++ +D+DD
Sbjct: 91 NKGDDGDDDNEDDNEDDDNDDDDDDDDDDDDD-----------------------DDDDD 127
Query: 112 DD-SDDSDDSD-DDSDGNV 128
DD DD++D D DD DG++
Sbjct: 128 DDGDDDNEDGDCDDDDGDL 146
Score = 33.9 bits (76), Expect = 0.045
Identities = 15/43 (34%), Positives = 23/43 (52%)
Query: 102 KMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVD 144
KM +D+DD + DD+ D +D D + G D D ++ E D
Sbjct: 65 KMHNHDDDDDAEDDDNGDDKEDGDDDNKGDDGDDDNEDDNEDD 107
Score = 28.5 bits (62), Expect = 1.9
Identities = 11/35 (31%), Positives = 18/35 (51%)
Query: 104 IEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDD 138
++ + DDDD + DD+ DD + + D DD
Sbjct: 64 LKMHNHDDDDDAEDDDNGDDKEDGDDDNKGDDGDD 98
>At2g11910 unknown protein
Length = 168
Score = 59.3 bits (142), Expect = 1e-09
Identities = 38/131 (29%), Positives = 61/131 (46%), Gaps = 16/131 (12%)
Query: 35 LTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIM 94
L VDQ + S N + ++ DDDD++D DE++ DE+ D G
Sbjct: 44 LIGLVAVDQRLSVKEGYKRGSRIEENKDASDSDDDDDDEDADEDDDDEDDANDEDFSG-- 101
Query: 95 RDDKGKGKMIEEEDEDDDD--------SDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLD 146
G+G EEE + +DD SDD DD D++ D + D++ ++ E D D
Sbjct: 102 ----GEGDEGEEEADPEDDPVTNGGGGSDDEDDDDEEGDNDDEDEDNEDEEEDDDEEDDD 157
Query: 147 NIL--PSRTRR 155
++ PS+ R+
Sbjct: 158 DVRQPPSKKRK 168
>At5g07170 putative protein
Length = 542
Score = 56.6 bits (135), Expect = 6e-09
Identities = 38/149 (25%), Positives = 65/149 (43%), Gaps = 24/149 (16%)
Query: 3 EPETNNHEEPTFPTKRKPDQ----EETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAV 58
+P + ++ TF T D + H+ + + + + + P +SSSD
Sbjct: 36 KPGLKDSQDSTFVTSVSYDMGNISSDDHYCGDDDENMKIAAAIPSSQDDPLIDDSSSDEE 95
Query: 59 PNNEPEN-NTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDS 117
++E + DDD DD D++ D++ DD +++D+DDDD DD
Sbjct: 96 DDSESTHCYAADDDADDTDDDEDDDD------------DD-------DDDDDDDDDDDDD 136
Query: 118 DDSDDDSDGNVSGSDSDFSDDPLAEVDLD 146
DD DDD + S + + DD L +D
Sbjct: 137 DDDDDDDESKDSEVEEEEGDDDLRMRKID 165
Score = 54.7 bits (130), Expect = 2e-08
Identities = 31/96 (32%), Positives = 46/96 (47%), Gaps = 10/96 (10%)
Query: 57 AVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDD 116
A+P+++ + DD D+ED+ S D DD ++ED+DDDD DD
Sbjct: 77 AIPSSQDDPLIDDSSSDEEDDSESTHCYAADDDA-----DDTD-----DDEDDDDDDDDD 126
Query: 117 SDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSR 152
DD DDD D + D + D + E + D+ L R
Sbjct: 127 DDDDDDDDDDDDDDDDDESKDSEVEEEEGDDDLRMR 162
Score = 40.8 bits (94), Expect = 4e-04
Identities = 29/109 (26%), Positives = 44/109 (39%), Gaps = 13/109 (11%)
Query: 65 NNTDDDDEDDEDEEN---------SDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSD 115
N + DD +D+EN S ++P++D +D + D+D DD+D
Sbjct: 57 NISSDDHYCGDDDENMKIAAAIPSSQDDPLIDDSSSD--EEDDSESTHCYAADDDADDTD 114
Query: 116 DSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRRTAQSGVRI 164
D D DDD D + D D DD + D D S +R+
Sbjct: 115 D--DEDDDDDDDDDDDDDDDDDDDDDDDDDDESKDSEVEEEEGDDDLRM 161
>At1g03530 unknown protein
Length = 801
Score = 56.6 bits (135), Expect = 6e-09
Identities = 33/115 (28%), Positives = 62/115 (53%), Gaps = 5/115 (4%)
Query: 40 TVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKG 99
+ + E+E ++ SSSD+ + E E++ D+ D+++ +E E VV ++ ++G
Sbjct: 244 SAESESETSSSSASSSDSSSSEEEESDEDESDKEENKKEEKFEHMVVGKEDDLAGELEEG 303
Query: 100 KGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTR 154
+ + ++EE+ DDD D+ DD DDD D D D ++ D D+ L +T+
Sbjct: 304 EIENLDEENGDDDIEDEDDDDDDDDD-----DDDDVNEMVAWSNDEDDDLGLQTK 353
Score = 41.2 bits (95), Expect = 3e-04
Identities = 30/125 (24%), Positives = 56/125 (44%), Gaps = 18/125 (14%)
Query: 34 KLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEP--------- 84
K++ D++++ S+++ + + D E+EE+ ++E
Sbjct: 224 KVSLAVDDDEKSDEAKGEMDSAESESETSSSSASSSDSSSSEEEESDEDESDKEENKKEE 283
Query: 85 -----VVDRKGKGIMRDDKGKGKMIEEEDEDDD----DSDDSDDSDDDSDGNVSGSDSDF 135
VV ++ ++G+ + ++EE+ DDD D DD DD DDD D N + S+
Sbjct: 284 KFEHMVVGKEDDLAGELEEGEIENLDEENGDDDIEDEDDDDDDDDDDDDDVNEMVAWSND 343
Query: 136 SDDPL 140
DD L
Sbjct: 344 EDDDL 348
Score = 34.7 bits (78), Expect = 0.026
Identities = 22/99 (22%), Positives = 46/99 (46%), Gaps = 19/99 (19%)
Query: 17 KRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDD--DDEDD 74
+ + D++E+ NK + V +E +L + + N + EN DD D++DD
Sbjct: 266 EEESDEDESDKEENKKEEKFEHMVVGKEDDLAGELEEGE--IENLDEENGDDDIEDEDDD 323
Query: 75 EDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDD 113
+D+++ D++ V +M+ +++DDD
Sbjct: 324 DDDDDDDDDDV---------------NEMVAWSNDEDDD 347
Score = 32.0 bits (71), Expect = 0.17
Identities = 21/83 (25%), Positives = 40/83 (47%), Gaps = 6/83 (7%)
Query: 68 DDDDEDDEDEENSDE-EPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDS-D 125
DDD++ DE + D E + D EEE+ D+D+SD ++ ++ +
Sbjct: 230 DDDEKSDEAKGEMDSAESESETSSSSASSSDSSSS---EEEESDEDESDKEENKKEEKFE 286
Query: 126 GNVSGSDSDFSDDPLAEVDLDNI 148
V G + D + + L E +++N+
Sbjct: 287 HMVVGKEDDLAGE-LEEGEIENL 308
Score = 28.5 bits (62), Expect = 1.9
Identities = 23/104 (22%), Positives = 46/104 (44%), Gaps = 6/104 (5%)
Query: 37 STTTVDQETELPTTVNSSSD-AVPNNEPENNTDDDDEDDEDE-----ENSDEEPVVDRKG 90
S+ V+ + + N + AV ++EP + + + E E + S P +D
Sbjct: 145 SSVEVEPNVCVEMSANGGDEPAVKDSEPVVSENSRSMEGETEPVVVSDASVACPTMDLDE 204
Query: 91 KGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSD 134
G+ + D+G IE E + D D+ D++ G + ++S+
Sbjct: 205 SGLKKTDEGLACSIEVGLEKVSLAVDDDEKSDEAKGEMDSAESE 248
>At5g63740 unknown protein
Length = 226
Score = 56.2 bits (134), Expect = 8e-09
Identities = 26/89 (29%), Positives = 46/89 (51%), Gaps = 8/89 (8%)
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDD 111
+ D + + + + D+D+++DEDE++ D++ D DD +E+D+D+
Sbjct: 64 DGDGDGDEDEDEDADADEDEDEDEDEDDDDDDDDDD--------DDDADDADDDEDDDDE 115
Query: 112 DDSDDSDDSDDDSDGNVSGSDSDFSDDPL 140
DD +D DD DDD D N D ++ L
Sbjct: 116 DDDEDEDDDDDDDDENDEECDDEYDSHRL 144
Score = 54.3 bits (129), Expect = 3e-08
Identities = 34/88 (38%), Positives = 44/88 (49%), Gaps = 16/88 (18%)
Query: 60 NNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDD 119
NN+ E + D D + DEDE DE+ D +D+ E+EDEDDDD DD DD
Sbjct: 56 NNDGEGDGDGDGDGDEDE---DEDADAD--------EDED-----EDEDEDDDDDDDDDD 99
Query: 120 SDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
DD D + D D DD + D D+
Sbjct: 100 DDDADDADDDEDDDDEDDDEDEDDDDDD 127
Score = 52.0 bits (123), Expect = 2e-07
Identities = 25/87 (28%), Positives = 42/87 (47%), Gaps = 20/87 (22%)
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDD 111
+ D + + + + D+D + DEDE+ ++E +++D+DD
Sbjct: 58 DGEGDGDGDGDGDEDEDEDADADEDEDEDEDED--------------------DDDDDDD 97
Query: 112 DDSDDSDDSDDDSDGNVSGSDSDFSDD 138
DD DD+DD+DDD D + D D DD
Sbjct: 98 DDDDDADDADDDEDDDDEDDDEDEDDD 124
Score = 43.5 bits (101), Expect = 6e-05
Identities = 22/69 (31%), Positives = 37/69 (52%), Gaps = 4/69 (5%)
Query: 76 DEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDF 135
D+ N+D E D G G D+ + + + ++++D+D D+ DD DDD D + D+D
Sbjct: 53 DDHNNDGEGDGDGDGDG----DEDEDEDADADEDEDEDEDEDDDDDDDDDDDDDADDADD 108
Query: 136 SDDPLAEVD 144
+D E D
Sbjct: 109 DEDDDDEDD 117
Score = 31.2 bits (69), Expect = 0.29
Identities = 27/98 (27%), Positives = 36/98 (36%), Gaps = 22/98 (22%)
Query: 71 DEDDEDEENS------DEEPVVDRKGKGIMR-----------DDKGKGKMIEEEDEDDDD 113
DED E+ E D V+ GI R D G+ + D D D+
Sbjct: 12 DEDKEEMEKRIRTVVIDSNLVLSNSTCGICRTAEDEYIKVYDDHNNDGEGDGDGDGDGDE 71
Query: 114 SDDSD-----DSDDDSDGNVSGSDSDFSDDPLAEVDLD 146
+D D D D+D D + D D DD + D D
Sbjct: 72 DEDEDADADEDEDEDEDEDDDDDDDDDDDDDADDADDD 109
>At1g47970 Unknown protein (T2J15.12)
Length = 198
Score = 55.8 bits (133), Expect = 1e-08
Identities = 31/109 (28%), Positives = 55/109 (50%), Gaps = 5/109 (4%)
Query: 31 KNPKLTSTTTVDQETELPTTVNSS-SDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRK 89
+ P++ + D + E V SD ++ E + DDD+E++ED+++ D+ V+
Sbjct: 14 EEPEIQVLSDDDSDEEQVKDVGEEVSDEDDDDGSEGDDDDDEEEEEDDDDDDDVQVLQSL 73
Query: 90 GKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDD 138
G ++ + + + +E+ DDD DD DD DDD D D D D+
Sbjct: 74 GGPPVQSAEDEDEEGDEDGNGDDDDDDGDDDDDDDD----DEDEDVEDE 118
Score = 50.1 bits (118), Expect = 6e-07
Identities = 34/111 (30%), Positives = 44/111 (39%), Gaps = 13/111 (11%)
Query: 49 TTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEED 108
T VN D EPE DD+ DE++ E V D D + EE+D
Sbjct: 2 TGVNDDDDKNFKEEPEIQVLSDDDSDEEQVKDVGEEVSDEDDDDGSEGDDDDDEEEEEDD 61
Query: 109 EDDDD-------------SDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLD 146
+DDDD S + +D + D DGN D D DD + D D
Sbjct: 62 DDDDDVQVLQSLGGPPVQSAEDEDEEGDEDGNGDDDDDDGDDDDDDDDDED 112
Score = 44.3 bits (103), Expect = 3e-05
Identities = 24/92 (26%), Positives = 47/92 (51%), Gaps = 2/92 (2%)
Query: 42 DQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKG 101
D++ E N D ++ +++ DD+DED EDE + E +V G+ D++
Sbjct: 83 DEDEEGDEDGNGDDDDDDGDDDDDDDDDEDEDVEDEGDLGTEYLVRPVGRA--EDEEDAS 140
Query: 102 KMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDS 133
EE+ ++D D+ +D ++D+ G S++
Sbjct: 141 DFEPEENGVEEDIDEGEDDENDNSGGAGKSEA 172
Score = 34.3 bits (77), Expect = 0.034
Identities = 20/63 (31%), Positives = 28/63 (43%), Gaps = 7/63 (11%)
Query: 56 DAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSD 115
DA EN ++D ++ ED+EN D G + K K EEDE+D +
Sbjct: 138 DASDFEPEENGVEEDIDEGEDDEN-------DNSGGAGKSEAPPKRKRAPEEDEEDSGDE 190
Query: 116 DSD 118
D D
Sbjct: 191 DDD 193
Score = 32.0 bits (71), Expect = 0.17
Identities = 19/76 (25%), Positives = 35/76 (46%), Gaps = 13/76 (17%)
Query: 73 DDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSD 132
+D+D++N EEP + ++ DD + DE+ + SD+D D G D
Sbjct: 5 NDDDDKNFKEEPEIQ-----VLSDD--------DSDEEQVKDVGEEVSDEDDDDGSEGDD 51
Query: 133 SDFSDDPLAEVDLDNI 148
D ++ + D D++
Sbjct: 52 DDDEEEEEDDDDDDDV 67
>At5g15140 putative aldose 1-epimerase - like protein
Length = 490
Score = 54.7 bits (130), Expect = 2e-08
Identities = 33/110 (30%), Positives = 52/110 (47%), Gaps = 14/110 (12%)
Query: 53 SSSDAVPNNEPENNTDDD----DEDDEDEENSDEEPVVDRKGKG----------IMRDDK 98
+ SD N++ E + D D+ E+++ DE+ VD+K G +D K
Sbjct: 61 AGSDDEDNDKKEKKKEHDVQKKDKQHENKDKDDEKKHVDKKKSGGHDKDDDDEKKHKDKK 120
Query: 99 GKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNI 148
G +++ +DD D DD DD DDD D V G D++ L E+ N+
Sbjct: 121 KDGHNDDDDSDDDTDDDDDDDDDDDDDDEVDGDDNEKEKIGLYELKKGNL 170
Score = 30.0 bits (66), Expect = 0.65
Identities = 14/65 (21%), Positives = 30/65 (45%)
Query: 17 KRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDED 76
+ K +E H+ K D++ + +D +++ ++ DDDD+DD+D
Sbjct: 87 ENKDKDDEKKHVDKKKSGGHDKDDDDEKKHKDKKKDGHNDDDDSDDDTDDDDDDDDDDDD 146
Query: 77 EENSD 81
++ D
Sbjct: 147 DDEVD 151
>At3g49140 putative protein
Length = 1229
Score = 54.3 bits (129), Expect = 3e-08
Identities = 27/64 (42%), Positives = 39/64 (60%), Gaps = 4/64 (6%)
Query: 69 DDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNV 128
D D + ED+E+ D+ D + G D++ ++E+EDEDDDD DD D+ DDDSD +
Sbjct: 936 DSDFETEDDESGDD----DSEDTGEDEDEEEWVAILEDEDEDDDDDDDDDEDDDDSDSDE 991
Query: 129 SGSD 132
S D
Sbjct: 992 SLGD 995
Score = 42.7 bits (99), Expect = 1e-04
Identities = 22/80 (27%), Positives = 45/80 (55%), Gaps = 6/80 (7%)
Query: 61 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS 120
++ + T+DD+ D+D E++ E+ + + I+ D+ +++D+DDDD DD D
Sbjct: 935 SDSDFETEDDESGDDDSEDTGEDED-EEEWVAILEDEDE-----DDDDDDDDDEDDDDSD 988
Query: 121 DDDSDGNVSGSDSDFSDDPL 140
D+S G+ + ++ S P+
Sbjct: 989 SDESLGDWANLETMRSCHPM 1008
Score = 32.7 bits (73), Expect = 0.100
Identities = 13/39 (33%), Positives = 25/39 (63%)
Query: 57 AVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMR 95
A+ +E E++ DDDD+D++D+++ +E + D MR
Sbjct: 965 AILEDEDEDDDDDDDDDEDDDDSDSDESLGDWANLETMR 1003
Score = 30.0 bits (66), Expect = 0.65
Identities = 13/43 (30%), Positives = 24/43 (55%)
Query: 105 EEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
E+++ DDDS+D+ + +D+ + D D DD + D D+
Sbjct: 942 EDDESGDDDSEDTGEDEDEEEWVAILEDEDEDDDDDDDDDEDD 984
Score = 29.3 bits (64), Expect = 1.1
Identities = 20/87 (22%), Positives = 38/87 (42%), Gaps = 21/87 (24%)
Query: 31 KNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKG 90
K +L + D ETE + + S+ +E E ED++++++ D++
Sbjct: 927 KEMELMGLSDSDFETEDDESGDDDSEDTGEDEDEEEWVAILEDEDEDDDDDDD------- 979
Query: 91 KGIMRDDKGKGKMIEEEDEDDDDSDDS 117
++ED+DD DSD+S
Sbjct: 980 --------------DDEDDDDSDSDES 992
>At4g26600 unknown protein
Length = 671
Score = 53.9 bits (128), Expect = 4e-08
Identities = 37/122 (30%), Positives = 62/122 (50%), Gaps = 9/122 (7%)
Query: 17 KRKPDQEETHHLPNKNPKLT--STTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDD 74
K+K +T L NK K + Q+ L S+ P +PE +TD+++E++
Sbjct: 8 KKKGSNSQTPPL-NKQTKASPLKKAAKTQKPPLKKQRKCISEKKPLKKPEVSTDEEEEEE 66
Query: 75 EDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDD--SDGNVSGSD 132
E+E+ SDE G + D +G ++D+DDDD DD DD D + ++ + GSD
Sbjct: 67 ENEQ-SDEG---SESGSDLFSDGDEEGNNDSDDDDDDDDDDDDDDEDAEPLAEDFLDGSD 122
Query: 133 SD 134
++
Sbjct: 123 NE 124
Score = 41.2 bits (95), Expect = 3e-04
Identities = 32/141 (22%), Positives = 56/141 (39%), Gaps = 21/141 (14%)
Query: 16 TKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSS------SDAVPNNEPENNTD- 68
T++ P +++ + K P + D+E E S SD + + E N D
Sbjct: 34 TQKPPLKKQRKCISEKKPLKKPEVSTDEEEEEEENEQSDEGSESGSDLFSDGDEEGNNDS 93
Query: 69 ---DDDEDDEDEENSDEEPVV---------DRKGKGIMRDDKGKGKMIEEEDE--DDDDS 114
DDD+DD+D+++ D EP+ + G D G +E + D
Sbjct: 94 DDDDDDDDDDDDDDEDAEPLAEDFLDGSDNEEVTMGSDLDSDSGGSKLERKSRAIDRKRK 153
Query: 115 DDSDDSDDDSDGNVSGSDSDF 135
+ D+DD+ N+ +F
Sbjct: 154 KEVQDADDEFKMNIKEKPDEF 174
>At5g37430 contains similarity to unknown protein (pir||C71432)
Length = 607
Score = 53.5 bits (127), Expect = 5e-08
Identities = 29/85 (34%), Positives = 47/85 (55%), Gaps = 15/85 (17%)
Query: 42 DQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKG 101
++E +LP T D + ++ + + DDD+ D+D+++ D++ D DD G
Sbjct: 537 EEEKQLPET---EFDWLNLHDDDGDDGDDDDGDDDDDDGDDDGDDD--------DDDGD- 584
Query: 102 KMIEEEDEDDDDSDDSDDSDDDSDG 126
+ +D+DDDD DD DD DDD DG
Sbjct: 585 ---DGDDDDDDDGDDGDDDDDDDDG 606
Score = 50.4 bits (119), Expect = 5e-07
Identities = 27/73 (36%), Positives = 35/73 (46%), Gaps = 19/73 (26%)
Query: 66 NTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSD 125
N DDD DD D+++ D++ DD G +D+ DDD DD DD DDD D
Sbjct: 551 NLHDDDGDDGDDDDGDDD------------DDDG-------DDDGDDDDDDGDDGDDDDD 591
Query: 126 GNVSGSDSDFSDD 138
+ D D DD
Sbjct: 592 DDGDDGDDDDDDD 604
Score = 41.6 bits (96), Expect = 2e-04
Identities = 21/54 (38%), Positives = 27/54 (49%), Gaps = 2/54 (3%)
Query: 94 MRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
+ DD G +++D DDDD D DD DDD D G D D D + D D+
Sbjct: 552 LHDDDGDDG--DDDDGDDDDDDGDDDGDDDDDDGDDGDDDDDDDGDDGDDDDDD 603
Score = 40.4 bits (93), Expect = 5e-04
Identities = 20/61 (32%), Positives = 27/61 (43%)
Query: 77 EENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFS 136
+E + P + + DD G + +D+DDD DD DD DDD D D D
Sbjct: 536 KEEEKQLPETEFDWLNLHDDDGDDGDDDDGDDDDDDGDDDGDDDDDDGDDGDDDDDDDGD 595
Query: 137 D 137
D
Sbjct: 596 D 596
Score = 26.2 bits (56), Expect = 9.3
Identities = 9/31 (29%), Positives = 20/31 (64%)
Query: 60 NNEPENNTDDDDEDDEDEENSDEEPVVDRKG 90
+++ ++ D DD+DD+D ++ D++ D G
Sbjct: 577 DDDDDDGDDGDDDDDDDGDDGDDDDDDDDGG 607
>At2g21420 Mutator-like transposase
Length = 468
Score = 51.6 bits (122), Expect = 2e-07
Identities = 24/71 (33%), Positives = 33/71 (45%), Gaps = 25/71 (35%)
Query: 68 DDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGN 127
DDDD+DD+D+++ D +D+DDDD DD DD DD+ DG
Sbjct: 391 DDDDDDDDDDDDDD-------------------------DDDDDDDDDDDDDEDDEDDGY 425
Query: 128 VSGSDSDFSDD 138
+ D D D
Sbjct: 426 IDSDDDDVDGD 436
Score = 49.7 bits (117), Expect = 8e-07
Identities = 25/74 (33%), Positives = 42/74 (55%), Gaps = 19/74 (25%)
Query: 61 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS 120
++ +++ DDDD+DD+D+++ D++ DD ++ED++DD DSDD
Sbjct: 391 DDDDDDDDDDDDDDDDDDDDDDD------------DD-------DDEDDEDDGYIDSDDD 431
Query: 121 DDDSDGNVSGSDSD 134
D D D N GS+ D
Sbjct: 432 DVDGDDNDDGSELD 445
Score = 46.6 bits (109), Expect = 7e-06
Identities = 19/43 (44%), Positives = 26/43 (60%)
Query: 106 EEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNI 148
++D+DDDD DD DD DDD D + D D DD + D D++
Sbjct: 391 DDDDDDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYIDSDDDDV 433
Score = 45.4 bits (106), Expect = 1e-05
Identities = 25/72 (34%), Positives = 40/72 (54%), Gaps = 20/72 (27%)
Query: 59 PNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDD--DSDD 116
P + +++ DDDD+DD+D+++ D++ D +E+DEDD DSDD
Sbjct: 388 PGCDDDDDDDDDDDDDDDDDDDDDDDDDD-----------------DEDDEDDGYIDSDD 430
Query: 117 SD-DSDDDSDGN 127
D D DD+ DG+
Sbjct: 431 DDVDGDDNDDGS 442
Score = 44.3 bits (103), Expect = 3e-05
Identities = 18/43 (41%), Positives = 26/43 (59%)
Query: 105 EEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
+++D+DDDD DD DD DDD D + D + D +VD D+
Sbjct: 395 DDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYIDSDDDDVDGDD 437
Score = 40.8 bits (94), Expect = 4e-04
Identities = 19/75 (25%), Positives = 37/75 (49%), Gaps = 19/75 (25%)
Query: 48 PTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEE 107
P + D +++ +++ DDDD+DD+D+E+ +++ +D
Sbjct: 388 PGCDDDDDDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYID-------------------S 428
Query: 108 DEDDDDSDDSDDSDD 122
D+DD D DD+DD +
Sbjct: 429 DDDDVDGDDNDDGSE 443
Score = 30.0 bits (66), Expect = 0.65
Identities = 14/37 (37%), Positives = 18/37 (47%)
Query: 115 DDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPS 151
DD DD DDD D + D D DD + + D + S
Sbjct: 392 DDDDDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYIDS 428
Score = 26.2 bits (56), Expect = 9.3
Identities = 13/34 (38%), Positives = 16/34 (46%)
Query: 114 SDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
S DD DDD D + D D DD + D D+
Sbjct: 387 SPGCDDDDDDDDDDDDDDDDDDDDDDDDDDDEDD 420
>At5g44280 putative protein
Length = 486
Score = 51.2 bits (121), Expect = 3e-07
Identities = 29/105 (27%), Positives = 53/105 (49%), Gaps = 16/105 (15%)
Query: 42 DQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKG 101
D+E + PN+ E++ DD E+DE+EE EE D++ +
Sbjct: 45 DEEVKHDEAEEDQEVVKPNDAEEDDDGDDAEEDEEEEVEAEE------------DEEAE- 91
Query: 102 KMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLD 146
EEE+E++++ ++ +DS + S ++SG S+F + L E+ D
Sbjct: 92 ---EEEEEEEEEEEEEEDSKERSPSSISGDQSEFMEIDLGEIRKD 133
Score = 33.5 bits (75), Expect = 0.058
Identities = 22/115 (19%), Positives = 48/115 (41%), Gaps = 12/115 (10%)
Query: 46 ELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIE 105
E+P + D PE D ++D+ EE +E V + D+ +
Sbjct: 12 EIPDVADQPRDRF---NPEATQDLQEKDETKEEKEGDEEV---------KHDEAEEDQEV 59
Query: 106 EEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRRTAQS 160
+ D ++ DD DD+++D + V + + +++ E + + ++ R+ S
Sbjct: 60 VKPNDAEEDDDGDDAEEDEEEEVEAEEDEEAEEEEEEEEEEEEEEEDSKERSPSS 114
Score = 26.6 bits (57), Expect = 7.1
Identities = 17/82 (20%), Positives = 34/82 (40%), Gaps = 9/82 (10%)
Query: 60 NNEPENNTD---DDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDD 116
N E +N ++ DDD DD + D++ D +G + + + K
Sbjct: 269 NMEQQNASEAHEDDDNDDNNNRGRDKDSSSDERGTEVRQKKRRK------RSTSRSTQHP 322
Query: 117 SDDSDDDSDGNVSGSDSDFSDD 138
S + ++GN + +D++ D
Sbjct: 323 SSSGANKNNGNCADNDTEVYRD 344
Score = 26.6 bits (57), Expect = 7.1
Identities = 16/64 (25%), Positives = 29/64 (45%), Gaps = 8/64 (12%)
Query: 83 EPVVDRKGKG-----IMRDDK---GKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSD 134
E +V R+ G +MR + G + + + ++ ++ + DD+ D N G D D
Sbjct: 236 EALVKRRSLGKEAAVLMRSPRIASGSRRRRNSRNMEQQNASEAHEDDDNDDNNNRGRDKD 295
Query: 135 FSDD 138
S D
Sbjct: 296 SSSD 299
>At4g12610 putative protein
Length = 649
Score = 51.2 bits (121), Expect = 3e-07
Identities = 30/89 (33%), Positives = 47/89 (52%), Gaps = 13/89 (14%)
Query: 53 SSSDAVPNNEPENNTD----------DDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGK 102
+ D N+PE D + +D++DEEN +EE + + GK + + GK
Sbjct: 294 TDDDEAVGNDPEEREDLLAPEIPAPPEIKQDEDDEENEEEEGGLSKSGKELKKL-LGKAN 352
Query: 103 MIEEEDEDDDDSDDSDDSDDDSDGNVSGS 131
++E DEDDD DDSDD ++ + G V+ S
Sbjct: 353 GLDESDEDDD--DDSDDEEETNYGTVTNS 379
Score = 38.1 bits (87), Expect = 0.002
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 7/86 (8%)
Query: 60 NNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDD 119
+ E N +D DED+E+E + ++RK DD +G + D DDDD + DD
Sbjct: 230 DEEEGNVSDRGDEDEEEEASRKSRLGLNRKSND---DDDEEGPRGGDLDMDDDDIEKGDD 286
Query: 120 SDDD----SDGNVSGSDSDFSDDPLA 141
+ + D G+D + +D LA
Sbjct: 287 WEHEEIFTDDDEAVGNDPEEREDLLA 312
Score = 29.3 bits (64), Expect = 1.1
Identities = 19/81 (23%), Positives = 34/81 (41%), Gaps = 8/81 (9%)
Query: 72 EDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSD--------DSDDSDDD 123
E D ++E+ R K D+ +G + + DED+++ + +DDD
Sbjct: 205 EVDNEKESGGTSGGGGRGRKKSSGGDEEEGNVSDRGDEDEEEEASRKSRLGLNRKSNDDD 264
Query: 124 SDGNVSGSDSDFSDDPLAEVD 144
+ G D D DD + + D
Sbjct: 265 DEEGPRGGDLDMDDDDIEKGD 285
>At2g27470 putative CCAAT-binding transcription factor subunit
Length = 275
Score = 51.2 bits (121), Expect = 3e-07
Identities = 38/147 (25%), Positives = 64/147 (42%), Gaps = 24/147 (16%)
Query: 11 EPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDD 70
+P KRK ++ T K+ T D+ETE T + + + E EN D++
Sbjct: 130 KPKETKKRKQEEPSTQKGARKSKIDEETKRNDEETENDNTEEENGN---DEEDENGNDEE 186
Query: 71 DEDDE--------DEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS-- 120
DE+D+ DEEN DE + G + + + +EE + ++S + D S
Sbjct: 187 DENDDENTEENGNDEENDDEN--TEENGNDEENEKEDEENSMEENGNESEESGNEDHSME 244
Query: 121 ---------DDDSDGNVSGSDSDFSDD 138
+++ DG+VSGS + D
Sbjct: 245 ENGSGVGEDNENEDGSVSGSGEEVESD 271
Score = 40.8 bits (94), Expect = 4e-04
Identities = 24/110 (21%), Positives = 55/110 (49%), Gaps = 22/110 (20%)
Query: 40 TVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKG 99
T ++ E P+T + + + E + N ++ + D+ +EEN ++E D+ G
Sbjct: 134 TKKRKQEEPSTQKGARKSKIDEETKRNDEETENDNTEEENGNDE-----------EDENG 182
Query: 100 KGKMIEEEDEDDDDS-------DDSDDSDDDSDGNVSGSDSDFSDDPLAE 142
+EEDE+DD++ +++DD + + +GN ++ + ++ + E
Sbjct: 183 N----DEEDENDDENTEENGNDEENDDENTEENGNDEENEKEDEENSMEE 228
Score = 38.9 bits (89), Expect = 0.001
Identities = 26/116 (22%), Positives = 53/116 (45%), Gaps = 17/116 (14%)
Query: 3 EPETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNE 62
E E N EE + + ++ + N N D+E + T + +D E
Sbjct: 170 EEENGNDEEDENGNDEEDENDDENTEENGN---------DEENDDENTEENGNDEENEKE 220
Query: 63 PENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDK-------GKGKMIEEEDEDD 111
E N+ +++ +E EE+ +E+ ++ G G+ D++ G G+ +E ++ED+
Sbjct: 221 DEENSMEEN-GNESEESGNEDHSMEENGSGVGEDNENEDGSVSGSGEEVESDEEDE 275
>At2g22080 En/Spm-like transposon protein
Length = 140
Score = 51.2 bits (121), Expect = 3e-07
Identities = 27/97 (27%), Positives = 47/97 (47%), Gaps = 28/97 (28%)
Query: 60 NNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDD 119
++EPE + +DDEDD++ EN DE+ E+ED+D++DD +
Sbjct: 72 DDEPEGDDGNDDEDDDNHENDDED-----------------------EEEDEDENDDGGE 108
Query: 120 SDDDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRR 156
DDD D V + + +D D + + P + R++
Sbjct: 109 EDDDEDAEVEEEEEEEDED-----DEEALQPPKKRKK 140
Score = 50.1 bits (118), Expect = 6e-07
Identities = 34/100 (34%), Positives = 51/100 (51%), Gaps = 15/100 (15%)
Query: 56 DAVPNNEPENNTDDDDEDDEDEENSDEE---------PVVDR--KGKGIMRDDKGKGKMI 104
DAV N NN D D+ EDEE S ++ PV D G+ DD+ +G
Sbjct: 23 DAVEGNG--NNDDSGDDGGEDEEGSADDAEGKETKKGPVSDPDLNGEAGDNDDEPEGDDG 80
Query: 105 EEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVD 144
++++DD+ +D +D ++D D N G + D DD AEV+
Sbjct: 81 NDDEDDDNHENDDEDEEEDEDENDDGGEED--DDEDAEVE 118
Score = 43.1 bits (100), Expect = 7e-05
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 2/63 (3%)
Query: 55 SDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDS 114
+D P + N+ +DDD + D+E DEE D G DD ++ EEE+E+D+D
Sbjct: 71 NDDEPEGDDGNDDEDDDNHENDDE--DEEEDEDENDDGGEEDDDEDAEVEEEEEEEDEDD 128
Query: 115 DDS 117
+++
Sbjct: 129 EEA 131
Score = 35.8 bits (81), Expect = 0.012
Identities = 28/94 (29%), Positives = 42/94 (43%), Gaps = 16/94 (17%)
Query: 23 EETHHLPNKNPKLTSTT-TVDQETELPTTVNSSSDAVPNNEPENNTDDDDE-DDEDEENS 80
+ET P +P L D E E + D N+ E+ +D+DE DD EE+
Sbjct: 52 KETKKGPVSDPDLNGEAGDNDDEPEGDDGNDDEDDDNHENDDEDEEEDEDENDDGGEEDD 111
Query: 81 DEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDS 114
DE+ V+ + EEEDEDD+++
Sbjct: 112 DEDAEVEEEE--------------EEEDEDDEEA 131
>At3g14670 hypothetical protein
Length = 232
Score = 50.8 bits (120), Expect = 4e-07
Identities = 40/149 (26%), Positives = 64/149 (42%), Gaps = 22/149 (14%)
Query: 4 PETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPT---TVNSSSDAVPN 60
P T + T T P E +NP + V++E ++PT TV + N
Sbjct: 18 PGTKPIDAVTKATTEPPMTTEEPSASKQNPVVIEGRGVEEE-QIPTIITTVVEEGEKSDN 76
Query: 61 NEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDS 120
NE EN+ D+ E+ E+EE+ +EE + K EEE+++++ + S
Sbjct: 77 NEEENSEKDEKEESEEEESEEEEKEEEEK---------------EEEEKEEEGNVAGGGS 121
Query: 121 DDDSD---GNVSGSDSDFSDDPLAEVDLD 146
DDS G S SD + D+ +D
Sbjct: 122 SDDSSRTLGKESSSDENMDDETAVGKQVD 150
Score = 30.4 bits (67), Expect = 0.49
Identities = 34/159 (21%), Positives = 54/159 (33%), Gaps = 21/159 (13%)
Query: 3 EPETNNHEEPTFPTKRKPDQEETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNE 62
E NN EE + +++ +EE K + ++E N
Sbjct: 72 EKSDNNEEENSEKDEKEESEEEESEEEEKEEEEKEEEEKEEE---------------GNV 116
Query: 63 PENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSD-DSD 121
+ DD +E+S +E + D G D K+ E E+D D + D D +
Sbjct: 117 AGGGSSDDSSRTLGKESSSDENMDDETAVGKQVDIPAAMKINEMGQENDGDPKEKDGDLE 176
Query: 122 DDSDGNVSGSDSDFSDDPLAEVDLDNILPSRTRRRTAQS 160
D D + D + E D P R R Q+
Sbjct: 177 KDGDQEKDPKEKDPKEKDPKEKD-----PKRRTRMVLQN 210
>At1g48400
Length = 513
Score = 50.4 bits (119), Expect = 5e-07
Identities = 25/87 (28%), Positives = 43/87 (48%), Gaps = 25/87 (28%)
Query: 41 VDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGK 100
V+ L V+++ +++ +++ DDDD+DD+D+++ D
Sbjct: 266 VEARLSLRLWVSTNDYDYSDDDDDDDDDDDDDDDDDDDDDD------------------- 306
Query: 101 GKMIEEEDEDDDDSDDSDDSDDDSDGN 127
+D+DDDD DD DD DDD DG+
Sbjct: 307 ------DDDDDDDDDDDDDDDDDDDGD 327
Score = 49.7 bits (117), Expect = 8e-07
Identities = 25/86 (29%), Positives = 40/86 (46%), Gaps = 24/86 (27%)
Query: 58 VPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDS 117
V N+ + + DDDD+DD+D+++ D++ D+DDDD DD
Sbjct: 276 VSTNDYDYSDDDDDDDDDDDDDDDDD------------------------DDDDDDDDDD 311
Query: 118 DDSDDDSDGNVSGSDSDFSDDPLAEV 143
DD DDD D + + +P A +
Sbjct: 312 DDDDDDDDDDDDDDGDYYIVEPKAPI 337
Score = 43.1 bits (100), Expect = 7e-05
Identities = 19/41 (46%), Positives = 24/41 (58%)
Query: 107 EDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
+D+DDDD DD DD DDD D + D D DD + D D+
Sbjct: 285 DDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 325
Score = 38.5 bits (88), Expect = 0.002
Identities = 18/40 (45%), Positives = 21/40 (52%)
Query: 108 DEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
D DDD DD DD DDD D + D D DD + D D+
Sbjct: 282 DYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 321
Score = 35.8 bits (81), Expect = 0.012
Identities = 17/44 (38%), Positives = 21/44 (47%)
Query: 104 IEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDN 147
+ D D D DD DD DDD D + D D DD + D D+
Sbjct: 276 VSTNDYDYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 319
Score = 32.0 bits (71), Expect = 0.17
Identities = 12/47 (25%), Positives = 27/47 (56%)
Query: 37 STTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEE 83
ST D + + D +++ +++ DDDD+DD+D+++ D++
Sbjct: 277 STNDYDYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 323
Score = 30.0 bits (66), Expect = 0.65
Identities = 10/38 (26%), Positives = 25/38 (65%)
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRK 89
+ D +++ +++ DDDD+DD+D+++ + +V+ K
Sbjct: 297 DDDDDDDDDDDDDDDDDDDDDDDDDDDDDGDYYIVEPK 334
>At5g22640 unknown protein
Length = 871
Score = 50.1 bits (118), Expect = 6e-07
Identities = 32/97 (32%), Positives = 50/97 (50%), Gaps = 14/97 (14%)
Query: 41 VDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDE-----ENSD-------EEPVVDR 88
+ QE EL +A+ + + E + +E+ + E E+ D EE VV
Sbjct: 609 IQQELELVEAEICLEEAIEDMDEELKKKEQEEEKKTEMGLTEEDEDVLVPVYKEEKVVTA 668
Query: 89 KGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSD 125
K K ++++K + K +++DEDDDD DD DD DDD D
Sbjct: 669 KEK--IQENKQEEKYKDDDDEDDDDGDDDDDDDDDDD 703
Score = 32.3 bits (72), Expect = 0.13
Identities = 26/125 (20%), Positives = 42/125 (32%), Gaps = 31/125 (24%)
Query: 23 EETHHLPNKNPKLTSTTTVDQETELPTTVNSSSDAVPNNEP--ENNTDDDDEDDEDEENS 80
EE + K T +++ ++ V V E EN ++ +DD+D
Sbjct: 631 EELKKKEQEEEKKTEMGLTEEDEDVLVPVYKEEKVVTAKEKIQENKQEEKYKDDDD---- 686
Query: 81 DEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPL 140
E+D+D DD DD DD DD + +D + P
Sbjct: 687 -------------------------EDDDDGDDDDDDDDDDDLGPSSFGSADKGRRNSPF 721
Query: 141 AEVDL 145
+ L
Sbjct: 722 SSSSL 726
Score = 30.4 bits (67), Expect = 0.49
Identities = 33/140 (23%), Positives = 49/140 (34%), Gaps = 38/140 (27%)
Query: 18 RKPDQEETHHLPNKNPKLTSTTTVDQETELPT-----TVNSSSDAVPNNEPENNTDDDDE 72
+K +QEE K ++ T D++ +P V + N + E DDDDE
Sbjct: 634 KKKEQEE-----EKKTEM-GLTEEDEDVLVPVYKEEKVVTAKEKIQENKQEEKYKDDDDE 687
Query: 73 DDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSD 132
DD+D + ++D+DDDD D S +D S
Sbjct: 688 DDDDGD---------------------------DDDDDDDDDDLGPSSFGSADKGRRNSP 720
Query: 133 SDFSDDPLAEVDLDNILPSR 152
S A L + SR
Sbjct: 721 FSSSSLSFASCTLFPAVQSR 740
>At5g42280 putative protein
Length = 694
Score = 48.9 bits (115), Expect = 1e-06
Identities = 29/87 (33%), Positives = 41/87 (46%), Gaps = 22/87 (25%)
Query: 62 EPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSD 121
E ++N DDDD+DD+D++N D+ VVD D+DDDD DD D D
Sbjct: 158 EDDDNDDDDDDDDDDDDNDDD--VVD--------------------DDDDDDDDDDDVVD 195
Query: 122 DDSDGNVSGSDSDFSDDPLAEVDLDNI 148
DD + SD + +V L +
Sbjct: 196 DDYGDSACLSDGNHLKCKCCQVPLQKV 222
Score = 43.1 bits (100), Expect = 7e-05
Identities = 26/95 (27%), Positives = 45/95 (47%), Gaps = 29/95 (30%)
Query: 50 TVNSSSDAVPNNEPENNTDDD-----DEDDEDEENSDEEPVVDRKGKGIMRDDKGKGKMI 104
+V+ + D P N + ++ ++ D+D ED++N D+
Sbjct: 128 SVDPTFDFSPRNWKDKSSTEESIYEVDDDCEDDDNDDD---------------------- 165
Query: 105 EEEDEDDDDSDDS--DDSDDDSDGNVSGSDSDFSD 137
+++D+DDDD+DD DD DDD D + D D+ D
Sbjct: 166 DDDDDDDDDNDDDVVDDDDDDDDDDDDVVDDDYGD 200
Score = 40.0 bits (92), Expect = 6e-04
Identities = 16/35 (45%), Positives = 22/35 (62%)
Query: 104 IEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDD 138
++++ EDDD+ DD DD DDD D + D D DD
Sbjct: 153 VDDDCEDDDNDDDDDDDDDDDDNDDDVVDDDDDDD 187
Score = 35.8 bits (81), Expect = 0.012
Identities = 18/53 (33%), Positives = 27/53 (49%)
Query: 97 DKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSDFSDDPLAEVDLDNIL 149
DK + E +DD + DD+DD DDD D + D DD + D D+++
Sbjct: 142 DKSSTEESIYEVDDDCEDDDNDDDDDDDDDDDDNDDDVVDDDDDDDDDDDDVV 194
Score = 29.3 bits (64), Expect = 1.1
Identities = 12/46 (26%), Positives = 26/46 (56%)
Query: 52 NSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVDRKGKGIMRDD 97
++ D +++ ++N DD +DD+D+++ D++ V D G D
Sbjct: 161 DNDDDDDDDDDDDDNDDDVVDDDDDDDDDDDDVVDDDYGDSACLSD 206
Score = 28.1 bits (61), Expect = 2.5
Identities = 14/57 (24%), Positives = 30/57 (52%), Gaps = 2/57 (3%)
Query: 31 KNPKLTSTTTVDQETELPTTVNSSSDAVPNNEPENNTDDDDEDDEDEENSDEEPVVD 87
K+ T + + + + N D +++ +N+ D D+DD+D++ D++ VVD
Sbjct: 141 KDKSSTEESIYEVDDDCEDDDNDDDDDDDDDDDDNDDDVVDDDDDDDD--DDDDVVD 195
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.298 0.124 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,467,288
Number of Sequences: 26719
Number of extensions: 337764
Number of successful extensions: 13727
Number of sequences better than 10.0: 1204
Number of HSP's better than 10.0 without gapping: 783
Number of HSP's successfully gapped in prelim test: 427
Number of HSP's that attempted gapping in prelim test: 2418
Number of HSP's gapped (non-prelim): 4689
length of query: 172
length of database: 11,318,596
effective HSP length: 92
effective length of query: 80
effective length of database: 8,860,448
effective search space: 708835840
effective search space used: 708835840
T: 11
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC133780.10


