

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133572.9 + phase: 0
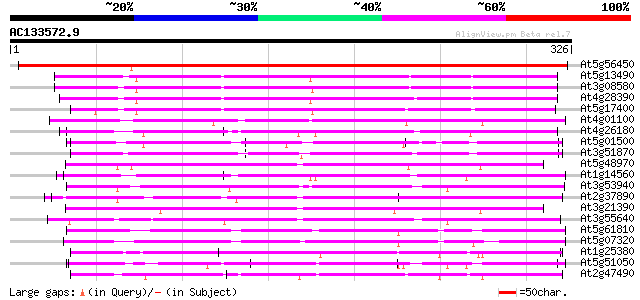
(326 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g56450 ADP/ATP translocase-like protein 503 e-143
At5g13490 adenosine nucleotide translocator 214 4e-56
At3g08580 adenylate translocator 213 1e-55
At4g28390 ADP,ATP carrier-like protein 208 3e-54
At5g17400 ADP/ATP translocase-like protein 199 2e-51
At4g01100 putative carrier protein 133 1e-31
At4g26180 putative mitochondrial carrier protein 133 1e-31
At5g01500 unknown protein 130 7e-31
At3g51870 carrier like protein 130 7e-31
At5g48970 mitochondrial carrier protein-like 130 1e-30
At1g14560 mitochondrial carrier like protein 130 1e-30
At3g53940 unknown protein 125 3e-29
At2g37890 mitochondrial carrier like protein 122 3e-28
At3g21390 unknown protein 116 1e-26
At3g55640 Ca-dependent solute carrier - like protein 113 1e-25
At5g61810 peroxisomal Ca-dependent solute carrier - like protein 112 2e-25
At5g07320 peroxisomal Ca-dependent solute carrier-like protein 107 7e-24
At1g25380 unknown protein 107 1e-23
At5g51050 calcium-binding transporter-like protein 103 1e-22
At2g47490 mitochondrial carrier like protein 102 2e-22
>At5g56450 ADP/ATP translocase-like protein
Length = 330
Score = 503 bits (1295), Expect = e-143
Identities = 246/323 (76%), Positives = 285/323 (88%), Gaps = 4/323 (1%)
Query: 6 EDPEEKALNLKRKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNL 65
ED E+ + N + + +FQ+DL+AGAVMGG VHTIVAPIERAKLLLQTQESN+
Sbjct: 6 EDEEDPSRNRRNQSPLSLPQTLKHFQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNI 65
Query: 66 AIVA----SGRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKS 121
AIV +G+R+FKGMFD I RTVREEGV+SLWRGNGSSVLRYYPSVALNFSLKDLY+S
Sbjct: 66 AIVGDEGHAGKRRFKGMFDFIFRTVREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRS 125
Query: 122 ILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIH 181
ILR +S ++IFSGA ANF+AG+AAGCT+LI+VYPLDIAHTRLAADIG+ E RQFRGIH
Sbjct: 126 ILRNSSSQENHIFSGALANFMAGSAAGCTALIVVYPLDIAHTRLAADIGKPEARQFRGIH 185
Query: 182 HFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMV 241
HFL+TI +KDGVRGIYRGLPASLHG++IHRGLYFGGFDT+KE+ SE++KPELALWKRW +
Sbjct: 186 HFLSTIHKKDGVRGIYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPELALWKRWGL 245
Query: 242 AQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNV 301
AQAVTTSAGL SYPLDTVRRR+MMQSGMEHP+Y STLDCW+KIYR+EGL SFYRGA+SN+
Sbjct: 246 AQAVTTSAGLASYPLDTVRRRIMMQSGMEHPMYRSTLDCWKKIYRSEGLASFYRGALSNM 305
Query: 302 FRSTGAAAILVLYDEVKKFMNLG 324
FRSTG+AAILV YDEVK+F+N G
Sbjct: 306 FRSTGSAAILVFYDEVKRFLNWG 328
>At5g13490 adenosine nucleotide translocator
Length = 385
Score = 214 bits (546), Expect = 4e-56
Identities = 120/298 (40%), Positives = 175/298 (58%), Gaps = 12/298 (4%)
Query: 27 FNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGR--RKFKGMFDCIIR 84
F NF D M G V T APIER KLL+Q Q+ L +GR +KG+ DC R
Sbjct: 81 FTNFAIDFMMGGVSAAVSKTAAAPIERVKLLIQNQDEMLK---AGRLTEPYKGIRDCFGR 137
Query: 85 TVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAG 144
T+R+EG+ SLWRGN ++V+RY+P+ ALNF+ KD +K + + D + + N +G
Sbjct: 138 TIRDEGIGSLWRGNTANVIRYFPTQALNFAFKDYFKRLF-NFKKDKDGYWKWFAGNLASG 196
Query: 145 AAAGCTSLILVYPLDIAHTRLAADIGRTE----VRQFRGIHHFLATIFQKDGVRGIYRGL 200
AAG +SL+ VY LD A TRLA D + RQF G+ + DG+ G+YRG
Sbjct: 197 GAAGASSLLFVYSLDYARTRLANDSKSAKKGGGERQFNGLVDVYKKTLKSDGIAGLYRGF 256
Query: 201 PASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVR 260
S G++++RGLYFG +D++K +L + + + + + +T AGL SYP+DTVR
Sbjct: 257 NISCAGIIVYRGLYFGLYDSVKPVLLTGDLQD-SFFASFALGWLITNGAGLASYPIDTVR 315
Query: 261 RRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
RRMMM SG E Y S+ D + +I + EG S ++GA +N+ R+ A +L YD+++
Sbjct: 316 RRMMMTSG-EAVKYKSSFDAFSQIVKKEGAKSLFKGAGANILRAVAGAGVLAGYDKLQ 372
>At3g08580 adenylate translocator
Length = 381
Score = 213 bits (542), Expect = 1e-55
Identities = 118/298 (39%), Positives = 177/298 (58%), Gaps = 12/298 (4%)
Query: 27 FNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGR--RKFKGMFDCIIR 84
F NF D + G V T APIER KLL+Q Q+ ++ +GR +KG+ DC R
Sbjct: 77 FTNFALDFLMGGVSAAVSKTAAAPIERVKLLIQNQDE---MIKAGRLSEPYKGIGDCFGR 133
Query: 85 TVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAG 144
T+++EG SLWRGN ++V+RY+P+ ALNF+ KD +K + + D + + N +G
Sbjct: 134 TIKDEGFGSLWRGNTANVIRYFPTQALNFAFKDYFKRLF-NFKKDRDGYWKWFAGNLASG 192
Query: 145 AAAGCTSLILVYPLDIAHTRLAADIGRTEV----RQFRGIHHFLATIFQKDGVRGIYRGL 200
AAG +SL+ VY LD A TRLA D + RQF G+ + DG+ G+YRG
Sbjct: 193 GAAGASSLLFVYSLDYARTRLANDAKAAKKGGGGRQFDGLVDVYRKTLKTDGIAGLYRGF 252
Query: 201 PASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVR 260
S G++++RGLYFG +D++K +L + + + + + +T AGL SYP+DTVR
Sbjct: 253 NISCVGIIVYRGLYFGLYDSVKPVLLTGDLQD-SFFASFALGWVITNGAGLASYPIDTVR 311
Query: 261 RRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
RRMMM SG E Y S+LD +++I + EG S ++GA +N+ R+ A +L YD+++
Sbjct: 312 RRMMMTSG-EAVKYKSSLDAFKQILKNEGAKSLFKGAGANILRAVAGAGVLSGYDKLQ 368
>At4g28390 ADP,ATP carrier-like protein
Length = 379
Score = 208 bits (530), Expect = 3e-54
Identities = 116/294 (39%), Positives = 174/294 (58%), Gaps = 11/294 (3%)
Query: 30 FQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGR--RKFKGMFDCIIRTVR 87
F D + G V T APIER KLL+Q Q+ ++ +GR +KG+ DC RTV+
Sbjct: 79 FLIDFLMGGVSAAVSKTAAAPIERVKLLIQNQDE---MIKAGRLSEPYKGISDCFARTVK 135
Query: 88 EEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAA 147
+EG+++LWRGN ++V+RY+P+ ALNF+ KD +K + D + + N +G AA
Sbjct: 136 DEGMLALWRGNTANVIRYFPTQALNFAFKDYFKRLF-NFKKEKDGYWKWFAGNLASGGAA 194
Query: 148 GCTSLILVYPLDIAHTRLAADIGRTE---VRQFRGIHHFLATIFQKDGVRGIYRGLPASL 204
G +SL+ VY LD A TRLA D + RQF G+ DG+ G+YRG S
Sbjct: 195 GASSLLFVYSLDYARTRLANDAKAAKKGGQRQFNGMVDVYKKTIASDGIVGLYRGFNISC 254
Query: 205 HGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMM 264
G+V++RGLYFG +D++K ++ + + L +++ +T AGL SYP+DTVRRRMM
Sbjct: 255 VGIVVYRGLYFGLYDSLKPVVLVDGLQDSFL-ASFLLGWGITIGAGLASYPIDTVRRRMM 313
Query: 265 MQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
M SG E Y S+L + +I + EG S ++GA +N+ R+ A +L YD+++
Sbjct: 314 MTSG-EAVKYKSSLQAFSQIVKNEGAKSLFKGAGANILRAVAGAGVLAGYDKLQ 366
>At5g17400 ADP/ATP translocase-like protein
Length = 306
Score = 199 bits (505), Expect = 2e-51
Identities = 114/292 (39%), Positives = 172/292 (58%), Gaps = 17/292 (5%)
Query: 36 AGAVMGGAVHTIV----APIERAKLLLQTQESNLAIVASGR--RKFKGMFDCIIRTVREE 89
A VMGGA + APIER KLLLQ Q ++ +G R + G+ +C R REE
Sbjct: 12 ADFVMGGAAAIVAKSAAAPIERVKLLLQNQGE---MIKTGHLIRPYTGLGNCFTRIYREE 68
Query: 90 GVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGC 149
GV+S WRGN ++V+RY+P+ A NF+ K +K++L G + D + N +G+AAG
Sbjct: 69 GVLSFWRGNQANVIRYFPTQASNFAFKGYFKNLL-GCSKEKDGYLKWFAGNVASGSAAGA 127
Query: 150 TSLILVYPLDIAHTRLAADIGRTEV---RQFRGIHHFLATIFQKDGVRGIYRGLPASLHG 206
T+ + +Y LD A TRL D V RQF+G+ DG++G+YRG S+ G
Sbjct: 128 TTSLFLYHLDYARTRLGTDAKECSVNGKRQFKGMIDVYRKTLSSDGIKGLYRGFGVSIVG 187
Query: 207 MVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQ 266
+ ++RG+YFG +DTIK ++ S E +++ ++TTSAG+++YP DT+RRRMM+
Sbjct: 188 ITLYRGMYFGMYDTIKPIVLVGSL-EGNFLASFLLGWSITTSAGVIAYPFDTLRRRMMLT 246
Query: 267 SGMEHPV-YNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEV 317
SG PV Y +T+ R+I ++EG + YRG +N+ A +L YD++
Sbjct: 247 SG--QPVKYRNTIHALREILKSEGFYALYRGVTANMLLGVAGAGVLAGYDQL 296
>At4g01100 putative carrier protein
Length = 352
Score = 133 bits (335), Expect = 1e-31
Identities = 100/326 (30%), Positives = 150/326 (45%), Gaps = 37/326 (11%)
Query: 24 SSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCII 83
S F + + L AG V GG T VAP+ER K+LLQ Q + K+ G +
Sbjct: 32 SYAFKSICKSLFAGGVAGGVSRTAVAPLERMKILLQVQNPH-------NIKYSGTVQGLK 84
Query: 84 RTVREEGVISLWRGNGSSVLRYYPSVALNFSLKD------LYKSILRGGNSNPDNIFSGA 137
R EG+ L++GNG++ R P+ A+ F + LY R GN N
Sbjct: 85 HIWRTEGLRGLFKGNGTNCARIVPNSAVKFFSYEQASNGILYMYRQRTGNENAQLT---P 141
Query: 138 SANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIY 197
AGA AG ++ YP+D+ RL + Q+RGI H LAT+ +++G R +Y
Sbjct: 142 LLRLGAGATAGIIAMSATYPMDMVRGRLTVQTANSPY-QYRGIAHALATVLREEGPRALY 200
Query: 198 RGLPASLHGMVIHRGLYFGGFDTIKEMLSEES------KPELALWKRWMVAQAVTTSAGL 251
RG S+ G+V + GL F ++++K+ L +E+ EL + R T
Sbjct: 201 RGWLPSVIGVVPYVGLNFSVYESLKDWLVKENPYGLVENNELTVVTRLTCGAIAGTVGQT 260
Query: 252 VSYPLDTVRRRMMMQSGMEHPV-------------YNSTLDCWRKIYRTEGLISFYRGAV 298
++YPLD +RRRM M + Y +D +RK R EG + Y+G V
Sbjct: 261 IAYPLDVIRRRMQMVGWKDASAIVTGEGRSTASLEYTGMVDAFRKTVRHEGFGALYKGLV 320
Query: 299 SNVFRSTGAAAI-LVLYDEVKKFMNL 323
N + + AI V Y+ VK + +
Sbjct: 321 PNSVKVVPSIAIAFVTYEMVKDVLGV 346
>At4g26180 putative mitochondrial carrier protein
Length = 325
Score = 133 bits (334), Expect = 1e-31
Identities = 98/300 (32%), Positives = 157/300 (51%), Gaps = 27/300 (9%)
Query: 30 FQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFK--GMFDCIIRTVR 87
F ++L+AG V GG T VAP+ER K+L QT+ R +FK G+ I + +
Sbjct: 17 FAKELIAGGVTGGIAKTAVAPLERIKILFQTR----------RDEFKRIGLVGSINKIGK 66
Query: 88 EEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAA 147
EG++ +RGNG+SV R P AL++ + Y+ + G PD G + VAG+ A
Sbjct: 67 TEGLMGFYRGNGASVARIVPYAALHYMAYEEYRRWIIFGF--PDTT-RGPLLDLVAGSFA 123
Query: 148 GCTSLILVYPLDIAHTRLA--ADIGRTEVRQ--FRGIHHFLATIFQKDGVRGIYRGLPAS 203
G T+++ YPLD+ T+LA + V Q +RGI + +++ G RG+YRG+ S
Sbjct: 124 GGTAVLFTYPLDLVRTKLAYQTQVKAIPVEQIIYRGIVDCFSRTYRESGARGLYRGVAPS 183
Query: 204 LHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAG-LVSYPLDTVRRR 262
L+G+ + GL F ++ +K + E K +++L +V +V G ++YPLD VRR+
Sbjct: 184 LYGIFPYAGLKFYFYEEMKRHVPPEHKQDISL---KLVCGSVAGLLGQTLTYPLDVVRRQ 240
Query: 263 MMMQ---SGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVK 318
M ++ S ++ T+ KI R EG + G N + + AI +YD +K
Sbjct: 241 MQVERLYSAVKEETRRGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSVAIGFTVYDIMK 300
Score = 42.7 bits (99), Expect = 3e-04
Identities = 27/91 (29%), Positives = 45/91 (48%), Gaps = 3/91 (3%)
Query: 34 LMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVIS 93
L+ G+V G T+ P++ + +Q + A+ RR G + + REEG
Sbjct: 217 LVCGSVAGLLGQTLTYPLDVVRRQMQVERLYSAVKEETRR---GTMQTLFKIAREEGWKQ 273
Query: 94 LWRGNGSSVLRYYPSVALNFSLKDLYKSILR 124
L+ G + L+ PSVA+ F++ D+ K LR
Sbjct: 274 LFSGLSINYLKVVPSVAIGFTVYDIMKLHLR 304
>At5g01500 unknown protein
Length = 415
Score = 130 bits (328), Expect = 7e-31
Identities = 94/292 (32%), Positives = 143/292 (48%), Gaps = 30/292 (10%)
Query: 36 AGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFK---GMFDCIIRTVREEGVI 92
AGA G A ++ AP++R KLL+QT V +G++ K G + I +EEG+
Sbjct: 121 AGAFAGAAAKSVTAPLDRIKLLMQTHG-----VRAGQQSAKKAIGFIEAITLIGKEEGIK 175
Query: 93 SLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSL 152
W+GN V+R P A+ + YK + RG + AGA AG TS
Sbjct: 176 GYWKGNLPQVIRIVPYSAVQLFAYETYKKLFRGKDGQ-----LSVLGRLGAGACAGMTST 230
Query: 153 ILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRG 212
++ YPLD+ RLA + G +R + + +++GV Y GL SL + +
Sbjct: 231 LITYPLDVLRLRLAVEPG------YRTMSQVALNMLREEGVASFYNGLGPSLLSIAPYIA 284
Query: 213 LYFGGFDTIKEMLSE--ESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSGME 270
+ F FD +K+ L E + K + +L +VA A+ T YPLDT+RR+M ++
Sbjct: 285 INFCVFDLVKKSLPEKYQQKTQSSLLTA-VVAAAIATG---TCYPLDTIRRQMQLKG--- 337
Query: 271 HPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKFM 321
Y S LD + I EG++ YRG V N +S ++I L +D VKK +
Sbjct: 338 -TPYKSVLDAFSGIIAREGVVGLYRGFVPNALKSMPNSSIKLTTFDIVKKLI 388
Score = 58.2 bits (139), Expect = 6e-09
Identities = 45/197 (22%), Positives = 83/197 (41%), Gaps = 22/197 (11%)
Query: 34 LMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVIS 93
L AGA G I P++ +L L + ++ M + +REEGV S
Sbjct: 219 LGAGACAGMTSTLITYPLDVLRLRLAVEPG-----------YRTMSQVALNMLREEGVAS 267
Query: 94 LWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLI 153
+ G G S+L P +A+NF + DL K L P+ ++ + A +
Sbjct: 268 FYNGLGPSLLSIAPYIAINFCVFDLVKKSL------PEKYQQKTQSSLLTAVVAAAIATG 321
Query: 154 LVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGL 213
YPLD ++ + + ++ + + I ++GV G+YRG + + + +
Sbjct: 322 TCYPLDTIRRQM-----QLKGTPYKSVLDAFSGIIAREGVVGLYRGFVPNALKSMPNSSI 376
Query: 214 YFGGFDTIKEMLSEESK 230
FD +K++++ K
Sbjct: 377 KLTTFDIVKKLIAASEK 393
Score = 51.6 bits (122), Expect = 6e-07
Identities = 43/186 (23%), Positives = 85/186 (45%), Gaps = 10/186 (5%)
Query: 138 SANFVAGAAAGCTSLILVYPLD---IAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVR 194
+A F AGA AG + + PLD + G+ ++ G + I +++G++
Sbjct: 116 AALFFAGAFAGAAAKSVTAPLDRIKLLMQTHGVRAGQQSAKKAIGFIEAITLIGKEEGIK 175
Query: 195 GIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSY 254
G ++G + +V + + ++T K++ + +L++ R ++ L++Y
Sbjct: 176 GYWKGNLPQVIRIVPYSAVQLFAYETYKKLFRGKD-GQLSVLGRLGAGACAGMTSTLITY 234
Query: 255 PLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVL 313
PLD +R R+ ++ G Y + + R EG+ SFY G ++ AI +
Sbjct: 235 PLDVLRLRLAVEPG-----YRTMSQVALNMLREEGVASFYNGLGPSLLSIAPYIAINFCV 289
Query: 314 YDEVKK 319
+D VKK
Sbjct: 290 FDLVKK 295
>At3g51870 carrier like protein
Length = 381
Score = 130 bits (328), Expect = 7e-31
Identities = 91/287 (31%), Positives = 139/287 (47%), Gaps = 20/287 (6%)
Query: 36 AGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVISLW 95
AGA+ G A T+ AP++R KLL+QT L +K G + I +EEGV W
Sbjct: 93 AGALAGAAAKTVTAPLDRIKLLMQTHGIRLG--QQSAKKAIGFIEAITLIAKEEGVKGYW 150
Query: 96 RGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLILV 155
+GN V+R P A+ + YK++ +G + I AGA AG TS +L
Sbjct: 151 KGNLPQVIRVLPYSAVQLLAYESYKNLFKGKDDQLSVI-----GRLAAGACAGMTSTLLT 205
Query: 156 YPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYF 215
YPLD+ RLA + ++R + ++ + +G+ Y GL SL G+ + + F
Sbjct: 206 YPLDVLRLRLAVE------PRYRTMSQVALSMLRDEGIASFYYGLGPSLVGIAPYIAVNF 259
Query: 216 GGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYN 275
FD +K+ L EE + + + A A L YPLDTVRR+M M+ Y
Sbjct: 260 CIFDLVKKSLPEEYRKKAQ--SSLLTAVLSAGIATLTCYPLDTVRRQMQMRG----TPYK 313
Query: 276 STLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKFM 321
S + + I +GLI YRG + N ++ ++I L +D VK+ +
Sbjct: 314 SIPEAFAGIIDRDGLIGLYRGFLPNALKTLPNSSIRLTTFDMVKRLI 360
Score = 50.1 bits (118), Expect = 2e-06
Identities = 43/186 (23%), Positives = 85/186 (45%), Gaps = 10/186 (5%)
Query: 138 SANFVAGAAAGCTSLILVYPLDIAHTRLAAD---IGRTEVRQFRGIHHFLATIFQKDGVR 194
+A F AGA AG + + PLD + +G+ ++ G + I +++GV+
Sbjct: 88 AAIFAAGALAGAAAKTVTAPLDRIKLLMQTHGIRLGQQSAKKAIGFIEAITLIAKEEGVK 147
Query: 195 GIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSY 254
G ++G + ++ + + +++ K + + +L++ R ++ L++Y
Sbjct: 148 GYWKGNLPQVIRVLPYSAVQLLAYESYKNLFKGKD-DQLSVIGRLAAGACAGMTSTLLTY 206
Query: 255 PLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRG-AVSNVFRSTGAAAILVL 313
PLD +R R+ ++ P Y + + R EG+ SFY G S V + A +
Sbjct: 207 PLDVLRLRLAVE-----PRYRTMSQVALSMLRDEGIASFYYGLGPSLVGIAPYIAVNFCI 261
Query: 314 YDEVKK 319
+D VKK
Sbjct: 262 FDLVKK 267
>At5g48970 mitochondrial carrier protein-like
Length = 339
Score = 130 bits (327), Expect = 1e-30
Identities = 87/305 (28%), Positives = 143/305 (46%), Gaps = 30/305 (9%)
Query: 33 DLMAGAVMGGAVHTIVAPIERAKLLLQTQ---ESNLAIVA---SGRRKFKGMFDCIIRTV 86
D AGA+ GG ++ +P++ K+ Q Q ++ +V SG K+ GM
Sbjct: 21 DASAGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQATKDIF 80
Query: 87 REEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAA 146
REEG WRGN ++L P ++ F++ KS G D+I +FV+GA
Sbjct: 81 REEGFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTEDHIHLSPYLSFVSGAL 140
Query: 147 AGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHG 206
AGC + + YP D+ T LA+ + E + + + I Q G+RG+Y GL +L
Sbjct: 141 AGCAATLGSYPFDLLRTILAS---QGEPKVYPTMRSAFVDIIQSRGIRGLYNGLTPTLVE 197
Query: 207 MVIHRGLYFGGFDTIKEMLSEESK------------PELALWKRWMVAQAVTTSAGLVSY 254
+V + GL FG +D K + + ++ L+ ++ ++ TSA LV +
Sbjct: 198 IVPYAGLQFGTYDMFKRWMMDWNRYKLSSKIPINVDTNLSSFQLFICGLGAGTSAKLVCH 257
Query: 255 PLDTVRRRMMMQSGMEHP---------VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRST 305
PLD V++R ++ HP Y + LD R+I +EG Y+G V + ++
Sbjct: 258 PLDVVKKRFQIEGLQRHPRYGARVERRAYRNMLDGLRQIMISEGWHGLYKGIVPSTVKAA 317
Query: 306 GAAAI 310
A A+
Sbjct: 318 PAGAV 322
>At1g14560 mitochondrial carrier like protein
Length = 331
Score = 130 bits (326), Expect = 1e-30
Identities = 93/312 (29%), Positives = 159/312 (50%), Gaps = 34/312 (10%)
Query: 32 RDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGV 91
+ L+AG G T VAP+ER K+LLQT+ ++ K G+ + + ++ +G
Sbjct: 25 KTLIAGGAAGAIAKTAVAPLERIKILLQTRTNDF--------KTLGVSQSLKKVLQFDGP 76
Query: 92 ISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTS 151
+ ++GNG+SV+R P AL++ ++Y+ + N + SG + VAG+AAG T+
Sbjct: 77 LGFYKGNGASVIRIIPYAALHYMTYEVYRDWILEKNLP---LGSGPIVDLVAGSAAGGTA 133
Query: 152 LILVYPLDIAHTRLAADIGRTE----------VRQ--FRGIHHFLATIFQKDGVRGIYRG 199
++ YPLD+A T+LA + T RQ + GI LA +++ G RG+YRG
Sbjct: 134 VLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQPTYSGIKEVLAMAYKEGGPRGLYRG 193
Query: 200 LPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAG-LVSYPLDT 258
+ +L G++ + GL F ++ +K + EE + + + + A+ G ++YPLD
Sbjct: 194 IGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSV---RMHLPCGALAGLFGQTITYPLDV 250
Query: 259 VRRRMM------MQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-L 311
VRR+M M S + Y +T D I RT+G + G N + + AI
Sbjct: 251 VRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYIKIVPSVAIGF 310
Query: 312 VLYDEVKKFMNL 323
+Y+ +K +M +
Sbjct: 311 TVYESMKSWMRI 322
Score = 42.7 bits (99), Expect = 3e-04
Identities = 25/97 (25%), Positives = 47/97 (47%)
Query: 28 NNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVR 87
N+ + L GA+ G TI P++ + +Q + +++K FD + VR
Sbjct: 225 NSVRMHLPCGALAGLFGQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVR 284
Query: 88 EEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILR 124
+G L+ G + ++ PSVA+ F++ + KS +R
Sbjct: 285 TQGWKQLFAGLSINYIKIVPSVAIGFTVYESMKSWMR 321
>At3g53940 unknown protein
Length = 365
Score = 125 bits (314), Expect = 3e-29
Identities = 94/302 (31%), Positives = 154/302 (50%), Gaps = 26/302 (8%)
Query: 34 LMAGAVMGGAVHTIVAPIERAKLLLQTQ--ESNLAIVASGRRKFKGMFDCIIRTVREEGV 91
L+AG + G T AP+ R +L Q Q +S AI++S ++ R V+EEG
Sbjct: 73 LLAGGIAGAFSKTCTAPLARLTILFQIQGMQSEAAILSS-----PNIWHEASRIVKEEGF 127
Query: 92 ISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGG---NSNPDNIFSGASANFVAGAAAG 148
+ W+GN +V P A+NF + YK+ L S N S +FV+G AG
Sbjct: 128 RAFWKGNLVTVAHRLPYGAVNFYAYEEYKTFLHSNPVLQSYKGNAGVDISVHFVSGGLAG 187
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMV 208
T+ YPLD+ TRL+A R + ++G+ H TI +++G+ G+Y+GL A+L G+
Sbjct: 188 LTAASATYPLDLVRTRLSAQ--RNSI-YYQGVGHAFRTICREEGILGLYKGLGATLLGVG 244
Query: 209 IHRGLYFGGFDTIKEM-LSEESKPELALWKRWMVAQAVTTSAGLVS----YPLDTVRRRM 263
+ F ++T K LS A +V+ + +G+VS +PLD VRRRM
Sbjct: 245 PSLAISFAAYETFKTFWLSHRPNDSNA-----VVSLGCGSLSGIVSSTATFPLDLVRRRM 299
Query: 264 MMQ-SGMEHPVYNSTL-DCWRKIYRTEGLISFYRGAVSNVFRST-GAAAILVLYDEVKKF 320
++ +G VY + L ++ I++TEG+ YRG + ++ G + ++E+KK
Sbjct: 300 QLEGAGGRARVYTTGLFGTFKHIFKTEGMRGLYRGIIPEYYKVVPGVGIAFMTFEELKKL 359
Query: 321 MN 322
++
Sbjct: 360 LS 361
>At2g37890 mitochondrial carrier like protein
Length = 337
Score = 122 bits (306), Expect = 3e-28
Identities = 87/308 (28%), Positives = 149/308 (48%), Gaps = 23/308 (7%)
Query: 25 SRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQ--ESNLAIVASGRRKFKGMFDCI 82
++ FQ +L+AG + G T AP+ R +L Q Q +S A+++ + +
Sbjct: 37 AKLGTFQ-NLLAGGIAGAISKTCTAPLARLTILFQLQGMQSEGAVLSRPNLRREAS---- 91
Query: 83 IRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNP------DNIFSG 136
R + EEG + W+GN +V+ P A+NF + Y NSNP N
Sbjct: 92 -RIINEEGYRAFWKGNLVTVVHRIPYTAVNFYAYEKYNLFF---NSNPVVQSFIGNTSGN 147
Query: 137 ASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGI 196
+FV+G AG T+ YPLD+ TRLAA + ++GI H TI +++G+ G+
Sbjct: 148 PIVHFVSGGLAGITAATATYPLDLVRTRLAA---QRNAIYYQGIEHTFRTICREEGILGL 204
Query: 197 YRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPL 256
Y+GL A+L G+ + F ++++K + L + + +YPL
Sbjct: 205 YKGLGATLLGVGPSLAINFAAYESMKLFWHSHRPNDSDLVVSLVSGGLAGAVSSTATYPL 264
Query: 257 DTVRRRMMMQ-SGMEHPVYNSTL-DCWRKIYRTEGLISFYRGAVSNVFRST-GAAAILVL 313
D VRRRM ++ +G VYN+ L ++ I+++EG YRG + ++ G + +
Sbjct: 265 DLVRRRMQVEGAGGRARVYNTGLFGTFKHIFKSEGFKGIYRGILPEYYKVVPGVGIVFMT 324
Query: 314 YDEVKKFM 321
YD +++ +
Sbjct: 325 YDALRRLL 332
Score = 73.2 bits (178), Expect = 2e-13
Identities = 50/207 (24%), Positives = 94/207 (45%), Gaps = 13/207 (6%)
Query: 21 SFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFD 80
SF + N ++G + G T P++ + L Q + + ++G+
Sbjct: 139 SFIGNTSGNPIVHFVSGGLAGITAATATYPLDLVRTRLAAQRNAIY--------YQGIEH 190
Query: 81 CIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASAN 140
REEG++ L++G G+++L PS+A+NF+ + K N ++ +
Sbjct: 191 TFRTICREEGILGLYKGLGATLLGVGPSLAINFAAYESMKLFWHSHRPNDSDLV----VS 246
Query: 141 FVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFR-GIHHFLATIFQKDGVRGIYRG 199
V+G AG S YPLD+ R+ + R + G+ IF+ +G +GIYRG
Sbjct: 247 LVSGGLAGAVSSTATYPLDLVRRRMQVEGAGGRARVYNTGLFGTFKHIFKSEGFKGIYRG 306
Query: 200 LPASLHGMVIHRGLYFGGFDTIKEMLS 226
+ + +V G+ F +D ++ +L+
Sbjct: 307 ILPEYYKVVPGVGIVFMTYDALRRLLT 333
>At3g21390 unknown protein
Length = 335
Score = 116 bits (291), Expect = 1e-26
Identities = 79/304 (25%), Positives = 138/304 (44%), Gaps = 29/304 (9%)
Query: 33 DLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTV----RE 88
D AG V G + +P++ K+ Q Q A A + K ++ + RT RE
Sbjct: 18 DASAGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTKDIFRE 77
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
EG+ WRGN ++L P ++ F++ KS G + ++ ++++GA AG
Sbjct: 78 EGLSGFWRGNVPALLMVVPYTSIQFAVLHKVKSFAAGSSKAENHAQLSPYLSYISGALAG 137
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMV 208
C + + YP D+ T LA+ + E + + + +I Q G++G+Y GL +L ++
Sbjct: 138 CAATVGSYPFDLLRTVLAS---QGEPKVYPNMRSAFLSIVQTRGIKGLYAGLSPTLIEII 194
Query: 209 IHRGLYFGGFDTIK-------------EMLSEESKPELALWKRWMVAQAVTTSAGLVSYP 255
+ GL FG +DT K S L+ ++ ++ A T + LV +P
Sbjct: 195 PYAGLQFGTYDTFKRWSMVYNKRYRSSSSSSTNPSDSLSSFQLFLCGLASGTVSKLVCHP 254
Query: 256 LDTVRRRMMMQSGMEHP---------VYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTG 306
LD V++R ++ HP Y + D +I R+EG Y+G V + ++
Sbjct: 255 LDVVKKRFQVEGLQRHPKYGARVELNAYKNMFDGLGQILRSEGWHGLYKGIVPSTIKAAP 314
Query: 307 AAAI 310
A A+
Sbjct: 315 AGAV 318
Score = 30.0 bits (66), Expect = 1.8
Identities = 29/111 (26%), Positives = 48/111 (43%), Gaps = 9/111 (8%)
Query: 16 KRKRNSFSSSR-----FNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVAS 70
KR R+S SSS ++FQ + G G + P++ K Q +
Sbjct: 216 KRYRSSSSSSTNPSDSLSSFQL-FLCGLASGTVSKLVCHPLDVVKKRFQVEGLQRHPKYG 274
Query: 71 GR---RKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDL 118
R +K MFD + + +R EG L++G S ++ P+ A+ F +L
Sbjct: 275 ARVELNAYKNMFDGLGQILRSEGWHGLYKGIVPSTIKAAPAGAVTFVAYEL 325
>At3g55640 Ca-dependent solute carrier - like protein
Length = 332
Score = 113 bits (283), Expect = 1e-25
Identities = 91/315 (28%), Positives = 151/315 (47%), Gaps = 27/315 (8%)
Query: 23 SSSRFNNFQRD-------LMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKF 75
SS R QR L+AG + G T AP+ R +L Q Q + A+ RK
Sbjct: 20 SSHRLTQDQRSHIESASQLLAGGLAGAFSKTCTAPLSRLTILFQVQ--GMHTNAAALRKP 77
Query: 76 KGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSIL---RGGNSNPDN 132
+ + R + EEG+ + W+GN ++ P ++NF + YK + G ++ +
Sbjct: 78 SILHEAS-RILNEEGLKAFWKGNLVTIAHRLPYSSVNFYAYEHYKKFMYMVTGMENHKEG 136
Query: 133 IFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDG 192
I S +FVAG AG T+ YPLD+ TRLAA +T+V + GI H L +I +G
Sbjct: 137 ISSNLFVHFVAGGLAGITAASATYPLDLVRTRLAA---QTKVIYYSGIWHTLRSITTDEG 193
Query: 193 VRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLV 252
+ G+Y+GL +L G+ + F +++++ + + MV+ A + +G+
Sbjct: 194 ILGLYKGLGTTLVGVGPSIAISFSVYESLRSYWRSTRPHDSPI----MVSLACGSLSGIA 249
Query: 253 S----YPLDTVRRRMMMQS-GMEHPVYNS-TLDCWRKIYRTEGLISFYRGAVSNVFRST- 305
S +PLD VRRR ++ G VY + L ++I +TEG YRG + ++
Sbjct: 250 SSTATFPLDLVRRRKQLEGIGGRAVVYKTGLLGTLKRIVQTEGARGLYRGILPEYYKVVP 309
Query: 306 GAAAILVLYDEVKKF 320
G + Y+ +K +
Sbjct: 310 GVGICFMTYETLKLY 324
>At5g61810 peroxisomal Ca-dependent solute carrier - like protein
Length = 478
Score = 112 bits (281), Expect = 2e-25
Identities = 85/297 (28%), Positives = 142/297 (47%), Gaps = 34/297 (11%)
Query: 34 LMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVIS 93
L+AG + G T AP++R K+ LQ Q +NL +V + I + RE+ ++
Sbjct: 208 LLAGGIAGAVSRTATAPLDRLKVALQVQRTNLGVVPT-----------IKKIWREDKLLG 256
Query: 94 LWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLI 153
+RGNG +V + P A+ F+ ++ K I+ G + + G S +AG AG +
Sbjct: 257 FFRGNGLNVAKVAPESAIKFAAYEMLKPIIGGADGD-----IGTSGRLLAGGLAGAVAQT 311
Query: 154 LVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGL 213
+YP+D+ TRL + + + I+ ++G R YRGL SL G++ + G+
Sbjct: 312 AIYPMDLVKTRLQTFVSEVGTPK---LWKLTKDIWIQEGPRAFYRGLCPSLIGIIPYAGI 368
Query: 214 YFGGFDTIKEM-----LSEESKP-ELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQS 267
++T+K++ L + ++P L M + A+ S YPL +R RM S
Sbjct: 369 DLAAYETLKDLSRAHFLHDTAEPGPLIQLGCGMTSGALGASC---VYPLQVIRTRMQADS 425
Query: 268 GMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKFMNL 323
S + K R EGL FYRG N F+ +A+I ++Y+ +KK + L
Sbjct: 426 SK-----TSMGQEFLKTLRGEGLKGFYRGIFPNFFKVIPSASISYLVYEAMKKNLAL 477
>At5g07320 peroxisomal Ca-dependent solute carrier-like protein
Length = 479
Score = 107 bits (268), Expect = 7e-24
Identities = 85/301 (28%), Positives = 142/301 (46%), Gaps = 38/301 (12%)
Query: 32 RDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGV 91
R L+AG + G T AP++R K++LQ Q R G+ I + RE+ +
Sbjct: 207 RLLLAGGLAGAVSRTATAPLDRLKVVLQVQ-----------RAHAGVLPTIKKIWREDKL 255
Query: 92 ISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTS 151
+ +RGNG +V++ P A+ F ++ K ++ G + + G S +AG AG +
Sbjct: 256 MGFFRGNGLNVMKVAPESAIKFCAYEMLKPMIGGEDGD-----IGTSGRLMAGGMAGALA 310
Query: 152 LILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHR 211
+YP+D+ TRL + +E + + I+ ++G R Y+GL SL G+V +
Sbjct: 311 QTAIYPMDLVKTRLQTCV--SEGGKAPKLWKLTKDIWVREGPRAFYKGLFPSLLGIVPYA 368
Query: 212 GLYFGGFDTIKEM-----LSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQ 266
G+ ++T+K++ L + L M + A+ S YPL VR RM
Sbjct: 369 GIDLAAYETLKDLSRTYILQDTEPGPLIQLSCGMTSGALGASC---VYPLQVVRTRMQAD 425
Query: 267 SG---MEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKFMN 322
S M+ N+ + EGL FYRG + N+ + AA+I ++Y+ +KK M
Sbjct: 426 SSKTTMKQEFMNT--------MKGEGLRGFYRGLLPNLLKVVPAASITYIVYEAMKKNMA 477
Query: 323 L 323
L
Sbjct: 478 L 478
>At1g25380 unknown protein
Length = 363
Score = 107 bits (266), Expect = 1e-23
Identities = 79/291 (27%), Positives = 141/291 (48%), Gaps = 14/291 (4%)
Query: 36 AGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVISLW 95
AGA G T V P++ K LQ A ASG+R + + ++EEG ++
Sbjct: 23 AGATAGAIAATFVCPLDVIKTRLQVLGLPEA-PASGQRGGV-IITSLKNIIKEEGYRGMY 80
Query: 96 RGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLILV 155
RG +++ P+ A+ FS+ K +L+ + +N +A A AG + I
Sbjct: 81 RGLSPTIIALLPNWAVYFSVYGKLKDVLQSSDGK-----LSIGSNMIAAAGAGAATSIAT 135
Query: 156 YPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYF 215
PL + TRL R V ++ + + I ++GVRG+Y G+ SL G V H + F
Sbjct: 136 NPLWVVKTRLMTQGIRPGVVPYKSVMSAFSRICHEEGVRGLYSGILPSLAG-VSHVAIQF 194
Query: 216 GGFDTIKEMLSEESKPELALWKRWMVAQAVTTS---AGLVSYPLDTVRRRMMMQSGMEH- 271
++ IK+ +++ + VA A + + A +++YP + +R ++ Q + +
Sbjct: 195 PAYEKIKQYMAKMDNTSVENLSPGNVAIASSIAKVIASILTYPHEVIRAKLQEQGQIRNA 254
Query: 272 -PVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKF 320
Y+ +DC K++R+EG+ YRG +N+ R+T +A I Y+ + +F
Sbjct: 255 ETKYSGVIDCITKVFRSEGIPGLYRGCATNLLRTTPSAVITFTTYEMMLRF 305
Score = 72.0 bits (175), Expect = 4e-13
Identities = 56/206 (27%), Positives = 100/206 (48%), Gaps = 8/206 (3%)
Query: 122 ILRGGNSNPD-NIFSGASANFVAGAAAGCTSLILVYPLDIAHTRL-AADIGRTEVRQFRG 179
++ GNS D +AN AGA AG + V PLD+ TRL + RG
Sbjct: 1 MIEHGNSTFDYRSIREVAANAGAGATAGAIAATFVCPLDVIKTRLQVLGLPEAPASGQRG 60
Query: 180 --IHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWK 237
I L I +++G RG+YRGL ++ ++ + +YF + +K++L + S +L++
Sbjct: 61 GVIITSLKNIIKEEGYRGMYRGLSPTIIALLPNWAVYFSVYGKLKDVL-QSSDGKLSIGS 119
Query: 238 RWMVAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV--YNSTLDCWRKIYRTEGLISFYR 295
+ A + + + PL V+ R+M Q G+ V Y S + + +I EG+ Y
Sbjct: 120 NMIAAAGAGAATSIATNPLWVVKTRLMTQ-GIRPGVVPYKSVMSAFSRICHEEGVRGLYS 178
Query: 296 GAVSNVFRSTGAAAILVLYDEVKKFM 321
G + ++ + A Y+++K++M
Sbjct: 179 GILPSLAGVSHVAIQFPAYEKIKQYM 204
>At5g51050 calcium-binding transporter-like protein
Length = 487
Score = 103 bits (257), Expect = 1e-22
Identities = 81/298 (27%), Positives = 142/298 (47%), Gaps = 33/298 (11%)
Query: 35 MAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVISL 94
+AG + G A T AP++R K+LLQ Q+++ I R K ++ ++ GV
Sbjct: 213 IAGGIAGAASRTATAPLDRLKVLLQIQKTDARI----REAIKLIW-------KQGGVRGF 261
Query: 95 WRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLIL 154
+RGNG ++++ P A+ F +L+K+ + G N D G + AG AG +
Sbjct: 262 FRGNGLNIVKVAPESAIKFYAYELFKNAI-GENMGEDKADIGTTVRLFAGGMAGAVAQAS 320
Query: 155 VYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLY 214
+YPLD+ TRL + V R + I +G R Y+GL SL G++ + G+
Sbjct: 321 IYPLDLVKTRLQTYTSQAGVAVPR-LGTLTKDILVHEGPRAFYKGLFPSLLGIIPYAGID 379
Query: 215 FGGFDTIKEM----LSEESKPELALWKRWMVAQAVTTSAGLVS----YPLDTVRRRMMMQ 266
++T+K++ + ++++P +V T +G + YPL VR RM +
Sbjct: 380 LAAYETLKDLSRTYILQDAEPGP------LVQLGCGTISGALGATCVYPLQVVRTRMQAE 433
Query: 267 SGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKFMNL 323
S +R+ EG + Y+G + N+ + AA+I ++Y+ +KK + L
Sbjct: 434 RAR-----TSMSGVFRRTISEEGYRALYKGLLPNLLKVVPAASITYMVYEAMKKSLEL 486
Score = 48.5 bits (114), Expect = 5e-06
Identities = 47/185 (25%), Positives = 78/185 (41%), Gaps = 16/185 (8%)
Query: 141 FVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGL 200
F+AG AG S PLD ++ I +T+ R I + I+++ GVRG +RG
Sbjct: 212 FIAGGIAGAASRTATAPLD--RLKVLLQIQKTDAR----IREAIKLIWKQGGVRGFFRGN 265
Query: 201 PASLHGMVIHRGLYFGGFDTIKEMLSE---ESKPELALWKRWMVAQAVTTSAGLVSYPLD 257
++ + + F ++ K + E E K ++ R A YPLD
Sbjct: 266 GLNIVKVAPESAIKFYAYELFKNAIGENMGEDKADIGTTVRLFAGGMAGAVAQASIYPLD 325
Query: 258 TVRRRMM---MQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVL 313
V+ R+ Q+G+ P + + I EG +FY+G ++ A I L
Sbjct: 326 LVKTRLQTYTSQAGVAVPRLGTLT---KDILVHEGPRAFYKGLFPSLLGIIPYAGIDLAA 382
Query: 314 YDEVK 318
Y+ +K
Sbjct: 383 YETLK 387
Score = 46.2 bits (108), Expect = 2e-05
Identities = 49/197 (24%), Positives = 84/197 (41%), Gaps = 24/197 (12%)
Query: 34 LMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVIS 93
L AG + G + P++ K LQT S + R D ++ EG +
Sbjct: 307 LFAGGMAGAVAQASIYPLDLVKTRLQTYTSQAGVAVP--RLGTLTKDILV----HEGPRA 360
Query: 94 LWRGNGSSVLRYYPSVALNF----SLKDLYKS-ILRGGNSNPDNIFSGASANFVAGAAAG 148
++G S+L P ++ +LKDL ++ IL+ P G +G
Sbjct: 361 FYKGLFPSLLGIIPYAGIDLAAYETLKDLSRTYILQDAEPGP-------LVQLGCGTISG 413
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMV 208
VYPL + TR+ A+ RT + G+ F TI ++G R +Y+GL +L +V
Sbjct: 414 ALGATCVYPLQVVRTRMQAERARTSM---SGV--FRRTI-SEEGYRALYKGLLPNLLKVV 467
Query: 209 IHRGLYFGGFDTIKEML 225
+ + ++ +K+ L
Sbjct: 468 PAASITYMVYEAMKKSL 484
>At2g47490 mitochondrial carrier like protein
Length = 312
Score = 102 bits (255), Expect = 2e-22
Identities = 81/292 (27%), Positives = 135/292 (45%), Gaps = 16/292 (5%)
Query: 36 AGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKG--MFDCIIRTVREEGVIS 93
AGA G T V P++ K Q + G KG + + + + EG+
Sbjct: 19 AGAAAGVVAATFVCPLDVIKTRFQVH----GLPKLGDANIKGSLIVGSLEQIFKREGMRG 74
Query: 94 LWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLI 153
L+RG +V+ + A+ F++ D KS L SN + GA N +A + AG + I
Sbjct: 75 LYRGLSPTVMALLSNWAIYFTMYDQLKSFLC---SNDHKLSVGA--NVLAASGAGAATTI 129
Query: 154 LVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGL 213
PL + TRL R + ++ L I ++G+RG+Y GL +L G + H +
Sbjct: 130 ATNPLWVVKTRLQTQGMRVGIVPYKSTFSALRRIAYEEGIRGLYSGLVPALAG-ISHVAI 188
Query: 214 YFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTS---AGLVSYPLDTVRRRMMMQSGME 270
F ++ IK L+++ + VA A + + A ++YP + VR R+ Q
Sbjct: 189 QFPTYEMIKVYLAKKGDKSVDNLNARDVAVASSIAKIFASTLTYPHEVVRARLQEQGHHS 248
Query: 271 HPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKFM 321
Y+ DC +K++ +G FYRG +N+ R+T AA I ++ V +F+
Sbjct: 249 EKRYSGVRDCIKKVFEKDGFPGFYRGCATNLLRTTPAAVITFTSFEMVHRFL 300
Score = 71.6 bits (174), Expect = 5e-13
Identities = 56/201 (27%), Positives = 93/201 (45%), Gaps = 10/201 (4%)
Query: 127 NSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAA----DIGRTEVRQFRGIHH 182
NS+P N N AGAAAG + V PLD+ TR +G ++ I
Sbjct: 4 NSHPPNS-KNVLCNAAAGAAAGVVAATFVCPLDVIKTRFQVHGLPKLGDANIKGSL-IVG 61
Query: 183 FLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVA 242
L IF+++G+RG+YRGL ++ ++ + +YF +D +K L +L++ + A
Sbjct: 62 SLEQIFKREGMRGLYRGLSPTVMALLSNWAIYFTMYDQLKSFLCSNDH-KLSVGANVLAA 120
Query: 243 QAVTTSAGLVSYPLDTVRRRMMMQSGMEHPV--YNSTLDCWRKIYRTEGLISFYRGAVSN 300
+ + + PL V+ R+ Q GM + Y ST R+I EG+ Y G V
Sbjct: 121 SGAGAATTIATNPLWVVKTRLQTQ-GMRVGIVPYKSTFSALRRIAYEEGIRGLYSGLVPA 179
Query: 301 VFRSTGAAAILVLYDEVKKFM 321
+ + A Y+ +K ++
Sbjct: 180 LAGISHVAIQFPTYEMIKVYL 200
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,908,523
Number of Sequences: 26719
Number of extensions: 278352
Number of successful extensions: 1073
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 625
Number of HSP's gapped (non-prelim): 149
length of query: 326
length of database: 11,318,596
effective HSP length: 100
effective length of query: 226
effective length of database: 8,646,696
effective search space: 1954153296
effective search space used: 1954153296
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC133572.9


