

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133571.8 - phase: 0
(495 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
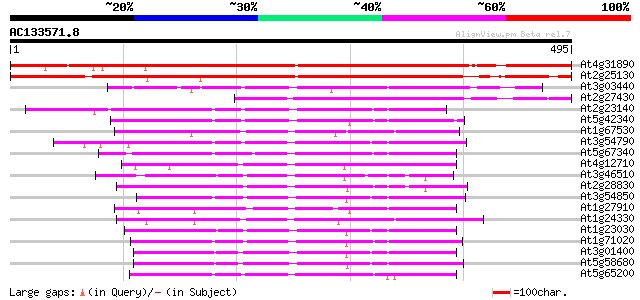
Score E
Sequences producing significant alignments: (bits) Value
At4g31890 unknown protein 533 e-151
At2g25130 unknown protein 498 e-141
At3g03440 unknown protein 135 7e-32
At2g27430 unknown protein 131 1e-30
At2g23140 hypothetical protein 125 5e-29
At5g42340 arm repeat containing protein 121 8e-28
At1g67530 unknown protein 121 8e-28
At3g54790 unknown protein 120 1e-27
At5g67340 unknown proteins 120 2e-27
At4g12710 unknown protein 118 9e-27
At3g46510 arm repeat containing protein homolog 117 1e-26
At2g28830 unknown protein 117 1e-26
At3g54850 putative protein 115 4e-26
At1g27910 unknown protein 115 4e-26
At1g24330 hypothetical protein 114 2e-25
At1g23030 unknown protein 112 5e-25
At1g71020 unknown protein 107 2e-23
At3g01400 unknown protein 104 1e-22
At5g58680 unknown protein 99 7e-21
At5g65200 unknown protein 98 9e-21
>At4g31890 unknown protein
Length = 518
Score = 533 bits (1372), Expect = e-151
Identities = 315/532 (59%), Positives = 378/532 (70%), Gaps = 51/532 (9%)
Query: 1 MAKCHRSNVDSIVLHHRHHAKNTTAGNFRF------SASSFRRIIFDALSCGGASRHHRH 54
MAKCHR+N+ S++L +++ + F S S+FRR I DA+SCGG+SR+ RH
Sbjct: 1 MAKCHRNNIGSLILDRAPSTSSSSTSGYHFRLWSTFSRSTFRRKIVDAVSCGGSSRY-RH 59
Query: 55 YHREEEISSVASTATVKE----------------LRGEVQEE---KTEKLLDLLNIQVHE 95
REE+ T T K L G EE K+EKL DLLN+ E
Sbjct: 60 ELREEDDEGSYVTVTAKSTVASSKDAKANTIGAALNGVAFEETSKKSEKLCDLLNLAEVE 119
Query: 96 TNAESKKKEETLTEMKHVVKDLR-----------GEDSTKRRIAAARVRSLTKEDSEARG 144
+ E+ KKEE L +K VV++L+ G D K+ AA+ VR L KEDSEAR
Sbjct: 120 ADVETMKKEEALEVLKRVVRELQSAAAAREDNDDGGDCRKKITAASEVRLLAKEDSEARV 179
Query: 145 SLAMLGAISPLVGMLD-SEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIES 203
+LAMLGAI PLV M+D S + +QI SLYALLNLGI NDANKAAIVK GAVHKMLKLIES
Sbjct: 180 TLAMLGAIPPLVSMIDDSRIVDAQIASLYALLNLGIGNDANKAAIVKAGAVHKMLKLIES 239
Query: 204 PCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDAL 263
P D ++EA+VANFLGLSALDSNKPIIGSSGAI FLV+ L+NLD +S SSQ ++DAL
Sbjct: 240 PNTPDQEIAEAVVANFLGLSALDSNKPIIGSSGAIIFLVKTLQNLDETS--SSQAREDAL 297
Query: 264 RALYNLSINQTNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRKAISAVKD 323
RALYNLSI Q N+SF+LETDL+ +L+N++ DMEVSER+L+ILSNLV+ PEGRKAI V D
Sbjct: 298 RALYNLSIYQPNVSFILETDLITYLLNTLGDMEVSERILAILSNLVAVPEGRKAIGLVCD 357
Query: 324 AITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQ 383
A VLVDVLNWTDSP CQEKA+YILM+MAHK Y DRQ MIEAGI S+LLELTL+G+ALAQ
Sbjct: 358 AFPVLVDVLNWTDSPGCQEKATYILMLMAHKGYGDRQVMIEAGIESALLELTLLGSALAQ 417
Query: 384 KRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSSSLVKTDGGGKECLMEEVNMM 443
KRASRIL+C R+DKGKQV S G+ G +SAPI + + + + E MM
Sbjct: 418 KRASRILECLRVDKGKQVLDST--GSCG-ALSAPIYGTRDNGLDHE--------ENDLMM 466
Query: 444 SDEKKAVKQLVQQSLQNNMMKIVKRANLRQDFVPSERFASLTSSSTSKSLPF 495
S+E+KAVKQLVQQSLQ+NM +IVKRANL QDFVPSE F SL+ SSTSKSLPF
Sbjct: 467 SEERKAVKQLVQQSLQSNMKRIVKRANLPQDFVPSEHFKSLSLSSTSKSLPF 518
>At2g25130 unknown protein
Length = 468
Score = 498 bits (1282), Expect = e-141
Identities = 296/504 (58%), Positives = 355/504 (69%), Gaps = 45/504 (8%)
Query: 1 MAKCHRSNVDSIVLHHRHHAKNTTAGNFRFSASSFRRIIFDALSCGGASRHHRHYHREEE 60
MAKCHR+NVD ++LH A +++ FS SS RRIIFDA+SCGG+SR+ R E++
Sbjct: 1 MAKCHRNNVDPLILHRIPSASSSSLSGNTFSGSSLRRIIFDAISCGGSSRYRRELREEDD 60
Query: 61 ISSVASTATVKELRGEVQEEKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGE 120
ST + GE K EKL DLLN+ V E+ E+KKKEETL +K VVKDL+ E
Sbjct: 61 EGCSKST-----IIGEDSMRKPEKLSDLLNLAVIESGVETKKKEETLEILKRVVKDLQAE 115
Query: 121 -----DSTKRRIAAA-RVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQ--IDSLY 172
++ +++IAAA VR L K+D EAR +LAMLGAI PLV M+D E I SLY
Sbjct: 116 AEAEAEAAEKKIAAASEVRLLAKDDIEARVTLAMLGAIPPLVSMIDDESQSEDALIASLY 175
Query: 173 ALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPII 232
ALLNLGI ND NKAAIVK G VHKMLKL+ES + +++EAIVANFLGLSALDSNKPII
Sbjct: 176 ALLNLGIGNDVNKAAIVKAGVVHKMLKLVESSKPPNQAIAEAIVANFLGLSALDSNKPII 235
Query: 233 GSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSI 292
GSSGAI FLV+ LKN + +S SSQ ++DALRALYNLSI N+SF+LETDL+ FL+N++
Sbjct: 236 GSSGAIIFLVKTLKNFEETS--SSQAREDALRALYNLSIYHQNVSFILETDLIPFLLNTL 293
Query: 293 EDMEVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMA 352
DMEVSER+L+IL+N+VS PEGRKAI V +A +LVDVLNW DS +CQEKA YILM+MA
Sbjct: 294 GDMEVSERILAILTNVVSVPEGRKAIGEVVEAFPILVDVLNWNDSIKCQEKAVYILMLMA 353
Query: 353 HKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFR-LDKGKQVSRSCDGGNLG 411
HK Y DR AMIEAGI SSLLELTLVG+ LAQKRASR+L+C R +DKGKQ
Sbjct: 354 HKGYGDRNAMIEAGIESSLLELTLVGSPLAQKRASRVLECLRVVDKGKQ----------- 402
Query: 412 LTVSAPICASSSSLVKTDGGGKECLMEEVNMMSDEKKAVKQLVQQSLQNNMMKIVKRANL 471
VSAPI +SS G+E M+DE+KAVKQLVQQSLQ+NM +IVKRANL
Sbjct: 403 --VSAPIYGTSSL-------GRE--RGHDLRMTDERKAVKQLVQQSLQSNMKRIVKRANL 451
Query: 472 RQDFVPSERFASLTSSSTSKSLPF 495
DFV TS SKSL F
Sbjct: 452 PHDFV-------TTSQHFSKSLTF 468
>At3g03440 unknown protein
Length = 408
Score = 135 bits (339), Expect = 7e-32
Identities = 111/392 (28%), Positives = 200/392 (50%), Gaps = 40/392 (10%)
Query: 87 DLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSL 146
+LL+ V +++ S + + ++ V+ +R ED R AA +R LTK R
Sbjct: 44 ELLHTAVSVSSSVSVSSSSSAS-IQRVLSLIRSEDCDSRLFAAKEIRRLTKTSHRCRRHF 102
Query: 147 AMLGAISPLVGML--DSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESP 204
+ A+ PLV ML DS + H + +L ALLNL + ++ NK +I++ GA+ ++ ++S
Sbjct: 103 SQ--AVEPLVSMLRFDSPESHHEA-ALLALLNLAVKDEKNKVSIIEAGALEPIINFLQSN 159
Query: 205 CVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALR 264
++ E A+ L LSA +NKPIIG++G +P LV+++K+ S Q K DA+
Sbjct: 160 ---SPTLQEYASASLLTLSASANNKPIIGANGVVPLLVKVIKH------GSPQAKADAVM 210
Query: 265 ALYNLSINQTNISFVLET----DLVVFLINSIEDMEVSERVLSILSNL-VSSPEGRKAIS 319
AL NLS N+S +L T ++ L +S + + SE+ S++ L VS E R +
Sbjct: 211 ALSNLSTLPDNLSMILATKPLSPILNLLKSSKKSSKTSEKCCSLIEALMVSGEEARTGLV 270
Query: 320 AVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYAD-RQAMIEAGIVSSLLELTLVG 378
+ + + +V+VL S + +E A +L+ + + R+ ++ G++ LLELT+ G
Sbjct: 271 SDEGGVLAVVEVLE-NGSLQAREHAVGVLLTLCQSDRSKYREPILREGVIPGLLELTVQG 329
Query: 379 TALAQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSSSLVKTDGGGKECLME 438
T+ ++ +A R+L R + + D T+ + + S + D GK
Sbjct: 330 TSKSRIKAQRLLCLLRNSESPRSEVQPD------TIENIVSSLISHIDGDDQSGK----- 378
Query: 439 EVNMMSDEKKAVKQLVQQSLQNNMMKIVKRAN 470
KK + ++VQ S++ ++ + +RA+
Sbjct: 379 -------AKKMLAEMVQVSMEKSLRHLQERAS 403
Score = 40.4 bits (93), Expect = 0.002
Identities = 74/345 (21%), Positives = 142/345 (40%), Gaps = 47/345 (13%)
Query: 30 FSASSFRRIIFDALSCGGASRHHRHYHREEEISSVASTATVKELRGEVQEEKTEK-LLDL 88
F+A RR+ + C RH+ S A V LR + E E LL L
Sbjct: 83 FAAKEIRRLTKTSHRC------RRHF-------SQAVEPLVSMLRFDSPESHHEAALLAL 129
Query: 89 LNIQVHETNAESKKKEETLTE---MKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGS 145
LN+ V + +K + ++ E ++ ++ L+ T + A+A + +L+ + +
Sbjct: 130 LNLAVKD-----EKNKVSIIEAGALEPIINFLQSNSPTLQEYASASLLTLSASANN-KPI 183
Query: 146 LAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPC 205
+ G + LV ++ ++ D++ AL NL D N + I+ + +L L++S
Sbjct: 184 IGANGVVPLLVKVIKHGSPQAKADAVMALSNLSTLPD-NLSMILATKPLSPILNLLKSS- 241
Query: 206 VVDSSVSEAIVA--NFLGLSALDSNKPIIGSSGAIPFLVRILKN---------------- 247
S SE + L +S ++ ++ G + +V +L+N
Sbjct: 242 KKSSKTSEKCCSLIEALMVSGEEARTGLVSDEGGVLAVVEVLENGSLQAREHAVGVLLTL 301
Query: 248 --LDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIEDM-EVS-ERVLS 303
D S +++ + L L++ T+ S + L+ L NS EV + + +
Sbjct: 302 CQSDRSKYREPILREGVIPGLLELTVQGTSKSRIKAQRLLCLLRNSESPRSEVQPDTIEN 361
Query: 304 ILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYIL 348
I+S+L+S +G K + +V V QE+AS ++
Sbjct: 362 IVSSLISHIDGDDQSGKAKKMLAEMVQVSMEKSLRHLQERASTLV 406
>At2g27430 unknown protein
Length = 282
Score = 131 bits (329), Expect = 1e-30
Identities = 93/297 (31%), Positives = 155/297 (51%), Gaps = 23/297 (7%)
Query: 199 KLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQV 258
KL ++ V+D S A L LS+L + + + SS +PFL+ + NS + +
Sbjct: 9 KLPKNVEVLDQSTRHAFAELLLSLSSLTNTQLPVASSQILPFLMDTM----NSDSTDMKT 64
Query: 259 KQDALRALYNLSINQTNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRKAI 318
K+ L + NL + N ++ V L++ + ++SE+ L+ L LV + G+KA+
Sbjct: 65 KEICLATISNLCLVLENAGPLVLNGAVETLLSLMSTKDLSEKALASLGKLVVTQMGKKAM 124
Query: 319 SAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVG 378
L+++L W D P+CQE A+YILM++AH++++ R+ M +AGIV LLE++L+G
Sbjct: 125 EDCLLVSKGLIEILTWEDIPKCQEYAAYILMVLAHQSWSQREKMAKAGIVPVLLEVSLLG 184
Query: 379 TALAQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSSSLVKTDGGGKECLME 438
+ L QKRA ++LQ F+ ++ ++ G + G S + G
Sbjct: 185 SPLVQKRAVKLLQWFKDERNVRM-----GPHSGPQTGWVSPGMGSPMSPRSG-------- 231
Query: 439 EVNMMSDEKKAVKQLVQQSLQNNMMKIVKRANLRQDFVPSERFASLTSSSTSKSLPF 495
+ +K +K LV+QSL NM I +R NL + S R SL S++SKSL +
Sbjct: 232 -----EEGRKMMKNLVKQSLYKNMEMITRRGNLDME-SESCRLKSLIISTSSKSLTY 282
>At2g23140 hypothetical protein
Length = 924
Score = 125 bits (314), Expect = 5e-29
Identities = 109/384 (28%), Positives = 189/384 (48%), Gaps = 27/384 (7%)
Query: 15 HHRHHAKNTTAGNFRFSASSFRRIIFDALSCGGASRHHRHYHREEEISSVASTATVKEL- 73
HHR + +T N F + ++ S R +++ S AT ++L
Sbjct: 517 HHRSPSATSTVSNEEFPRADANENSEESAHATPYSSDASGEIRSGPLAATTSAATRRDLS 576
Query: 74 -----------RGEVQEEKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDS 122
RG+ +E+L I +N + E T++K +V++L+
Sbjct: 577 DFSPKFMDRRTRGQFWRRPSERLGS--RIVSAPSNETRRDLSEVETQVKKLVEELKSSSL 634
Query: 123 TKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIAND 182
+R A A +R L K + + R + GAI LV +L S D +Q +++ ALLNL I ND
Sbjct: 635 DTQRQATAELRLLAKHNMDNRIVIGNSGAIVLLVELLYSTDSATQENAVTALLNLSI-ND 693
Query: 183 ANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLV 242
NK AI GA+ ++ ++E+ S E A LS ++ NK IG SGAI LV
Sbjct: 694 NNKKAIADAGAIEPLIHVLENG---SSEAKENSAATLFSLSVIEENKIKIGQSGAIGPLV 750
Query: 243 RILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIEDME-VSERV 301
+L N + + K+DA AL+NLSI+Q N + ++++ V +LI+ ++ + ++
Sbjct: 751 DLLGN------GTPRGKKDAATALFNLSIHQENKAMIVQSGAVRYLIDLMDPAAGMVDKA 804
Query: 302 LSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQA 361
+++L+NL + PEGR AI + I +LV+V+ S +E A+ L+ ++ +
Sbjct: 805 VAVLANLATIPEGRNAIGQ-EGGIPLLVEVVE-LGSARGKENAAAALLQLSTNSGRFCNM 862
Query: 362 MIEAGIVSSLLELTLVGTALAQKR 385
+++ G V L+ L+ GT A+++
Sbjct: 863 VLQEGAVPPLVALSQSGTPRAREK 886
>At5g42340 arm repeat containing protein
Length = 656
Score = 121 bits (304), Expect = 8e-28
Identities = 95/314 (30%), Positives = 164/314 (51%), Gaps = 16/314 (5%)
Query: 90 NIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAML 149
N ++ E + E E+ +V+ L ++R + ++R L +E+ E R +A
Sbjct: 357 NFKIPEKEVSPDSQNEQKDEVSLLVEALSSSQLEEQRRSVKQMRLLARENPENRVLIANA 416
Query: 150 GAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDS 209
GAI LV +L D Q +++ LLNL I ++ NK I GA+ +++++E+ +
Sbjct: 417 GAIPLLVQLLSYPDSGIQENAVTTLLNLSI-DEVNKKLISNEGAIPNIIEILENG---NR 472
Query: 210 SVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNL 269
E A LS LD NK IG S IP LV +L++ + + K+DAL AL+NL
Sbjct: 473 EARENSAAALFSLSMLDENKVTIGLSNGIPPLVDLLQH------GTLRGKKDALTALFNL 526
Query: 270 SINQTNISFVLETDLVVFLINSIEDMEVS--ERVLSILSNLVSSPEGRKAISAVKDAITV 327
S+N N ++ +V L+N ++D + + LSIL L S PEGR+AI + I
Sbjct: 527 SLNSANKGRAIDAGIVQPLLNLLKDKNLGMIDEALSILLLLASHPEGRQAIGQL-SFIET 585
Query: 328 LVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRAS 387
LV+ + +P+ +E A+ +L+ + + A ++ G+ L+E+T GT AQ++A+
Sbjct: 586 LVEFIR-QGTPKNKECATSVLLELGSNNSSFILAALQFGVYEYLVEITTSGTNRAQRKAN 644
Query: 388 RILQCFRLDKGKQV 401
++Q + K +Q+
Sbjct: 645 ALIQL--ISKSEQI 656
>At1g67530 unknown protein
Length = 782
Score = 121 bits (304), Expect = 8e-28
Identities = 90/314 (28%), Positives = 165/314 (51%), Gaps = 21/314 (6%)
Query: 93 VHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAA-RVRSLTKEDSEARGSLAMLGA 151
V + + E L + ++ L E+ +++ ++R L K+D EAR + G
Sbjct: 408 VSDDDDEEDSDINVLERYQDLLAVLNEEEGLEKKCKVVEKIRLLLKDDEEARIFMGANGF 467
Query: 152 ISPLVGML----DSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVV 207
+ L+ L D + +Q AL NL + N+ NK ++ G + + K+I S
Sbjct: 468 VEALLRFLGSAVDDNNAAAQDSGAMALFNLAVNNNRNKELMLTSGVIRLLEKMISS---- 523
Query: 208 DSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALY 267
+ + A +L LS LD K +IGSS A+PFLV++L+ + +Q K DAL ALY
Sbjct: 524 -AESHGSATALYLNLSCLDEAKSVIGSSQAVPFLVQLLQ-----KEIETQCKLDALHALY 577
Query: 268 NLSINQTNISFVLETDLVV----FLINSIEDMEVSERVLSILSNLVSSPEGRKAISAVKD 323
NLS NI +L ++++ L ++ E++ + E+ L++L NL SS EG+ + +
Sbjct: 578 NLSTYSPNIPALLSSNIIKSLQGLLASTGENLWI-EKSLAVLLNLASSQEGKDEAVSSQG 636
Query: 324 AITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQ 383
I+ L VL+ D+ E QE+A L+I+ + + Q +++ G++ SL+ +++ GT +
Sbjct: 637 MISSLATVLDMGDTTE-QEQAVSCLLILCNGRESCIQMVLQEGVIPSLVSISVNGTPRGR 695
Query: 384 KRASRILQCFRLDK 397
+++ ++L FR ++
Sbjct: 696 EKSQKLLMLFREER 709
>At3g54790 unknown protein
Length = 760
Score = 120 bits (302), Expect = 1e-27
Identities = 108/384 (28%), Positives = 196/384 (50%), Gaps = 31/384 (8%)
Query: 39 IFDALSCGGASRHHRHYHREEEISSV----ASTATVKELRGEVQ---EEKTEKLLDLLNI 91
IF+ LS G + H R +SSV + T + + G Q E +K L+ N
Sbjct: 386 IFELLSPGQSYTHSRSESVCSVVSSVDYVPSVTHETESILGNHQSSSEMSPKKNLESSNN 445
Query: 92 QVHETNAESKKK----------EETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSE 141
HE +A + T + +V+DL+ + + AAA +R LT E
Sbjct: 446 VNHEHSAAKTYECSVHDLDDSGTMTTSHTIKLVEDLKSGSNKVKTAAAAEIRHLTINSIE 505
Query: 142 ARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLI 201
R + GAI+PL+ +L SE+ +Q ++ ALLNL I ++ NKA IV++GA+ ++ ++
Sbjct: 506 NRVHIGRCGAITPLLSLLYSEEKLTQEHAVTALLNLSI-SELNKAMIVEVGAIEPLVHVL 564
Query: 202 ESPCVVDSSVSEAIVANFLGLSALDSNKPIIG-SSGAIPFLVRILKNLDNSSKSSSQVKQ 260
+ + E A+ LS L N+ IG S+ AI LV +L K + + K+
Sbjct: 565 NTG---NDRAKENSAASLFSLSVLQVNRERIGQSNAAIQALVNLL------GKGTFRGKK 615
Query: 261 DALRALYNLSINQTNISFVLETDLVVFLINSIE-DMEVSERVLSILSNLVSSPEGRKAIS 319
DA AL+NLSI N + +++ V +L+ ++ D+E+ ++ +++L+NL + EGR+AI
Sbjct: 616 DAASALFNLSITHDNKARIVQAKAVKYLVELLDPDLEMVDKAVALLANLSAVGEGRQAI- 674
Query: 320 AVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGT 379
+ I +LV+ ++ S +E A+ +L+ + + +++ G + L+ L+ GT
Sbjct: 675 VREGGIPLLVETVD-LGSQRGKENAASVLLQLCLNSPKFCTLVLQEGAIPPLVALSQSGT 733
Query: 380 ALAQKRASRILQCFRLDKGKQVSR 403
A+++A ++L FR + ++ +
Sbjct: 734 QRAKEKAQQLLSHFRNQRDARMKK 757
>At5g67340 unknown proteins
Length = 707
Score = 120 bits (300), Expect = 2e-27
Identities = 96/317 (30%), Positives = 166/317 (52%), Gaps = 16/317 (5%)
Query: 79 EEKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKE 138
EE+ + ++ V ET + S + TE+K ++ DL+ +R A AR+R L +
Sbjct: 396 EERHWRHPGIIPATVRETGSSSSIE----TEVKKLIDDLKSSSLDTQREATARIRILARN 451
Query: 139 DSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKML 198
++ R +A AI LV +L S D Q D++ LLNL I ND NK+ I + GA+ ++
Sbjct: 452 STDNRIVIARCEAIPSLVSLLYSTDERIQADAVTCLLNLSI-NDNNKSLIAESGAIVPLI 510
Query: 199 KLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQV 258
++++ + ++ + A A LS ++ K IG +GAI LV +L S
Sbjct: 511 HVLKTGYLEEAKANSA--ATLFSLSVIEEYKTEIGEAGAIEPLVDLL------GSGSLSG 562
Query: 259 KQDALRALYNLSINQTNISFVLETDLVVFLINSIED-MEVSERVLSILSNLVSSPEGRKA 317
K+DA AL+NLSI+ N + V+E V +L+ ++ + E+ + +L+NL + EG+ A
Sbjct: 563 KKDAATALFNLSIHHENKTKVIEAGAVRYLVELMDPAFGMVEKAVVVLANLATVREGKIA 622
Query: 318 ISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLV 377
I + I VLV+V+ S +E A+ L+ + + +I G++ L+ LT
Sbjct: 623 IGE-EGGIPVLVEVVE-LGSARGKENATAALLQLCTHSPKFCNNVIREGVIPPLVALTKS 680
Query: 378 GTALAQKRASRILQCFR 394
GTA +++A +L+ F+
Sbjct: 681 GTARGKEKAQNLLKYFK 697
>At4g12710 unknown protein
Length = 402
Score = 118 bits (295), Expect = 9e-27
Identities = 96/313 (30%), Positives = 162/313 (51%), Gaps = 26/313 (8%)
Query: 99 ESKKKEETLTE---------MKHVVKDLRGEDSTKRRIAAARVRSLTKED---SEARGSL 146
E + +EET T + H+ K L D R AA +R L ++ S AR L
Sbjct: 27 EDEAEEETTTNTWKTTKELLILHLSKKLLHGDLDFRIEAAKEIRKLLRKSPVKSSARSKL 86
Query: 147 AMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCV 206
A G I PLV ML S ++ ++ SL ALLNL + N+ NK IVK GAV ++++++ +
Sbjct: 87 ADAGVIPPLVPMLFSSNVDARHASLLALLNLAVRNERNKIEIVKAGAVPPLIQILK---L 143
Query: 207 VDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRAL 266
++S+ E A L LSA +NK +I SSG P L+++L S + Q K DA+ AL
Sbjct: 144 HNASLRELATAAILTLSAAPANKAMIISSGVPPLLIQML------SSGTVQGKVDAVTAL 197
Query: 267 YNLSINQTNISFVLETDLVVFLINSIED----MEVSERVLSILSNLVS-SPEGRKAISAV 321
+NLS + + +L+ V LI+ +++ + +E+ +++ ++S S +GR AI++
Sbjct: 198 HNLSACKEYSAPILDAKAVYPLIHLLKECKKHSKFAEKATALVEMILSHSEDGRNAITSC 257
Query: 322 KDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTAL 381
+D I LV+ + + +L + R+ +++ G + LL T+ GT+
Sbjct: 258 EDGILTLVETVEDGSPLSIEHAVGALLSLCRSDRDKYRKLILKEGAIPGLLSSTVDGTSK 317
Query: 382 AQKRASRILQCFR 394
++ RA +L R
Sbjct: 318 SRDRARVLLDLLR 330
>At3g46510 arm repeat containing protein homolog
Length = 660
Score = 117 bits (293), Expect = 1e-26
Identities = 97/321 (30%), Positives = 165/321 (51%), Gaps = 27/321 (8%)
Query: 76 EVQEEKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSL 135
+++ K L + + AE+ K E+ + + + + +R AA +R L
Sbjct: 327 DIEPPKPPSSLRPRKVSSFSSPAEANKIEDLMWRLAY-------GNPEDQRSAAGEIRLL 379
Query: 136 TKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVH 195
K +++ R ++A GAI LVG+L + D Q S+ ALLNL I + NK AIV GA+
Sbjct: 380 AKRNADNRVAIAEAGAIPLLVGLLSTPDSRIQEHSVTALLNLSICEN-NKGAIVSAGAIP 438
Query: 196 KMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSS 255
++++++ + E A LS +D NK IG+ GAIP LV +L ++ +
Sbjct: 439 GIVQVLKKGSM---EARENAAATLFSLSVIDENKVTIGALGAIPPLVVLL------NEGT 489
Query: 256 SQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIED--MEVSERVLSILSNLVSSPE 313
+ K+DA AL+NL I Q N + ++ L + + + + L+IL+ L S PE
Sbjct: 490 QRGKKDAATALFNLCIYQGNKGKAIRAGVIPTLTRLLTEPGSGMVDEALAILAILSSHPE 549
Query: 314 GRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEA---GIVSS 370
G KAI DA+ LV+ + T SP +E A+ +L+ H D Q ++EA G++
Sbjct: 550 G-KAIIGSSDAVPSLVEFIR-TGSPRNRENAAAVLV---HLCSGDPQHLVEAQKLGLMGP 604
Query: 371 LLELTLVGTALAQKRASRILQ 391
L++L GT +++A+++L+
Sbjct: 605 LIDLAGNGTDRGKRKAAQLLE 625
>At2g28830 unknown protein
Length = 654
Score = 117 bits (293), Expect = 1e-26
Identities = 94/316 (29%), Positives = 164/316 (51%), Gaps = 20/316 (6%)
Query: 95 ETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISP 154
+ ++ S ++ +++ ++ L + RR AA +R L K+++ R ++A GAI
Sbjct: 342 KASSSSSAPDDEHNKIEELLLKLTSQQPEDRRSAAGEIRLLAKQNNHNRVAIAASGAIPL 401
Query: 155 LVGMLD-SEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSE 213
LV +L S D +Q ++ ++LNL I + + GAV ++ +++ + E
Sbjct: 402 LVNLLTISNDSRTQEHAVTSILNLSICQENKGKIVYSSGAVPGIVHVLQKGSM---EARE 458
Query: 214 AIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQ 273
A LS +D NK IG++GAIP LV +L S+ S + K+DA AL+NL I Q
Sbjct: 459 NAAATLFSLSVIDENKVTIGAAGAIPPLVTLL------SEGSQRGKKDAATALFNLCIFQ 512
Query: 274 TNISFVLETDLVVFLINSIEDME--VSERVLSILSNLVSSPEGRKAISAVKDAITVLVDV 331
N + LV L+ + + E + + LSIL+ L S P+G+ + A DA+ VLVD
Sbjct: 513 GNKGKAVRAGLVPVLMRLLTEPESGMVDESLSILAILSSHPDGKSEVGAA-DAVPVLVDF 571
Query: 332 LNWTDSPECQEKASYILMIMAHKAYADRQAMIEA---GIVSSLLELTLVGTALAQKRASR 388
+ + SP +E ++ +L+ H ++Q +IEA GI+ L+E+ GT +++A++
Sbjct: 572 IR-SGSPRNKENSAAVLV---HLCSWNQQHLIEAQKLGIMDLLIEMAENGTDRGKRKAAQ 627
Query: 389 ILQCFRLDKGKQVSRS 404
+L F +Q S
Sbjct: 628 LLNRFSRFNDQQKQHS 643
>At3g54850 putative protein
Length = 632
Score = 115 bits (289), Expect = 4e-26
Identities = 93/292 (31%), Positives = 158/292 (53%), Gaps = 14/292 (4%)
Query: 113 VVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLY 172
+++ L + ++R AA +R L K + + R +A GAI LV +L S D +Q S+
Sbjct: 350 LLEKLANGTTEQQRAAAGELRLLAKRNVDNRVCIAEAGAIPLLVELLSSPDPRTQEHSVT 409
Query: 173 ALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPII 232
ALLNL I N+ NK AIV GA+ +++++++ + E A LS +D NK I
Sbjct: 410 ALLNLSI-NEGNKGAIVDAGAITDIVEVLKNGSM---EARENAAATLFSLSVIDENKVAI 465
Query: 233 GSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSI 292
G++GAI L+ +L+ + + + K+DA A++NL I Q N S ++ +V L +
Sbjct: 466 GAAGAIQALISLLE------EGTRRGKKDAATAIFNLCIYQGNKSRAVKGGIVDPLTRLL 519
Query: 293 EDM--EVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMI 350
+D + + L+IL+ L ++ EG+ AI A ++I VLV+++ T SP +E A+ IL
Sbjct: 520 KDAGGGMVDEALAILAILSTNQEGKTAI-AEAESIPVLVEIIR-TGSPRNRENAAAILWY 577
Query: 351 MAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFRLDKGKQVS 402
+ E G +L ELT GT A+++A+ +L+ + +G V+
Sbjct: 578 LCIGNIERLNVAREVGADVALKELTENGTDRAKRKAASLLELIQQTEGVAVT 629
>At1g27910 unknown protein
Length = 768
Score = 115 bits (289), Expect = 4e-26
Identities = 87/316 (27%), Positives = 168/316 (52%), Gaps = 29/316 (9%)
Query: 93 VHETNAESKKKEETLTEMKH---VVKDLRGEDSTKRRIAAA-RVRSLTKEDSEARGSLAM 148
+ E ES+ +E+ +T ++ ++ L D+ +++ ++R L K+D EAR +
Sbjct: 403 IKEEACESEYQEDQVTLVERCTELLTTLTDVDTLRKKCRVVEQIRVLLKDDEEARILMGE 462
Query: 149 LGAISPLVGMLDS----EDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESP 204
G + L+ L S + +Q AL NL + N+ NK ++ G + + +++ +P
Sbjct: 463 NGCVEALLQFLGSALNENNASAQKVGAMALFNLAVDNNRNKELMLASGIIPLLEEMLCNP 522
Query: 205 CVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALR 264
S + A +L LS L+ KP+IGSS A+PF+V +L +++ Q K DAL
Sbjct: 523 HSHGS-----VTAIYLNLSCLEEAKPVIGSSLAVPFMVNLLW-----TETEVQCKVDALH 572
Query: 265 ALYNLSINQTNISFVLETDLVVFLINSIEDMEVS------ERVLSILSNLVSSPEGRKAI 318
+L++LS NI +L DLV N+++ + +S E+ L++L NLV + G+ +
Sbjct: 573 SLFHLSTYPPNIPCLLSADLV----NALQSLTISDEQRWTEKSLAVLLNLVLNEAGKDEM 628
Query: 319 SAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVG 378
+ ++ L +L+ T P QE+A +L+I+ + + + +++ G++ SL+ +++ G
Sbjct: 629 VSAPSLVSNLCTILD-TGEPNEQEQAVSLLLILCNHSEICSEMVLQEGVIPSLVSISVNG 687
Query: 379 TALAQKRASRILQCFR 394
T ++RA ++L FR
Sbjct: 688 TQRGRERAQKLLTLFR 703
>At1g24330 hypothetical protein
Length = 709
Score = 114 bits (284), Expect = 2e-25
Identities = 95/341 (27%), Positives = 163/341 (46%), Gaps = 28/341 (8%)
Query: 95 ETNAESKKKEETLTEMKHVVKDLRG-----------EDSTKRRIAAARVRSLTKEDSEAR 143
E+ + K+K E+ + L G ED K+ VR L K++ EAR
Sbjct: 338 ESERQQKEKNNAPDEVDSEINVLEGYQDILAIVDKEEDLAKKCKVVENVRILLKDNEEAR 397
Query: 144 GSLAMLGAISPLVGMLDS----EDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLK 199
+ G + + L+S + +Q AL NL + N+ NK ++ G + + K
Sbjct: 398 ILMGANGFVEAFLQFLESAVHDNNAAAQETGAMALFNLAVNNNRNKELMLTSGVIPLLEK 457
Query: 200 LIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVK 259
+I S A +L LS L+ KP+IGSS A+ F V +L + +Q K
Sbjct: 458 MISC-----SQSQGPATALYLNLSCLEKAKPVIGSSQAVSFFVNLL-----LQDTKTQCK 507
Query: 260 QDALRALYNLSINQTNISFVLETDLVVFL--INSIEDMEVSERVLSILSNLVSSPEGRKA 317
DAL ALYNLS NI +L ++++ L + S + E+ L++L NL SS EG++
Sbjct: 508 LDALHALYNLSTYSPNIPTLLSSNIIKSLQVLASTGNHLWIEKSLAVLLNLASSREGKEE 567
Query: 318 ISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLV 377
+ + I+ L VL+ D+ E QE+A L+I+ + + Q +++ G++ SL+ +++
Sbjct: 568 MITTQGMISTLATVLDTGDTVE-QEQAVSCLVILCTGSESCIQMVLQEGVIPSLVSISVN 626
Query: 378 GTALAQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPI 418
G+ + ++ ++L FR + + TVSAP+
Sbjct: 627 GSPRGRDKSQKLLMLFREQRHRDQPSPNKEEAPRKTVSAPM 667
>At1g23030 unknown protein
Length = 612
Score = 112 bits (280), Expect = 5e-25
Identities = 87/296 (29%), Positives = 151/296 (50%), Gaps = 15/296 (5%)
Query: 102 KKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDS 161
K ++ ++ +V+ L + RR A + +RSL+K ++ R +A GAI LV +L S
Sbjct: 325 KNSGDMSVIRALVQRLSSRSTEDRRNAVSEIRSLSKRSTDNRILIAEAGAIPVLVNLLTS 384
Query: 162 EDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLG 221
ED+ +Q +++ +LNL I + NK I+ GAV +++++ + + E A
Sbjct: 385 EDVATQENAITCVLNLSIYEN-NKELIMFAGAVTSIVQVLRAGTM---EARENAAATLFS 440
Query: 222 LSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLE 281
LS D NK IIG SGAIP LV +L+N + + K+DA AL+NL I N +
Sbjct: 441 LSLADENKIIIGGSGAIPALVDLLEN------GTPRGKKDAATALFNLCIYHGNKGRAVR 494
Query: 282 TDLVVFLINSIEDM---EVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSP 338
+V L+ + D + + L+ILS L ++ + + AI + + L+ +L TD
Sbjct: 495 AGIVTALVKMLSDSTRHRMVDEALTILSVLANNQDAKSAI-VKANTLPALIGILQ-TDQT 552
Query: 339 ECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFR 394
+E A+ IL+ + + + G V L++L+ GT +++A +L+ R
Sbjct: 553 RNRENAAAILLSLCKRDTEKLITIGRLGAVVPLMDLSKNGTERGKRKAISLLELLR 608
Score = 37.7 bits (86), Expect = 0.015
Identities = 28/106 (26%), Positives = 53/106 (49%), Gaps = 4/106 (3%)
Query: 54 HYHREEEISSVASTATVKELRGEVQEEKTEKLLDLLNIQVHETNAESK-KKEETLTEMKH 112
H ++ + + TA VK L + ++ L +L++ + +A+S K TL +
Sbjct: 486 HGNKGRAVRAGIVTALVKMLSDSTRHRMVDEALTILSVLANNQDAKSAIVKANTLPALIG 545
Query: 113 VVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGM 158
+ L+ + + R AAA + SL K D+E ++ LGA+ PL+ +
Sbjct: 546 I---LQTDQTRNRENAAAILLSLCKRDTEKLITIGRLGAVVPLMDL 588
>At1g71020 unknown protein
Length = 628
Score = 107 bits (266), Expect = 2e-23
Identities = 92/297 (30%), Positives = 152/297 (50%), Gaps = 16/297 (5%)
Query: 107 LTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSE-DLH 165
++ ++ +V L + RR A + +RSL+K ++ R +A GAI LV +L S+ D
Sbjct: 340 MSAIRALVCKLSSQSIEDRRTAVSEIRSLSKRSTDNRILIAEAGAIPVLVKLLTSDGDTE 399
Query: 166 SQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSAL 225
+Q +++ +LNL I + NK I+ GAV ++ ++ + + E A LS
Sbjct: 400 TQENAVTCILNLSI-YEHNKELIMLAGAVTSIVLVLRAGSM---EARENAAATLFSLSLA 455
Query: 226 DSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLV 285
D NK IIG+SGAI LV +L+ S + K+DA AL+NL I Q N + +V
Sbjct: 456 DENKIIIGASGAIMALVDLLQ------YGSVRGKKDAATALFNLCIYQGNKGRAVRAGIV 509
Query: 286 VFLINSIEDM---EVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQE 342
L+ + D +++ L+ILS L S+ + AI +AI L+D L D P +E
Sbjct: 510 KPLVKMLTDSSSERMADEALTILSVLASNQVAKTAILRA-NAIPPLIDCLQ-KDQPRNRE 567
Query: 343 KASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFRLDKGK 399
A+ IL+ + + ++ G V L+EL+ GT A+++A+ +L+ R K
Sbjct: 568 NAAAILLCLCKRDTEKLISIGRLGAVVPLMELSRDGTERAKRKANSLLELLRKSSRK 624
>At3g01400 unknown protein
Length = 355
Score = 104 bits (260), Expect = 1e-22
Identities = 80/288 (27%), Positives = 147/288 (50%), Gaps = 15/288 (5%)
Query: 110 MKHVVKDLRGEDST-KRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQI 168
+ H+V L S +++ AA +R L+K E R +A GAI PL+ ++ S DL Q
Sbjct: 64 INHLVSHLDSSYSIDEQKQAAMEIRLLSKNKPENRIKIAKAGAIKPLISLISSSDLQLQE 123
Query: 169 DSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSN 228
+ A+LNL + D NK +I GA+ +++ ++ + + E L LS ++ N
Sbjct: 124 YGVTAILNLSLC-DENKESIASSGAIKPLVRALK---MGTPTAKENAACALLRLSQIEEN 179
Query: 229 KPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFL 288
K IG SGAIP LV +L+ + K+DA ALY+L + N +++ ++ L
Sbjct: 180 KVAIGRSGAIPLLVNLLET------GGFRAKKDASTALYSLCSAKENKIRAVQSGIMKPL 233
Query: 289 INSIEDM--EVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASY 346
+ + D + ++ ++S L+S PE + AI + + VLV+++ + +E A
Sbjct: 234 VELMADFGSNMVDKSAFVMSLLMSVPESKPAI-VEEGGVPVLVEIVE-VGTQRQKEMAVS 291
Query: 347 ILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFR 394
IL+ + ++ R + G + L+ L+ GT+ A+++A +++ R
Sbjct: 292 ILLQLCEESVVYRTMVAREGAIPPLVALSQAGTSRAKQKAEALIELLR 339
>At5g58680 unknown protein
Length = 357
Score = 98.6 bits (244), Expect = 7e-21
Identities = 82/295 (27%), Positives = 148/295 (49%), Gaps = 16/295 (5%)
Query: 110 MKHVVKDLRGEDSTK-RRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQI 168
+++++ L S + ++ AA +R L+K E R LA GAI PLV ++ S DL Q
Sbjct: 62 IRNLITHLESSSSIEEQKQAAMEIRLLSKNKPENRIKLAKAGAIKPLVSLISSSDLQLQE 121
Query: 169 DSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSN 228
+ A+LNL + D NK IV GAV ++ + + + E L LS ++ N
Sbjct: 122 YGVTAVLNLSLC-DENKEMIVSSGAVKPLVNALR---LGTPTTKENAACALLRLSQVEEN 177
Query: 229 KPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFL 288
K IG SGAIP LV +L+N + K+DA ALY+L N + +E+ ++ L
Sbjct: 178 KITIGRSGAIPLLVNLLEN------GGFRAKKDASTALYSLCSTNENKTRAVESGIMKPL 231
Query: 289 INSIEDME--VSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASY 346
+ + D E + ++ +++ L+S+PE + A+ + + VLV+++ + +E +
Sbjct: 232 VELMIDFESDMVDKSAFVMNLLMSAPESKPAV-VEEGGVPVLVEIVE-AGTQRQKEISVS 289
Query: 347 ILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQK-RASRILQCFRLDKGKQ 400
IL+ + ++ R + G V L+ L+ + K +A +++ R + +Q
Sbjct: 290 ILLQLCEESVVYRTMVAREGAVPPLVALSQGSASRGAKVKAEALIELLRQPRQQQ 344
>At5g65200 unknown protein
Length = 556
Score = 98.2 bits (243), Expect = 9e-21
Identities = 85/299 (28%), Positives = 142/299 (47%), Gaps = 20/299 (6%)
Query: 106 TLTEMKHVVKD-LRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDL 164
T TE V+ + L+ + + +R +T+ + EAR SL +S L M+ S
Sbjct: 221 TATEEDEVIYNKLKSSEIFDQEQGLIMMRKMTRTNDEARVSLCSPRILSLLKNMIVSRYS 280
Query: 165 HSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSA 224
Q ++L +L+NL + + NK IV++G V ++ +++S E LS
Sbjct: 281 LVQTNALASLVNLSL-DKKNKLTIVRLGFVPILIDVLKSG---SREAQEHAAGTIFSLSL 336
Query: 225 LDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDL 284
D NK IG GA+ L+ L+ + S + + D+ ALY+L++NQTN S ++
Sbjct: 337 EDDNKMPIGVLGALQPLLHALR-----AAESDRTRHDSALALYHLTLNQTNRSKLVRLGA 391
Query: 285 VVFLINSIEDMEVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVL--NWTDSP---- 338
V L + + E + R L ++ NL EGR A+ +A+ +LV L WT+ P
Sbjct: 392 VPALFSMVRSGESASRALLVICNLACCSEGRSAMLDA-NAVAILVGKLREEWTEEPTEAR 450
Query: 339 ---ECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFR 394
+E L ++H++ + EA V L E+ GT A+++A +ILQ R
Sbjct: 451 SSSSARENCVAALFALSHESLRFKGLAKEARAVEVLKEVEERGTERAREKAKKILQLMR 509
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.129 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,542,434
Number of Sequences: 26719
Number of extensions: 379690
Number of successful extensions: 2061
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 1789
Number of HSP's gapped (non-prelim): 186
length of query: 495
length of database: 11,318,596
effective HSP length: 103
effective length of query: 392
effective length of database: 8,566,539
effective search space: 3358083288
effective search space used: 3358083288
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC133571.8


